Not So Dead Genes—Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts
Abstract
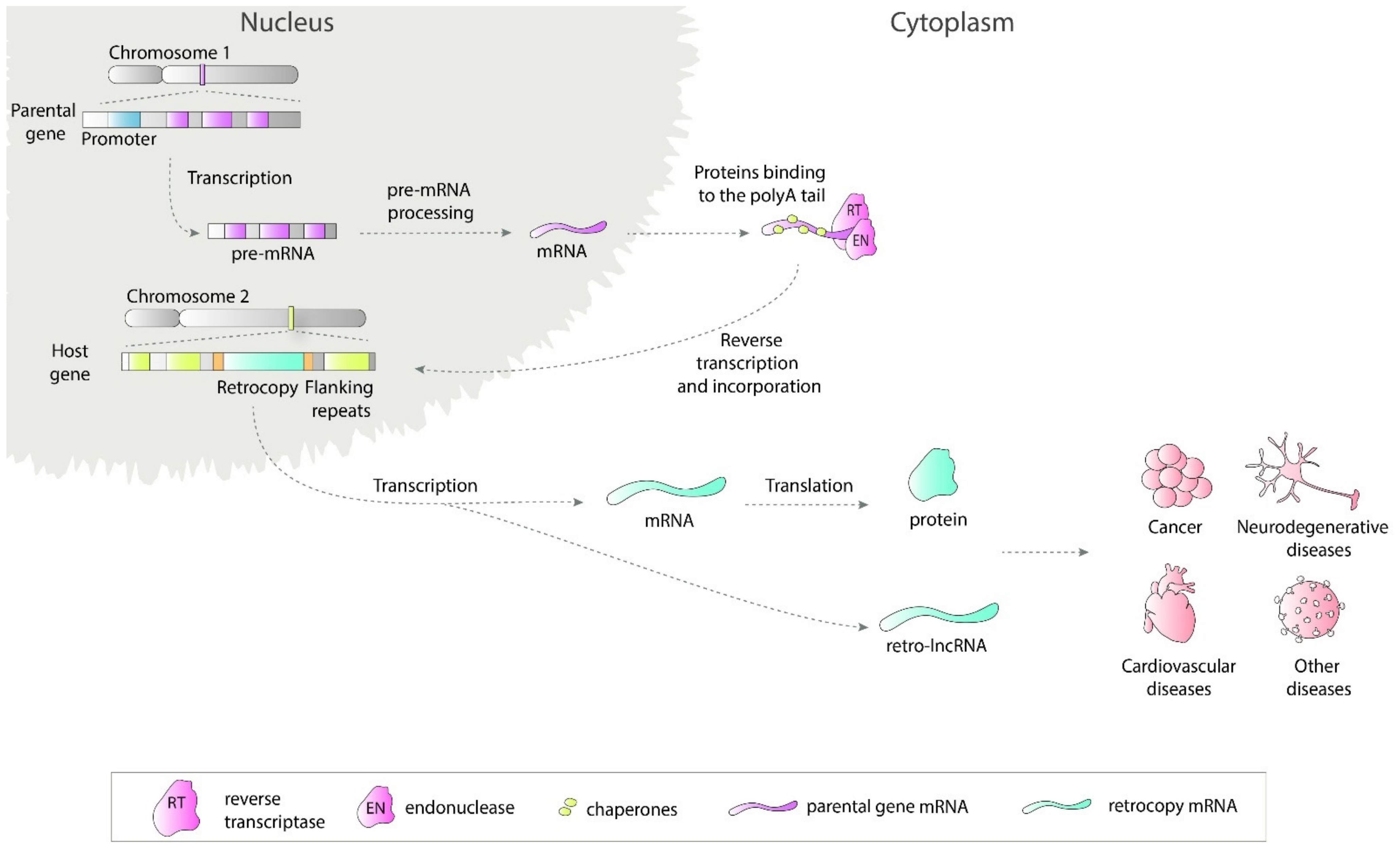
1. Introduction
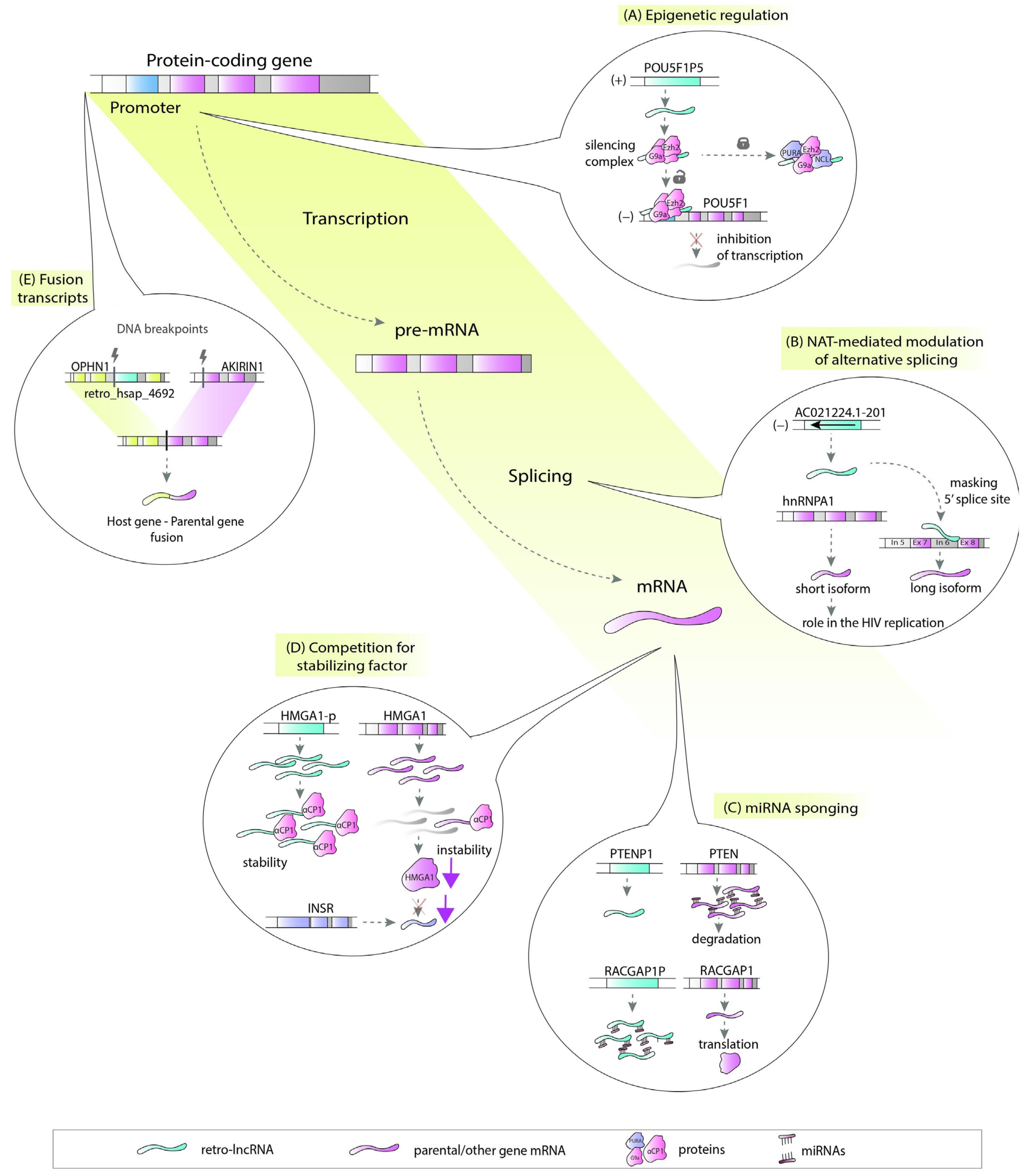
2. Retro-lncRNAs in Cancer
3. Retrocopies as lncRNAs in Neurodegenerative Disorders
4. Retro-lncRNAs and Other Diseases
4.1. Cardiovascular Diseases
4.2. Mental Disorders
4.3. Other Diseases
5. Noncoding Retrocopies as Putative Players in Pathogenic Processes
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Conflicts of Interest
References
- Brosius, J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene 1999, 238, 115–134. [Google Scholar] [CrossRef]
- Casola, C.; Betrán, E. the genomic impact of gene retrocopies: What have we learned from comparative genomics, population genomics, and transcriptomic analyses? Genome Biol. Evol. 2017, 9, 1351–1373. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, M.R.; Szcześniak, M.W.; Makałowska, I. Complex Analysis of Retroposed Genes’ Contribution to Human Genome, Proteome and Transcriptome. Genes 2020, 11, 542. [Google Scholar] [CrossRef] [PubMed]
- Carelli, F.N.; Hayakawa, T.; Go, Y.; Imai, H.; Warnefors, M.; Kaessmann, H. The life history of retrocopies illuminates the evolution of new mammalian genes. Genome Res. 2016, 26, 301–314. [Google Scholar] [CrossRef]
- Betran, E.; Wang, W.; Jin, L.; Long, M. Evolution of the phosphoglycerate mutase processed gene in human and chimpanzee revealing the origin of a new primate gene. Mol. Biol. Evol. 2002, 654–663. [Google Scholar] [CrossRef][Green Version]
- Vanin, E.F. Processed pseudogenes: Characteristics and evolution. Annu. Rev. Genet. 1985, 19, 253–272. [Google Scholar] [CrossRef]
- Esnault, C.; Maestre, J.; Heidmann, T. Human LINE retrotransposons generate processed pseudogenes. Nat. Genet. 2000, 363–367. [Google Scholar] [CrossRef]
- Mighell, A.J.; Smith, N.R.; Robinson, P.A.; Markham, A.F. Vertebrate pseudogenes. Febs Lett. 2000, 468, 109–114. [Google Scholar] [CrossRef]
- Betran, E. Retroposed new genes out of the X in drosophila. Genome Res. 2002, 12, 1854–1859. [Google Scholar] [CrossRef]
- Bai, Y.; Casola, C.; Feschotte, C.; Betrán, E. Comparative genomics reveals a constant rate of origination and convergent acquisition of functional retrogenes in drosophila. Genome Biol. 2007, 8, R11. [Google Scholar] [CrossRef]
- Haldane, J. The part played by recurent mutation in evolution. Am. Nat. 1933, 67, 5–19. [Google Scholar] [CrossRef]
- Fisher, R.A. The sheltering of lethals. Am. Nat. 1935, 69, 446–455. [Google Scholar] [CrossRef]
- Nei, M. Gene duplication and nucleotide substitution in evolution. Nature 1969, 221, 40–42. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fu, P. Pseudogene-Derived lncRNAs and their mirna sponging mechanism in human cancer. Front. Cell Dev. Biol. 2020, 8, 85. [Google Scholar] [CrossRef]
