Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Primary OSCC Cells
2.2. Culture of OSCC Cancer Stem Cells
2.3. Cell Viability Assay
2.4. Apoptosis Assessment by Annexin V/PI Staining
2.5. OSCCs and CSCs In Vitro Treatment for FTIRM Measurements
2.6. FTIRM Measurements and Data Analysis
2.7. Statistical Analysis
3. Results
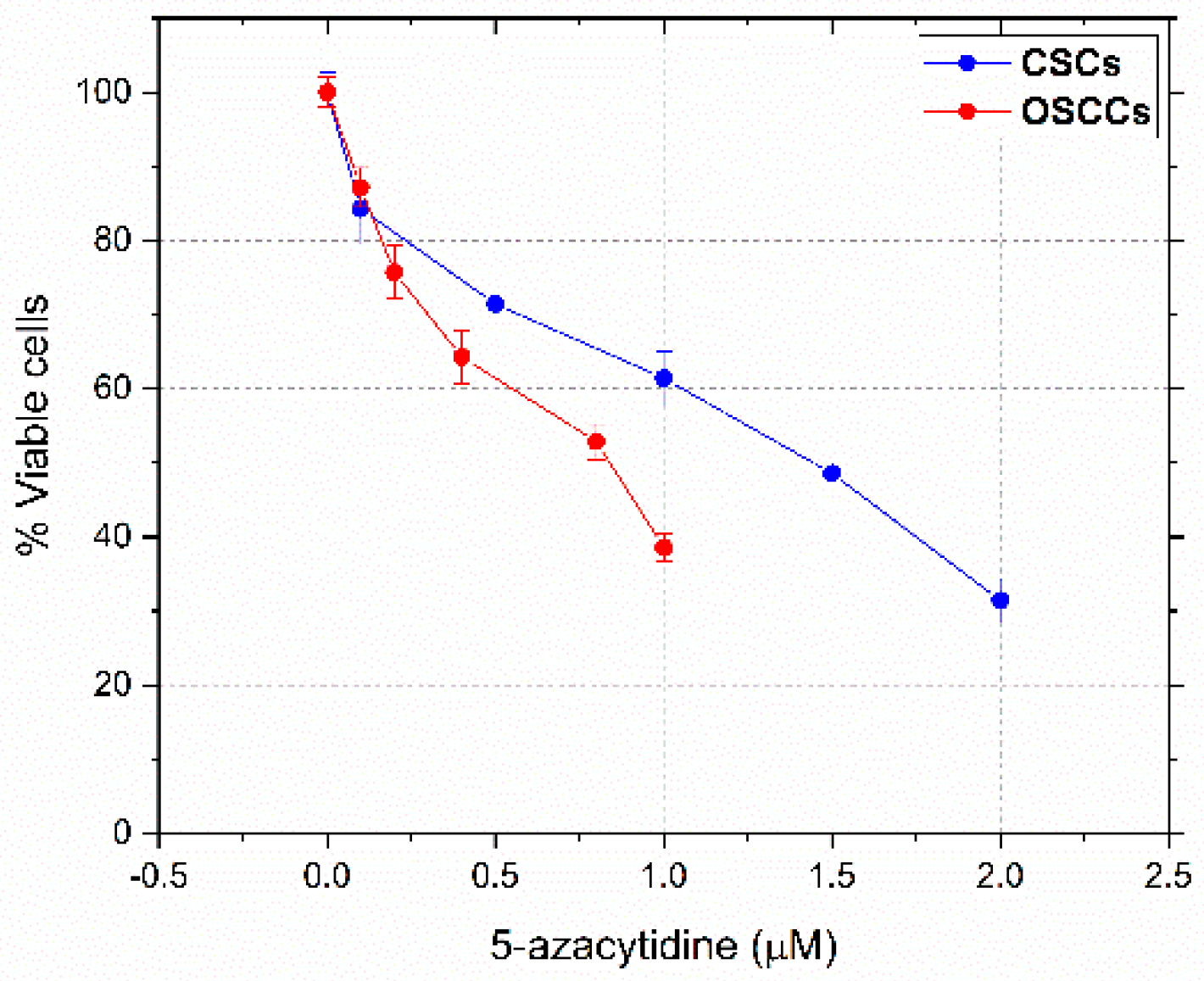
3.1. Dose-Response Curves and IC50 Values
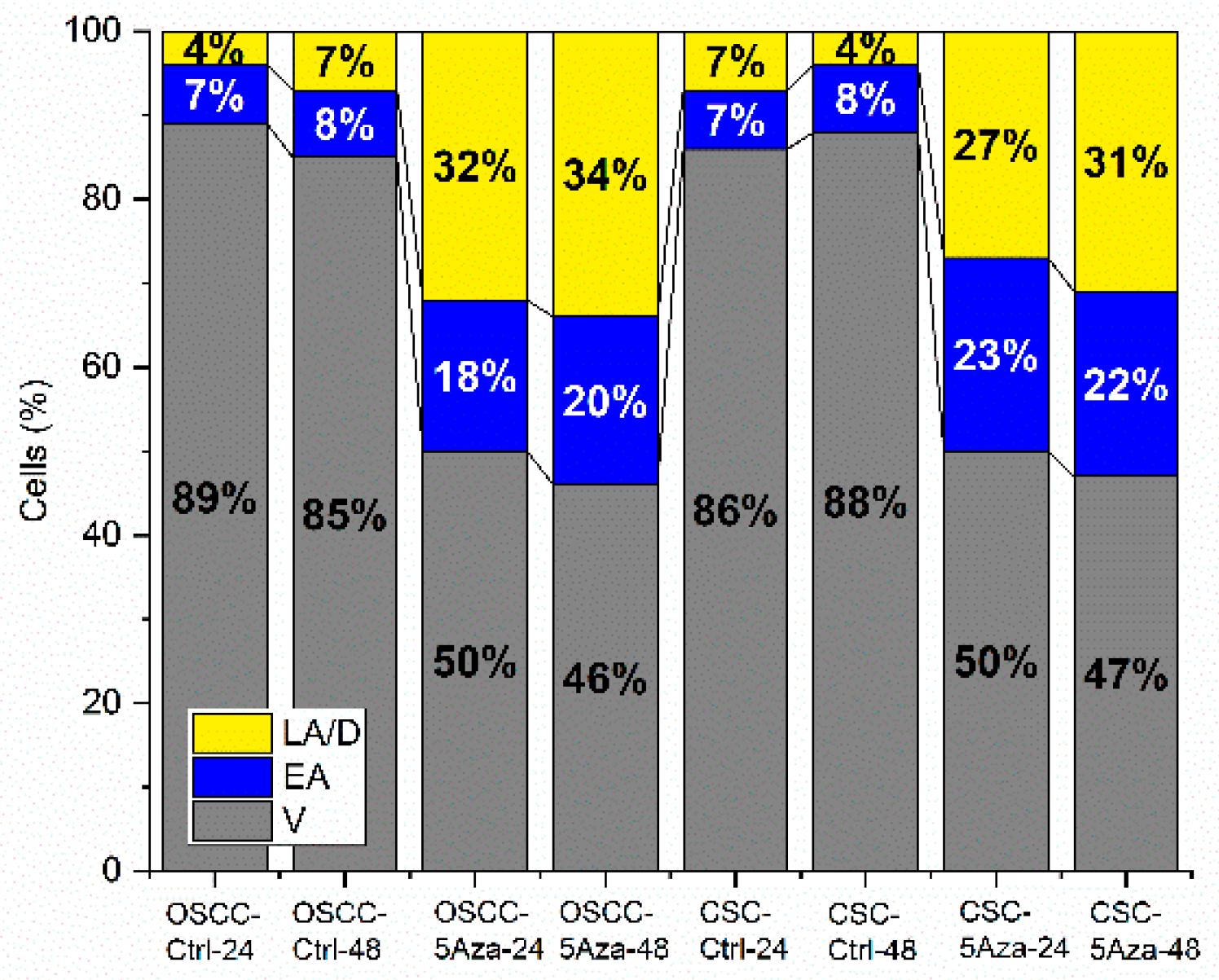
3.2. Apoptosis Rates
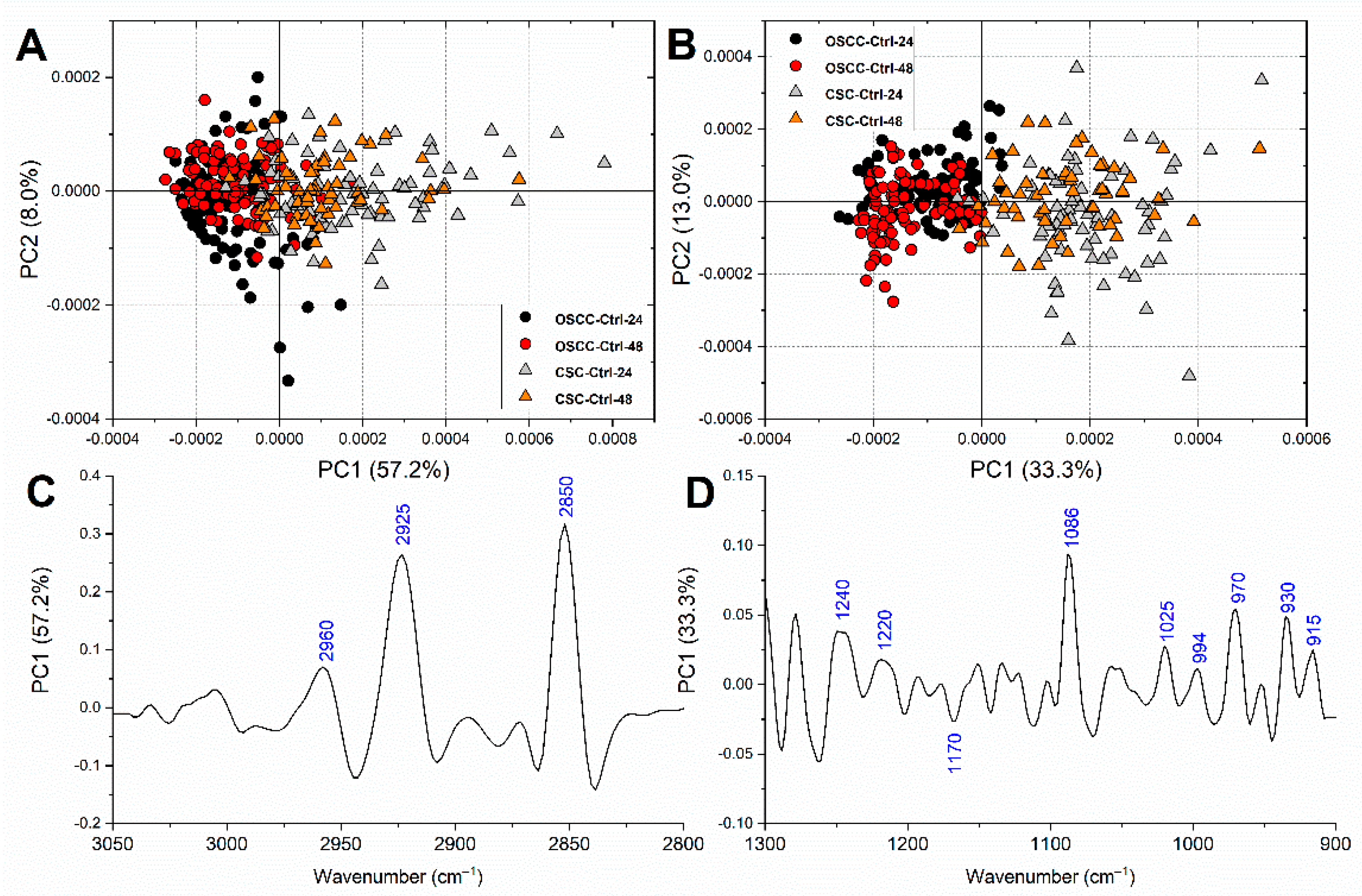
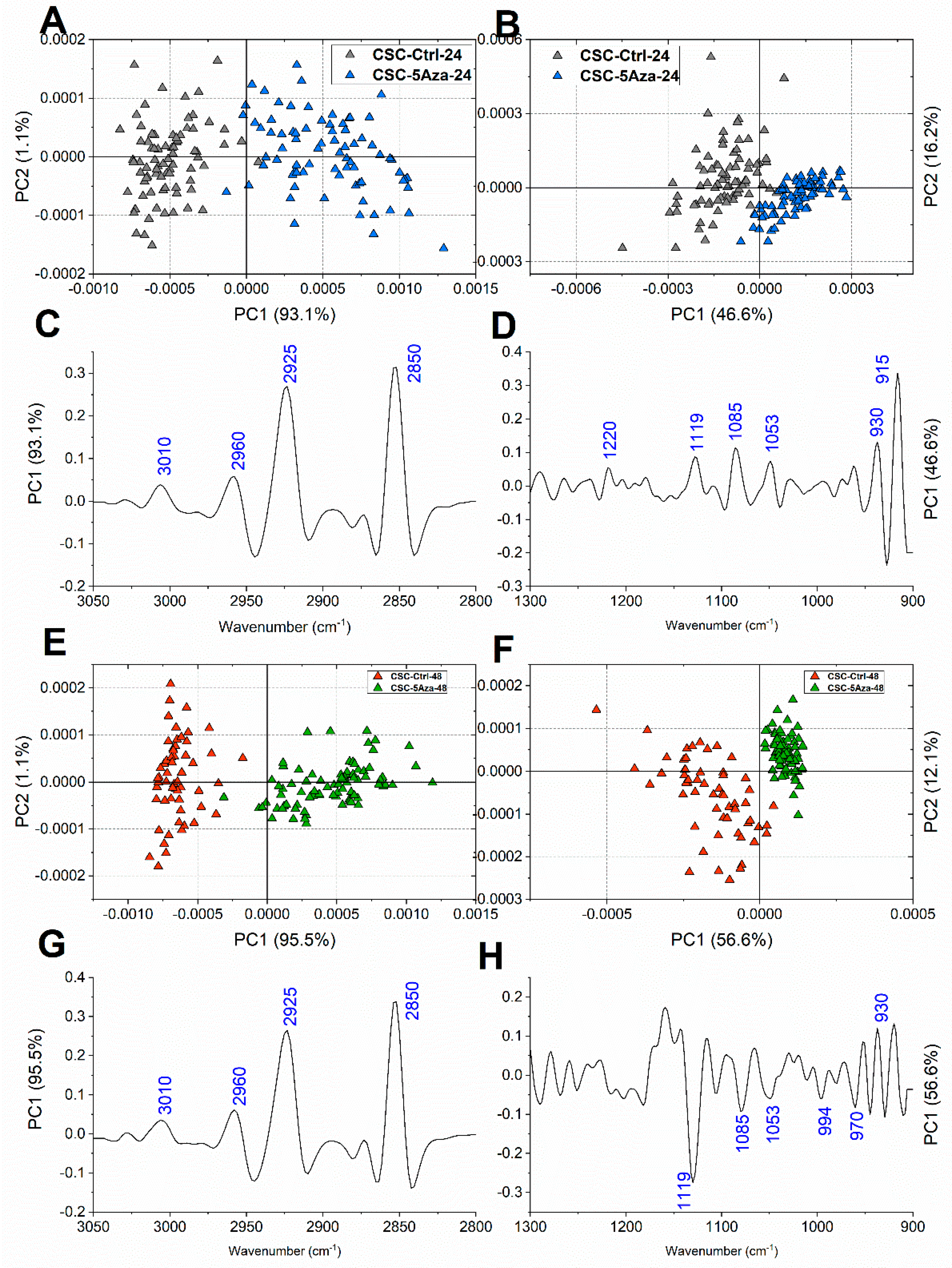
3.3. FTIRM Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, B.; Zhang, S.; Yue, K.; Wang, X.-D. The recurrence and survival of oral squamous cell carcinoma: A report of 275 cases. Chin. J. Cancer 2013, 32, 614–618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Argiris, A.; Karamouzis, M.V.; Raben, D.; Ferris, R.L. Head and neck cancer. Lancet 2008, 371, 1695–1709. [Google Scholar] [CrossRef]
- Peng, C.-H.; Liao, C.-T.; Peng, S.-C.; Chen, Y.-J.; Cheng, A.-J.; Juang, J.-L.; Tsai, C.-Y.; Chen, T.-C.; Chuang, Y.-J.; Tang, C.-Y.; et al. A Novel Molecular Signature Identified by Systems Genetics Approach Predicts Prognosis in Oral Squamous Cell Carcinoma. PLoS ONE 2011, 6, e23452. [Google Scholar] [CrossRef]
- de Camargo Cancela, M.; Voti, L.; Guerra-Yi, M.; Chapuis, F.; Mazuir, M.; Curado, M.P. Oral cavity cancer in developed and in developing countries: Population-based incidence. Head Neck 2010, 32, 357–367. [Google Scholar] [CrossRef]
- Brands, M.T.T.; Brennan, P.A.A.; Verbeek, A.L.M.L.M.; Merkx, M.A.W.A.W.; Geurts, S.M.E.M.E. Follow-up after curative treatment for oral squamous cell carcinoma. A critical appraisal of the guidelines and a review of the literature. Eur. J. Surg. Oncol. 2018, 44, 559–565. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Shi, S.; Yen, Y.; Brown, J.; Ta, J.Q.; Le, A.D. A subpopulation of CD133+ cancer stem-like cells characterized in human oral squamous cell carcinoma confer resistance to chemotherapy. Cancer Lett. 2010, 289, 151–160. [Google Scholar] [CrossRef] [PubMed]
- Rodini, C.O.; Lopes, N.M.; Lara, V.S.; Mackenzie, I.C. Oral cancer stem cells—Properties and consequences. J. Appl. Oral Sci. 2017, 25, 708–715. [Google Scholar] [CrossRef] [PubMed]