- Poliseno, L.; Salmena, L.; Zhang, J.; Carver, B.; Haveman, W.J.; Pandolfi, P.P. A coding-independent function of gene and pseudogene mRNAs regulates tumour biology. Nature 2010, 465, 1033–1038. [Google Scholar] [CrossRef]
- Ohno, S. Evolution by Gene Duplication; Springer: New York, NY, USA, 1970. [Google Scholar]
- Force, A.; Lynch, M.; Pickett, F.B.; Amores, A.; Yan, Y.L.; Postlethwait, J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 1999, 151, 1531–1545. [Google Scholar]
- Ciomborowska, J.; Rosikiewicz, W.; Szklarczyk, D.; Makałowski, W.; Makałowska, I. “Orphan” retrogenes in the human genome. Mol. Biol. Evol. 2013, 30, 384–396. [Google Scholar] [CrossRef]
- Krasnov, A.N. A retrocopy of a gene can functionally displace the source gene in evolution. Nucleic Acids Res. 2005, 33, 6654–6661. [Google Scholar] [CrossRef]
- Bai, Y.; Casola, C.; Betrán, E. Evolutionary origin of regulatory regions of retrogenes in drosophila. BMC Genom. 2008. [Google Scholar] [CrossRef]
- Sarda, S.; Hannenhalli, S. Orphan CpG islands as alternative promoters. Transcription 2018, 9, 171–176. [Google Scholar] [CrossRef]
- Devor, E.J. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. J. Hered. 2006, 97, 186–190. [Google Scholar] [CrossRef] [PubMed]
- Parker, H.G.; VonHoldt, B.M.; Quignon, P.; Margulies, E.H.; Shao, S.; Mosher, D.S.; Spady, T.C.; Elkahloun, A.; Cargill, M.; Jones, P.G.; et al. An expressed fgf4 retrogene is associated with breed-defining chondrodysplasia in domestic dogs. Science 2009, 325, 995–998. [Google Scholar] [CrossRef] [PubMed]
- Kubiak, M.R.; Makałowska, I. Protein-Coding Genes’ Retrocopies and Their Functions. Viruses 2017, 9, 80. [Google Scholar] [CrossRef]
- Abegglen, L.M.; Caulin, A.F.; Chan, A.; Lee, K.; Robinson, R.; Campbell, M.S.; Kiso, W.K.; Schmitt, D.L.; Waddell, P.J.; Bhaskara, S.; et al. Potential Mechanisms for Cancer Resistance in Elephants and Comparative Cellular Response to DNA Damage in Humans. JAMA 2015, 314, 1850–1860. [Google Scholar] [CrossRef]
- Sulak, M.; Fong, L.; Mika, K.; Chigurupati, S.; Yon, L.; Mongan, N.P.; Emes, R.D.; Lynch, V.J. TP53 copy number expansion is associated with the evolution of increased body size and an enhanced DNA damage response in elephants. eLife 2016, 5. [Google Scholar] [CrossRef]
- Rosikiewicz, W.; Kabza, M.; Kosinski, J.G.; Ciomborowska-Basheer, J.; Kubiak, M.R.; Makalowska, I. RetrogeneDB-a database of plant and animal retrocopies. Database J. Biol. Databases Curation 2017, 2017. [Google Scholar] [CrossRef] [PubMed]
- Glenfield, C.; McLysaght, A. pseudogenes provide evolutionary evidence for the competitive endogenous RNA hypothesis. Mol. Biol. Evol. 2018. [Google Scholar] [CrossRef]
- Bryzghalov, O.; Szcześniak, M.W.; Makałowska, I. Retroposition as a source of antisense long non-coding RNAs with possible regulatory functions. Acta Biochim. Pol. 2016, 63, 825–833. [Google Scholar] [CrossRef] [PubMed]
- Johnsson, P.; Ackley, A.; Vidarsdottir, L.; Lui, W.-O.; Corcoran, M.; Grandér, D.; Morris, K.V. A pseudogene long-noncoding-RNA network regulates PTEN transcription and translation in human cells. Nat. Struct. Mol. Biol. 2013, 20, 440–446. [Google Scholar] [CrossRef]
- Szcześniak, M.W.; Makałowska, I. lncRNA-RNA Interactions across the Human Transcriptome. PLoS ONE 2016, 11, e0150353. [Google Scholar] [CrossRef]
- Stambolic, V.; Suzuki, A.; de la Pompa, J.L.; Brothers, G.M.; Mirtsos, C.; Sasaki, T.; Ruland, J.; Penninger, J.M.; Siderovski, D.P.; Mak, T.W. Negative regulation of PKB/Akt-dependent cell survival by the tumor suppressor PTEN. Cell 1998, 95, 29–39. [Google Scholar] [CrossRef]
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. Mirbase: MicroRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144. [Google Scholar] [CrossRef]
- Chiefari, E.; Iiritano, S.; Paonessa, F.; Le Pera, I.; Arcidiacono, B.; Filocamo, M.; Foti, D.; Liebhaber, S.A.; Brunetti, A. Pseudogene-mediated posttranscriptional silencing of HMGA1 can result in insulin resistance and type 2 diabetes. Nat. Commun. 2010, 1, 40. [Google Scholar] [CrossRef]
- Bier, A.; Oviedo-Landaverde, I.; Zhao, J.; Mamane, Y.; Kandouz, M.; Batist, G. Connexin43 pseudogene in breast cancer cells offers a novel therapeutic target. Mol. Cancer Ther. 2009, 8, 786–793. [Google Scholar] [CrossRef]
- Kaer, K.; Branovets, J.; Hallikma, A.; Nigumann, P.; Speek, M. Intronic L1 retrotransposons and nested genes cause transcriptional interference by inducing intron retention, exonization and cryptic polyadenylation. PLoS ONE 2011, 6, e26099. [Google Scholar] [CrossRef]
- Shearwin, K.E.; Callen, B.P.; Egan, J.B. Transcriptional interference--a crash course. Trends Genet. TIG 2005, 21, 339–345. [Google Scholar] [CrossRef]
- Long, M.; Langley, C.H. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 1993, 260, 91–95. [Google Scholar] [CrossRef]
- Staszak, K.; Makałowska, I. Cancer, retrogenes, and evolution. Life 2021, 11, 72. [Google Scholar] [CrossRef]
- Pon, J.R.; Marra, M.A. Driver and passenger mutations in cancer. Annu. Rev. Pathol. 2015, 10, 25–50. [Google Scholar] [CrossRef]
- Andreozzi, M.; Quintavalle, C.; Benz, D.; Quagliata, L.; Matter, M.; Calabrese, D.; Tosti, N.; Ruiz, C.; Trapani, F.; Tornillo, L.; et al. HMGA1 expression in human hepatocellular carcinoma correlates with poor prognosis and promotes tumor growth and migration in in vitro models. Neoplasia 2016, 18, 724–731. [Google Scholar] [CrossRef]
- Palumbo Júnior, A.; de Sousa, V.P.L.; Esposito, F.; De Martino, M.; Forzati, F.; Moreira, F.C.d.B.; Simão, T.d.A.; Nasciutti, L.E.; Fusco, A.; Ribeiro Pinto, L.F.; et al. Overexpression of HMGA1 Figures as a potential prognostic factor in Endometrioid Endometrial Carcinoma (EEC). Genes 2019, 10, 372. [Google Scholar] [CrossRef] [PubMed]
- Esposito, F.; De Martino, M.; Petti, M.G.; Forzati, F.; Tornincasa, M.; Federico, A.; Arra, C.; Pierantoni, G.M.; Fusco, A. HMGA1 pseudogenes as candidate proto-oncogenic competitive endogenous RNAs. Oncotarget 2014, 5, 8341–8354. [Google Scholar] [CrossRef] [PubMed]