- Thakur, B.; Ray, P. Cisplatin triggers cancer stem cell enrichment in platinum-resistant cells through NF-κB-TNFα-PIK3CA loop. J. Exp. Clin. Cancer Res. 2017, 36, 164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castilho, R.; Squarize, C.; Almeida, L. Epigenetic Modifications and Head and Neck Cancer: Implications for Tumor Progression and Resistance to Therapy. Int. J. Mol. Sci. 2017, 18, 1506. [Google Scholar] [CrossRef]
- Freitas, D.P.; Teixeira, C.A.; Santos-Silva, F.; Vasconcelos, M.H.; Almeida, G.M. Therapy-induced enrichment of putative lung cancer stem-like cells. Int. J. Cancer 2014, 134, 1270–1278. [Google Scholar] [CrossRef]
- Shin, K.-H.; Kim, R.H. An Updated Review of Oral Cancer Stem Cells and Their Stemness Regulation. Crit. Rev. Oncog. 2018, 23, 189–200. [Google Scholar] [CrossRef]
- Usman, S.; Jamal, A.; Teh, M.-T.; Waseem, A. Major Molecular Signaling Pathways in Oral Cancer Associated With Therapeutic Resistance. Front. Oral Health 2021, 1, 1–19. [Google Scholar] [CrossRef]
- Kim, S.Y.; Han, Y.K.; Song, J.M.; Lee, C.H.; Kang, K.; Yi, J.M.; Park, H.R. Aberrantly hypermethylated tumor suppressor genes were identified in oral squamous cell carcinoma (OSCC). Clin. Epigenetics 2019, 11, 116. [Google Scholar] [CrossRef] [Green Version]
- do Amaral, G.C.L.S.; Planello, A.C.; Borgato, G.; de Lima, D.G.; Guimarães, G.N.; Marques, M.R.; de Souza, A.P. 5-Aza-CdR promotes partial MGMT demethylation and modifies expression of different genes in oral squamous cell carcinoma. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 2019, 127, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Stresemann, C.; Lyko, F. Modes of action of the DNA methyltransferase inhibitors azacytidine and decitabine. Int. J. Cancer 2008, 123, 8–13. [Google Scholar] [CrossRef]
- Christman, J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar] [CrossRef] [Green Version]
- Manzoni, E.F.M.; Pennarossa, G.; DeEguileor, M.; Tettamanti, G.; Gandolfi, F.; Brevini, T.A.L. 5-azacytidine affects TET2 and histone transcription and reshapes morphology of human skin fibroblasts. Sci. Rep. 2016, 6, 37017. [Google Scholar] [CrossRef]
- Sajadian, S.O.; Ehnert, S.; Vakilian, H.; Koutsouraki, E.; Damm, G.; Seehofer, D.; Thasler, W.; Dooley, S.; Baharvand, H.; Sipos, B.; et al. Induction of active demethylation and 5hmC formation by 5-azacytidine is TET2 dependent and suggests new treatment strategies against hepatocellular carcinoma. Clin. Epigenetics 2015, 7, 98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rocha, M.A.; Veronezi, G.M.B.; Felisbino, M.B.; Gatti, M.S.V.; Tamashiro, W.M.S.C.; Mello, M.L.S. Sodium valproate and 5-aza-2′-deoxycytidine differentially modulate DNA demethylation in G1 phase-arrested and proliferative HeLa cells. Sci. Rep. 2019, 9, 18236. [Google Scholar] [CrossRef] [PubMed]
- Toh, T.B.; Lim, J.J.; Chow, E.K.-H. Epigenetics in cancer stem cells. Mol. Cancer 2017, 16, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyng, F.; Ramos, I.; Ibrahim, O.; Byrne, H. Vibrational Microspectroscopy for Cancer Screening. Appl. Sci. 2015, 5, 23–35. [Google Scholar] [CrossRef]
- Butler, H.J.; Ashton, L.; Bird, B.; Cinque, G.; Curtis, K.; Dorney, J.; Esmonde-White, K.; Fullwood, N.J.; Gardner, B.; Martin-Hirsch, P.L.; et al. Using Raman spectroscopy to characterize biological materials. Nat. Protoc. 2016, 11, 664–687. [Google Scholar] [CrossRef] [Green Version]
- Baker, M.J.; Trevisan, J.; Bassan, P.; Bhargava, R.; Butler, H.J.; Dorling, K.M.; Fielden, P.R.; Fogarty, S.W.; Fullwood, N.J.; Heys, K.A.; et al. Using Fourier transform IR spectroscopy to analyze biological materials. Nat. Protoc. 2014, 9, 1771–1791. [Google Scholar] [CrossRef] [Green Version]
- Notarstefano, V.; Sabbatini, S.; Conti, C.; Pisani, M.; Astolfi, P.; Pro, C.; Rubini, C.; Vaccari, L.; Giorgini, E. Investigation of human pancreatic cancer tissues by Fourier Transform Infrared Hyperspectral Imaging. J. Biophotonics 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Gioacchini, G.; Notarstefano, V.; Sereni, E.; Zacà, C.; Coticchio, G.; Giorgini, E.; Vaccari, L.; Carnevali, O.; Borini, A. Does the molecular and metabolic profile of human granulosa cells correlate with oocyte fate? New insights by Fourier transform infrared microspectroscopy analysis. MHR Basic Sci. Reprod. Med. 2018, 24, 521–532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giorgini, E.; Sabbatini, S.; Rocchetti, R.; Notarstefano, V.; Rubini, C.; Conti, C.; Orilisi, G.; Mitri, E.; Bedolla, D.E.; Vaccari, L. In vitro FTIR microspectroscopy analysis of primary oral squamous carcinoma cells treated with cisplatin and 5-fluorouracil: A new spectroscopic approach for studying the drug–cell interaction. Analyst 2018, 143, 3317–3326. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Sabbatini, S.; Pro, C.; Belloni, A.; Orilisi, G.; Rubini, C.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Exploiting fourier transform infrared and Raman microspectroscopies on cancer stem cells from oral squamous cells carcinoma: New evidence of acquired cisplatin chemoresistance. Analyst 2020, 145, 8038–8049. [Google Scholar] [CrossRef] [PubMed]
- Pozzi, V.; Sartini, D.; Rocchetti, R.; Santarelli, A.; Rubini, C.; Morganti, S.; Giuliante, R.; Calabrese, S.; Di Ruscio, G.; Orlando, F.; et al. Identification and Characterization of Cancer Stem Cells from Head and Neck Squamous Cell Carcinoma Cell Lines. Cell. Physiol. Biochem. 2015, 36, 784–798. [Google Scholar] [CrossRef] [PubMed]
- Hayon, T.; Dvilansky, A.; Shpilberg, O.; Nathan, I. Appraisal of the MTT-based Assay as a Useful Tool for Predicting Drug Chemosensitivity in Leukemia. Leuk. Lymphoma 2003, 44, 1957–1962. [Google Scholar] [CrossRef]
- Giampieri, F.; Alvarez-Suarez, J.M.; Mazzoni, L.; Forbes-Hernandez, T.Y.; Gasparrini, M.; Gonzàlez-Paramàs, A.M.; Santos-Buelga, C.; Quiles, J.L.; Bompadre, S.; Mezzetti, B.; et al. An anthocyanin-rich strawberry extract protects against oxidative stress damage and improves mitochondrial functionality in human dermal fibroblasts exposed to an oxidizing agent. Food Funct. 2014, 5, 1939. [Google Scholar] [CrossRef]
- Vaccari, L.; Birarda, G.; Businaro, L.; Pacor, S.; Grenci, G. Infrared Microspectroscopy of Live Cells in Microfluidic Devices (MD-IRMS): Toward a Powerful Label-Free Cell-Based Assay. Anal. Chem. 2012, 84, 4768–4775. [Google Scholar] [CrossRef]
- Bonnier, F.; Byrne, H.J. Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems. Analyst 2012, 137, 322–332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Notarstefano, V.; Gioacchini, G.; Byrne, H.J.H.J.; Zacà, C.; Sereni, E.; Vaccari, L.; Borini, A.; Carnevali, O.; Giorgini, E. Vibrational characterization of granulosa cells from patients affected by unilateral ovarian endometriosis: New insights from infrared and Raman microspectroscopy. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2019, 212, 206–214. [Google Scholar] [CrossRef] [PubMed]