- Waters, A.M.; Der, C.J. KRAS: The critical driver and therapeutic target for pancreatic cancer. Cold Spring Harb. Perspect. Med. 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Grzeskowiak, C.L.; Kundu, S.T.; Mo, X.; Ivanov, A.A.; Zagorodna, O.; Lu, H.; Chapple, R.H.; Tsang, Y.H.; Moreno, D.; Mosqueda, M.; et al. In vivo screening identifies GATAD2B as a metastasis driver in KRAS-driven lung cancer. Nat. Commun. 2018, 9, 2732. [Google Scholar] [CrossRef]
- Miyoshi, N.; Fujino, S.; Ohue, M.; Yasui, M.; Takahashi, Y.; Sugimura, K.; Tomokuni, A.; Akita, H.; Kobayashi, S.; Takahashi, H.; et al. The POU5F1 gene expression in colorectal cancer: A novel prognostic marker. Surg. Today 2018, 48, 709–715. [Google Scholar] [CrossRef]
- Lathia, J.; Liu, H.; Matei, D. The clinical impact of cancer stem cells. Oncologist 2020, 25, 123–131. [Google Scholar] [CrossRef]
- Wang, L.; Guo, Z.-Y.; Zhang, R.; Xin, B.; Chen, R.; Zhao, J.; Wang, T.; Wen, W.-H.; Jia, L.-T.; Yao, L.-B.; et al. Pseudogene OCT4-pg4 functions as a natural micro RNA sponge to regulate OCT4 expression by competing for miR-145 in hepatocellular carcinoma. Carcinogenesis 2013, 34, 1773–1781. [Google Scholar] [CrossRef]
- Bai, M.; Yuan, M.; Liao, H.; Chen, J.; Xie, B.; Yan, D.; Xi, X.; Xu, X.; Zhang, Z.; Feng, Y. OCT4 pseudogene 5 upregulates OCT4 expression to promote proliferation by competing with miR-145 in endometrial carcinoma. Oncol. Rep. 2015, 33, 1745–1752. [Google Scholar] [CrossRef]
- Hawkins, P.G.; Morris, K.V. Transcriptional regulation of Oct4 by a long non-coding RNA antisense to Oct4-pseudogene 5. Transcription 2010, 1, 165–175. [Google Scholar] [CrossRef]
- Scarola, M.; Comisso, E.; Pascolo, R.; Chiaradia, R.; Marion, R.M.; Schneider, C.; Blasco, M.A.; Schoeftner, S.; Benetti, R. Epigenetic silencing of Oct4 by a complex containing SUV39H1 and Oct4 pseudogene lncRNA. Nat. Commun. 2015, 6, 7631. [Google Scholar] [CrossRef]
- Mei, D.; Song, H.; Wang, K.; Lou, Y.; Sun, W.; Liu, Z.; Ding, X.; Guo, J. Up-regulation of SUMO1 pseudogene 3 (SUMO1P3) in gastric cancer and its clinical association. Med. Oncol. 2013, 30, 709. [Google Scholar] [CrossRef]
- Jin, L.; Shen, K.; Chen, T.; Yu, W.; Zhang, H. SUMO-1 Gene Silencing Inhibits Proliferation and Promotes Apoptosis of Human Gastric Cancer SGC-7901 Cells. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2017, 41, 987–998. [Google Scholar] [CrossRef]
- Wang, M.-Y.; Chen, D.-P.; Qi, B.; Li, M.-Y.; Zhu, Y.-Y.; Yin, W.-J.; He, L.; Yu, Y.; Li, Z.-Y.; Lin, L.; et al. Pseudogene RACGAP1P activates RACGAP1/Rho/ERK signalling axis as a competing endogenous RNA to promote hepatocellular carcinoma early recurrence. Cell Death Dis. 2019, 10, 426. [Google Scholar] [CrossRef]