- Petibois, C.; Déléris, G. Chemical mapping of tumor progression by FT-IR imaging: Towards molecular histopathology. Trends Biotechnol. 2006, 24, 455–462. [Google Scholar] [CrossRef] [PubMed]
- Notarstefano, V.; Gioacchini, G.; Giorgini, E.; Montik, N.; Ciavattini, A.; Polidori, A.R.; Candela, F.A.; Vaccari, L.; Cignitti, M.; Carnevali, O. The Impact of Controlled Ovarian Stimulation Hormones on the Metabolic State and Endocannabinoid System of Human Cumulus Cells. Int. J. Mol. Sci. 2020, 21, 7124. [Google Scholar] [CrossRef]
- Notarstefano, V.; Sabbatini, S.; Sabbatini, M.; Arrais, A.; Belloni, A.; Pro, C.; Vaccari, L.; Osella, D.; Giorgini, E. Hyperspectral characterization of the MSTO-211H cell spheroid model: A FPA—FTIR imaging approach. Clin. Spectrosc. 2021, 3, 100011. [Google Scholar] [CrossRef]
- Ghosh, D.; Dey, S.K.; Saha, C. Mutation Induced Conformational Changes in Genomic DNA from Cancerous K562 Cells Influence Drug-DNA Binding Modes. PLoS ONE 2014, 9, e84880. [Google Scholar] [CrossRef] [Green Version]
- Whelan, D.R.; Bambery, K.R.; Heraud, P.; Tobin, M.J.; Diem, M.; McNaughton, D.; Wood, B.R. Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy. Nucleic Acids Res. 2011, 39, 5439–5448. [Google Scholar] [CrossRef] [Green Version]
- Banyay, M.; Sarkar, M.; Gräslund, A. A library of IR bands of nucleic acids in solution. Biophys. Chem. 2003, 104, 477–488. [Google Scholar] [CrossRef]
- Giorgini, E.; Randazzo, B.; Gioacchini, G.; Cardinaletti, G.; Vaccari, L.; Tibaldi, E.; Olivotto, I. New insights on the macromolecular building of rainbow trout (O. mykiss) intestine: FTIR Imaging and histological correlative study. Aquaculture 2018, 497, 1–9. [Google Scholar] [CrossRef]
- Vileno, B.; Jeney, S.; Sienkiewicz, A.; Marcoux, P.R.; Miller, L.M.; Forró, L. Evidence of lipid peroxidation and protein phosphorylation in cells upon oxidative stress photo-generated by fullerols. Biophys. Chem. 2010, 152, 164–169. [Google Scholar] [CrossRef] [Green Version]
- Talari, A.C.S.; Martinez, M.A.G.; Movasaghi, Z.; Rehman, S.; Rehman, I.U. Advances in Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectrosc. Rev. 2017, 52, 456–506. [Google Scholar] [CrossRef]