- Zhou, D.; Ren, K.; Wang, M.; Wang, J.; Li, E.; Hou, C.; Su, Y.; Jin, Y.; Zou, Q.; Zhou, P.; et al. Long non-coding RNA RACGAP1P promotes breast cancer invasion and metastasis via miR-345-5p/RACGAP1-mediated mitochondrial fission. Mol. Oncol. 2021, 15, 543–559. [Google Scholar] [CrossRef]
- Pliarchopoulou, K.; Kalogeras, K.T.; Kronenwett, R.; Wirtz, R.M.; Eleftheraki, A.G.; Batistatou, A.; Bobos, M.; Soupos, N.; Polychronidou, G.; Gogas, H.; et al. Prognostic significance of RACGAP1 mRNA expression in high-risk early breast cancer: A study in primary tumors of breast cancer patients participating in a randomized Hellenic Cooperative Oncology Group trial. Cancer Chemother. Pharmacol. 2013, 71, 245–255. [Google Scholar] [CrossRef]
- Wang, C.; Wang, W.; Liu, Y.; Yong, M.; Yang, Y.; Zhou, H. Rac GTPase activating protein 1 promotes oncogenic progression of epithelial ovarian cancer. Cancer Sci. 2018, 109, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Zhang, F.; Yu, M.; Zhao, P.; Ji, W.; Zhang, H.; Han, J.; Niu, R. Up-regulation of Anxa2 gene promotes proliferation and invasion of breast cancer MCF-7 cells. Cell Prolif. 2012, 45, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.-S.; Shi, L.-L.; Sun, F.; Zhang, Y.-F.; Chen, R.-W.; Yang, S.-L.; Hu, J.-L. High expression of ANXA2 pseudogene ANXA2P2 promotes an aggressive phenotype in hepatocellular carcinoma. Dis. Markers 2019, 2019, 9267046. [Google Scholar] [CrossRef] [PubMed]
- Cao, S.-W.; Huang, J.-L.; Chen, J.; Hu, Y.-W.; Hu, X.-M.; Ren, T.-Y.; Zheng, S.-H.; Lin, J.-D.; Tang, J.; Zheng, L.; et al. Long non-coding RNA UBE2CP3 promotes tumor metastasis by inducing epithelial-mesenchymal transition in hepatocellular carcinoma. Oncotarget 2017, 8, 65370–65385. [Google Scholar] [CrossRef]
- Peng, H.; Ishida, M.; Li, L.; Saito, A.; Kamiya, A.; Hamilton, J.P.; Fu, R.; Olaru, A.V.; An, F.; Popescu, I.; et al. Pseudogene INTS6P1 regulates its cognate gene INTS6 through competitive binding of miR-17-5p in hepatocellular carcinoma. Oncotarget 2015, 6, 5666–5677. [Google Scholar] [CrossRef]
- Hu, S.; Xu, L.; Li, L.; Luo, D.; Zhao, H.; Li, D.; Peng, B. Overexpression of lncRNA PTENP1 suppresses glioma cell proliferation and metastasis in vitro. Oncotargets Ther. 2019, 12, 147–156. [Google Scholar] [CrossRef]
- Vitiello, M.; Evangelista, M.; Zhang, Y.; Salmena, L.; Pandolfi, P.P.; Poliseno, L. PTENP1 is a ceRNA for PTEN: It’s CRISPR clear. J. Hematol. Oncol. 2020, 13, 73. [Google Scholar] [CrossRef]
- Rutnam, Z.J.; Du, W.W.; Yang, W.; Yang, X.; Yang, B.B. The pseudogene TUSC2P promotes TUSC2 function by binding multiple microRNAs. Nat. Commun. 2014, 5, 2914. [Google Scholar] [CrossRef]
- Wu, S.; Chen, S.; Lin, N.; Yang, J. Long non-coding RNA SUMO1P3 promotes hepatocellular carcinoma progression through activating Wnt/β-catenin signalling pathway by targeting miR-320a. J. Cell. Mol. Med. 2020, 24, 3108–3116. [Google Scholar] [CrossRef]