- Zucchiatti, P.; Mitri, E.; Kenig, S.; Billè, F.; Kourousias, G.; Bedolla, D.E.; Vaccari, L. Contribution of Ribonucleic Acid (RNA) to the Fourier Transform Infrared (FTIR) Spectrum of Eukaryotic Cells. Anal. Chem. 2016, 88, 12090–12098. [Google Scholar] [CrossRef] [PubMed]
- Mignolet, A.; Derenne, A.; Smolina, M.; Wood, B.R.; Goormaghtigh, E. FTIR spectral signature of anticancer drugs. Can drug mode of action be identified? Biochim. Biophys. Acta—Proteins Proteom. 2016, 1864, 85–101. [Google Scholar] [CrossRef]
- Gioacchini, G.; Giorgini, E.; Vaccari, L.; Ferraris, P.; Sabbatini, S.; Bianchi, V.; Borini, A.; Carnevali, O. A new approach to evaluate aging effects on human oocytes: Fourier transform infrared imaging spectroscopy study. Fertil. Steril. 2014, 101, 120–127. [Google Scholar] [CrossRef]
- Dovbeshko, G.I.; Chegel, V.I.; Gridina, N.Y.; Repnytska, O.P.; Shirshov, Y.M.; Tryndiak, V.P.; Todor, I.M.; Solyanik, G.I. Surface enhanced IR absorption of nucleic acids from tumor cells: FTIR reflectance study. Biopolymers 2002, 67, 470–486. [Google Scholar] [CrossRef] [PubMed]
- Wood, B.R. The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues. Chem. Soc. Rev. 2016, 45, 1980–1998. [Google Scholar] [CrossRef] [PubMed]
- Veronezi, G.M.B.; Felisbino, M.B.; Gatti, M.S.V.; Mello, M.L.S.; Vidal, B. de C. DNA Methylation Changes in Valproic Acid-Treated HeLa Cells as Assessed by Image Analysis, Immunofluorescence and Vibrational Microspectroscopy. PLoS ONE 2017, 12, e0170740. [Google Scholar] [CrossRef]
- De Carvalho, D.D.; Sharma, S.; You, J.S.; Su, S.-F.; Taberlay, P.C.; Kelly, T.K.; Yang, X.; Liang, G.; Jones, P.A. DNA Methylation Screening Identifies Driver Epigenetic Events of Cancer Cell Survival. Cancer Cell 2012, 21, 655–667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kiziltepe, T.; Hideshima, T.; Catley, L.; Raje, N.; Yasui, H.; Shiraishi, N.; Okawa, Y.; Ikeda, H.; Vallet, S.; Pozzi, S.; et al. 5-Azacytidine, a DNA methyltransferase inhibitor, induces ATR-mediated DNA double-strand break responses, apoptosis, and synergistic cytotoxicity with doxorubicin and bortezomib against multiple myeloma cells. Mol. Cancer Ther. 2007, 6, 1718–1727. [Google Scholar] [CrossRef] [Green Version]