- Qian, Y.-Y.; Li, K.; Liu, Q.-Y.; Liu, Z.-S. Long non-coding RNA PTENP1 interacts with miR-193a-3p to suppress cell migration and invasion through the PTEN pathway in hepatocellular carcinoma. Oncotarget 2017, 8, 107859–107869. [Google Scholar] [CrossRef]
- Liu, F.; Gong, R.; He, B.; Chen, F.; Hu, Z. TUSC2P suppresses the tumor function of esophageal squamous cell carcinoma by regulating TUSC2 expression and correlates with disease prognosis. BMC Cancer 2018, 18, 894. [Google Scholar] [CrossRef]
- Kong, Y.; Zhang, L.; Huang, Y.; He, T.; Zhang, L.; Zhao, X.; Zhou, X.; Zhou, D.; Yan, Y.; Zhou, J.; et al. Pseudogene PDIA3P1 promotes cell proliferation, migration and invasion, and suppresses apoptosis in hepatocellular carcinoma by regulating the p53 pathway. Cancer Lett. 2017, 407, 76–83. [Google Scholar] [CrossRef]
- Xu, T.; Li, D.; He, Y.; Zhang, F.; Qiao, M.; Chen, Y. The expression level of CSDAP1 in lung cancer and its clinical significance. Oncol. Lett. 2018, 16, 4361–4366. [Google Scholar] [CrossRef]
- Xu, H.; Chen, B.; Xing, J.; Wei, Z.; Liu, C.; Qiu, Y.; Lin, Y.; Ren, L. Upregulation of LGMNP1 confers radiotherapy resistance in glioblastoma. Oncol. Rep. 2019, 41, 3435–3443. [Google Scholar] [CrossRef]
- Lou, W.; Ding, B.; Fan, W. High Expression of pseudogene PTTG3P indicates a poor prognosis in human breast cancer. Mol. Ther. Oncolytics 2019, 14, 15–26. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, W.; Li, Y.; Sun, F.; Lin, J.; Li, L. CKS1BP7, a Pseudogene of CKS1B, is Co-Amplified with IGF1R in Breast Cancers. Pathol. Oncol. Res. POR 2018, 24, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Wang, C.; Zhang, J.; Zhou, Y.; Hu, W.; Guo, T. New insights into long noncoding RNAs and pseudogenes in prognosis of renal cell carcinoma. Cancer Cell Int. 2018, 18, 157. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Zhu, H.; Wu, X.; Xie, X.; Huang, G.; Xu, X.; Li, S.; Xing, C. Downregulated pseudogene CTNNAP1 promote tumor growth in human cancer by downregulating its cognate gene CTNNA1 expression. Oncotarget 2016, 7, 55518–55528. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yu, J. Long non-coding RNA transcribed from pseudogene PPIAP43 is associated with radiation sensitivity of small cell lung cancer cells. Oncol. Lett. 2019, 18, 4583–4592. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, T.; Yang, Z.; Jiang, C.; Seng, J. Long non-coding RNA FTH1P3 activates paclitaxel resistance in breast cancer through miR-206/ABCB1. J. Cell. Mol. Med. 2018, 22, 4068–4075. [Google Scholar] [CrossRef]
- Pan, Y.; Sun, C.; Huang, M.; Liu, Y.; Qi, F.; Liu, L.; Wen, J.; Liu, J.; Xie, K.; Ma, H.; et al. A genetic variant in pseudogene E2F3P1 contributes to prognosis of hepatocellular carcinoma. J. Biomed. Res. 2014, 28, 194–200. [Google Scholar] [CrossRef]
- Costa, V.; Esposito, R.; Aprile, M.; Ciccodicola, A. Non-coding RNA and pseudogenes in neurodegenerative diseases: “The (un)Usual Suspects”. Front. Genet. 2012, 3, 231. [Google Scholar] [CrossRef]