- Herbert, A.; Rich, A. The biology of left-handed Z-DNA. J. Biol. Chem. 1996, 271, 11595–11598. [Google Scholar] [CrossRef] [Green Version]
- Temiz, N.A.; Donohue, D.E.; Bacolla, A.; Luke, B.T.; Collins, J.R. The Role of Methylation in the Intrinsic Dynamics of B- and Z-DNA. PLoS ONE 2012, 7. [Google Scholar] [CrossRef] [PubMed]
- Rothenburg, S.; Koch-Nolte, F.; Haag, F. DNA methylation and Z-DNA formation as mediators of quantitative differences in the expression of alleles. Immunol. Rev. 2001, 184, 286–298. [Google Scholar] [CrossRef] [PubMed]
- Denbigh, J.L.; Perez-Guaita, D.; Vernooij, R.R.; Tobin, M.J.; Bambery, K.R.; Xu, Y.; Southam, A.D.; Khanim, F.L.; Drayson, M.T.; Lockyer, N.P.; et al. Probing the action of a novel anti-leukaemic drug therapy at the single cell level using modern vibrational spectroscopy techniques. Sci. Rep. 2017, 7, 2649. [Google Scholar] [CrossRef]
- Birarda, G.; Bedolla, D.E.; Mitri, E.; Pacor, S.; Grenci, G.; Vaccari, L. Apoptotic pathways of U937 leukemic monocytes investigated by infrared microspectroscopy and flow cytometry. Analyst 2014, 139, 3097–3106. [Google Scholar] [CrossRef] [Green Version]
- Benseny-Cases, N.; Klementieva, O.; Cotte, M.; Ferrer, I.; Cladera, J. Microspectroscopy (μFTIR) reveals co-localization of lipid oxidation and amyloid plaques in human Alzheimer disease brains. Anal. Chem. 2014, 86, 12047–12054. [Google Scholar] [CrossRef] [PubMed]
- Davis, P.K.; Johnson, G. V Energy metabolism and protein phosphorylation during apoptosis: A phosphorylation study of tau and high-molecular-weight tau in differentiated PC12 cells. Biochem. J. 1999, 340, 51–58. [Google Scholar] [CrossRef]
- Gruber, W.; Scheidt, T.; Aberger, F.; Huber, C.G. Understanding cell signaling in cancer stem cells for targeted therapy—can phosphoproteomics help to reveal the secrets? Cell Commun. Signal. 2017, 15, 12. [Google Scholar] [CrossRef] [Green Version]


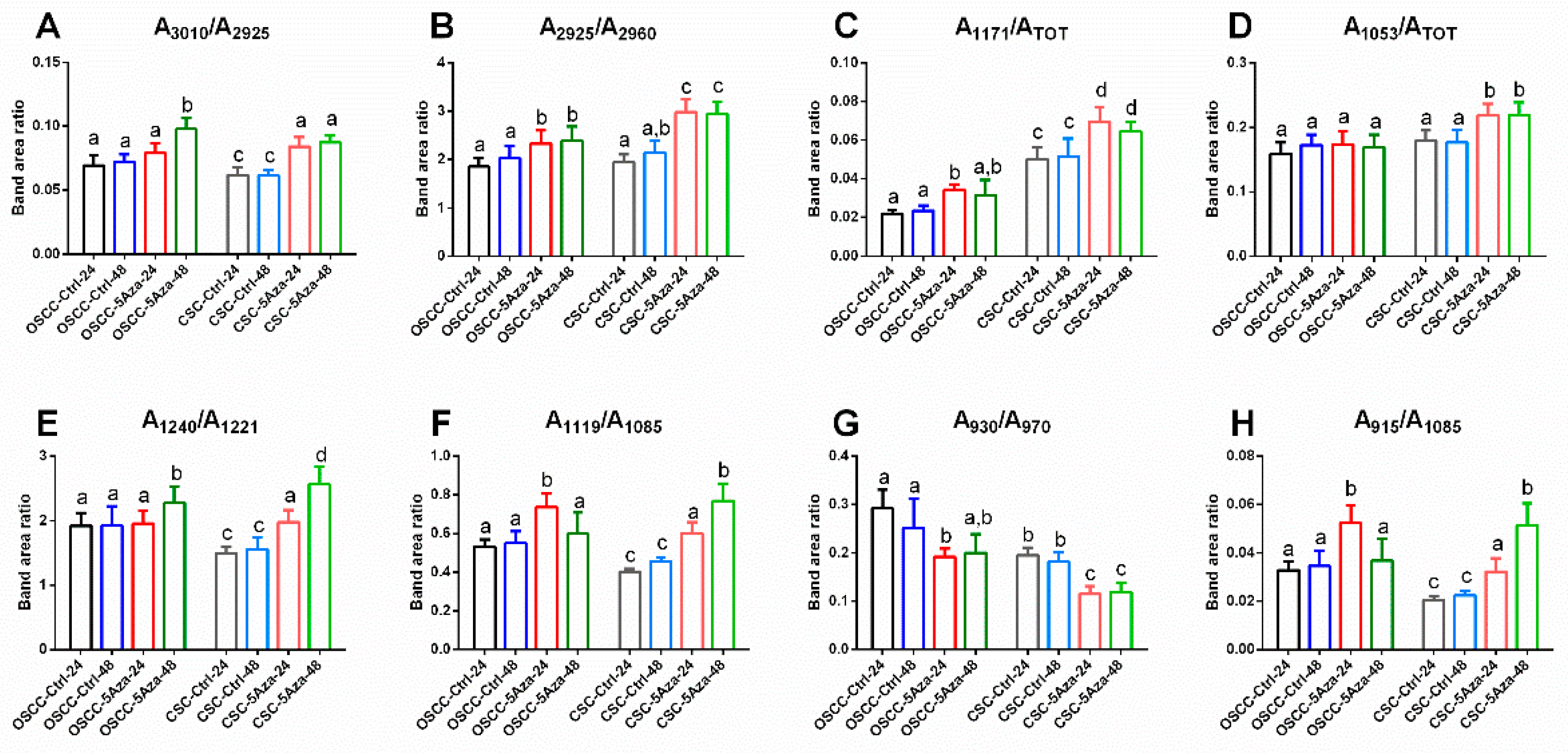
| Wavenumber (cm−1) | Vibrational Mode and Biochemical Assignment |
|---|---|
| ~3010 | Stretching vibration of =CH moiety [33,34] |
| ~2960,~2870 | Antisymmetric and symmetric stretching vibrations of CH3 groups of branched aliphatic chains of lipids [27,35,36] |
| ~2925, ~2850 | Antisymmetric and symmetric stretching vibrations of CH2 groups of linear aliphatic chains of lipids [27,35,36] |