- Yin, L.; Yao, J.; Deng, G.; Wang, X.; Cai, W.; Shen, J. Identification of candidate lncRNAs and circRNAs regulating WNT3/β-catenin signaling in essential hypertension. Aging 2020, 12, 8261–8288. [Google Scholar] [CrossRef]
- Wu, N.; Li, J.; Chen, X.; Xiang, Y.; Wu, L.; Li, C.; Zhang, H.; Tong, S.; Zhong, L.; Li, Y. Identification of long non-coding RNA and circular RNA expression profiles in atrial fibrillation. Heart Lung Circ. 2020, 29, e157–e167. [Google Scholar] [CrossRef]
- Lai, Y.; Li, J.; Zhong, L.; He, X.; Si, X.; Sun, Y.; Chen, Y.; Zhong, J.; Hu, Y.; Li, B.; et al. The pseudogene PTENP1 regulates smooth muscle cells as a competing endogenous RNA. Clin. Sci. 2019, 133, 1439–1455. [Google Scholar] [CrossRef]
- Bergman, O.; Karry, R.; Milhem, J.; Ben-Shachar, D. NDUFV2 pseudogene (NDUFV2P1) contributes to mitochondrial complex I deficits in schizophrenia. Mol. Psychiatry 2020, 25, 805–820. [Google Scholar] [CrossRef]
- Luo, T.; Ou, J.-N.; Cao, L.-F.; Peng, X.-Q.; Li, Y.-M.; Tian, Y.-Q. The autism-related lncRNA MSNP1AS regulates moesin protein to influence the RhoA, Rac1, and PI3K/Akt pathways and regulate the structure and survival of neurons. Autism Res. Off. J. Int. Soc. Autism Res. 2020, 13, 2073–2082. [Google Scholar] [CrossRef]
- Tong, J.; Yang, J.; Lv, H.; Lv, S.; Zhang, C.; Chen, Z.-J. Dysfunction of pseudogene PGK1P2 is involved in preeclampsia by acting as a competing endogenous RNA of PGK1. Pregnancy Hypertens. 2018, 13, 37–45. [Google Scholar] [CrossRef]
- Lv, H.; Tong, J.; Yang, J.; Lv, S.; Li, W.-P.; Zhang, C.; Chen, Z.-J. Dysregulated pseudogene HK2P1 may contribute to preeclampsia as a competing endogenous RNA for hexokinase 2 by impairing decidualization. Hypertension 2018, 71, 648–658. [Google Scholar] [CrossRef]
- Nuerzhati, Y.; Dong, R.; Song, Z.; Zheng, S. Role of the long non-coding RNA-Annexin A2 pseudogene 3/Annexin A2 signaling pathway in biliary atresia-associated hepatic injury. Int. J. Mol. Med. 2019, 43, 739–748. [Google Scholar] [CrossRef]
- Erkkinen, M.G.; Kim, M.-O.; Geschwind, M.D. Clinical neurology and epidemiology of the major neurodegenerative diseases. Cold Spring Harb. Perspect. Biol. 2018, 10. [Google Scholar] [CrossRef]
- Riva, P.; Ratti, A.; Venturin, M. The Long Non-Coding RNAs in neurodegenerative diseases: Novel mechanisms of pathogenesis. Curr. Alzheimer Res. 2016, 13, 1219–1231. [Google Scholar] [CrossRef]
- Yan, W.; Chen, Z.-Y.; Chen, J.-Q.; Chen, H.-M. LncRNA NEAT1 promotes autophagy in MPTP-induced Parkinson’s disease through stabilizing PINK1 protein. Biochem. Biophys. Res. Commun. 2018, 496, 1019–1024. [Google Scholar] [CrossRef]
- Mercer, T.R.; Dinger, M.E.; Sunkin, S.M.; Mehler, M.F.; Mattick, J.S. Specific expression of long noncoding RNAs in the mouse brain. Proc. Natl. Acad. Sci. USA 2008, 105, 716–721. [Google Scholar] [CrossRef]