| ~1240 | Antisymmetric stretching vibrations of phosphate moieties of A-form DNA [27,37,38,39] |
| ~1220 | Antisymmetric stretching vibrations of phosphate moieties of B-form DNA [27,37,38,39] |
| ~1170 | Stretching vibration of non-hydrogen bonds of C-OP groups mainly in proteins [40,41] |
| ~1119 | Stretching vibration of the skeletal structure around the C2−OH group of RNA and NTPs [27,33,39,42] |
| ~1086 | Symmetric stretching vibrations of phosphate moieties in nucleic acids [26,27,43] |
| ~1053 | Stretching vibration of C-OH groups in carbohydrates and glycosylated compounds [27,35,42] |
| ~1025 | Stretching vibration of CH2-OH moieties in carbohydrates and glycosylated compounds [26,27,44] |
| ~994 | C–C, C–O ring breathing of RNA ribose [26,27,42,43] |
| ~970 | Backbone vibrations of nucleic acids, mainly double-strand DNA [25,42,43] |
| ~930 | Left-handed helix DNA vibrations (Z-DNA) [33,39,45,46,47] |
| ~915 | Ribose-phosphate skeletal vibrations in RNA [39,47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Notarstefano, V.; Belloni, A.; Sabbatini, S.; Pro, C.; Orilisi, G.; Monterubbianesi, R.; Tosco, V.; Byrne, H.J.; Vaccari, L.; Giorgini, E. Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis. Cells 2021, 10, 2127. https://doi.org/10.3390/cells10082127
Notarstefano V, Belloni A, Sabbatini S, Pro C, Orilisi G, Monterubbianesi R, Tosco V, Byrne HJ, Vaccari L, Giorgini E. Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis. Cells. 2021; 10(8):2127. https://doi.org/10.3390/cells10082127
Chicago/Turabian StyleNotarstefano, Valentina, Alessia Belloni, Simona Sabbatini, Chiara Pro, Giulia Orilisi, Riccardo Monterubbianesi, Vincenzo Tosco, Hugh J. Byrne, Lisa Vaccari, and Elisabetta Giorgini. 2021. "Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis" Cells 10, no. 8: 2127. https://doi.org/10.3390/cells10082127
APA StyleNotarstefano, V., Belloni, A., Sabbatini, S., Pro, C., Orilisi, G., Monterubbianesi, R., Tosco, V., Byrne, H. J., Vaccari, L., & Giorgini, E. (2021). Cytotoxic Effects of 5-Azacytidine on Primary Tumour Cells and Cancer Stem Cells from Oral Squamous Cell Carcinoma: An In Vitro FTIRM Analysis. Cells, 10(8), 2127. https://doi.org/10.3390/cells10082127