- Kadakkuzha, B.M.; Liu, X.-A.; McCrate, J.; Shankar, G.; Rizzo, V.; Afinogenova, A.; Young, B.; Fallahi, M.; Carvalloza, A.C.; Raveendra, B.; et al. Transcriptome analyses of adult mouse brain reveal enrichment of lncRNAs in specific brain regions and neuronal populations. Front. Cell. Neurosci. 2015, 9, 63. [Google Scholar] [CrossRef]
- Fang, P.; Schachner, M.; Shen, Y.-Q. HMGB1 in development and diseases of the central nervous system. Mol. Neurobiol. 2012, 45, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, E.; Paudel, Y.N.; Piperi, C. Exploring the role of high-mobility group box 1 (HMGB1) protein in the pathogenesis of Huntington’s disease. J. Mol. Med. 2020, 98, 325–334. [Google Scholar] [CrossRef] [PubMed]
- Lelieveld, S.H.; Reijnders, M.R.F.; Pfundt, R.; Yntema, H.G.; Kamsteeg, E.-J.; de Vries, P.; de Vries, B.B.A.; Willemsen, M.H.; Kleefstra, T.; Löhner, K.; et al. Meta-analysis of 2,104 trios provides support for 10 new genes for intellectual disability. Nat. Neurosci. 2016, 19, 1194–1196. [Google Scholar] [CrossRef] [PubMed]
- Shoshan-Barmatz, V.; Nahon-Crystal, E.; Shteinfer-Kuzmine, A.; Gupta, R. VDAC1, mitochondrial dysfunction, and Alzheimer’s disease. Pharmacol. Res. 2018, 131, 87–101. [Google Scholar] [CrossRef]
- Gao, X.-L.; Tian, W.-J.; Liu, B.; Wu, J.; Xie, W.; Shen, Q. High-mobility group nucleosomal binding domain 2 protects against microcephaly by maintaining global chromatin accessibility during corticogenesis. J. Biol. Chem. 2020, 295, 468–480. [Google Scholar] [CrossRef]
- Matsumata, M.; Sakayori, N.; Maekawa, M.; Owada, Y.; Yoshikawa, T.; Osumi, N. The effects of Fabp7 and Fabp5 on postnatal hippocampal neurogenesis in the mouse. Stem Cells 2012, 30, 1532–1543. [Google Scholar] [CrossRef]
- López, L.; Zuluaga, M.J.; Lagos, P.; Agrati, D.; Bedó, G. The expression of Hypoxia-Induced Gene 1 (Higd1a) in the central nervous system of male and female rats differs according to age. J. Mol. Neurosci. 2018, 66, 462–473. [Google Scholar] [CrossRef]
- Csobonyeiova, M.; Polak, S.; Danisovic, L. recent overview of the use of iPSCs huntington’s disease modeling and therapy. Int. J. Mol. Sci. 2020, 21, 2239. [Google Scholar] [CrossRef]
- Yamanaka, S. Induction of pluripotent stem cells from mouse fibroblasts by four transcription factors. Cell Prolif. 2008, 41 (Suppl. 1), 51–56. [Google Scholar] [CrossRef]
- O’Connor, M.D.; Wederell, E.; Robertson, G.; Delaney, A.; Morozova, O.; Poon, S.S.S.; Yap, D.; Fee, J.; Zhao, Y.; McDonald, H.; et al. Retinoblastoma-binding proteins 4 and 9 are important for human pluripotent stem cell maintenance. Exp. Hematol. 2011, 39, 866–879.e861. [Google Scholar] [CrossRef]
- Nuzziello, N.; Liguori, M. The MicroRNA Centrism in the Orchestration of Neuroinflammation in Neurodegenerative Diseases. Cells 2019, 8, 1193. [Google Scholar] [CrossRef]
- Tsend-Ayush, E.; O’Sullivan, L.A.; Grützner, F.S.; Onnebo, S.M.N.; Lewis, R.S.; Delbridge, M.L.; Marshall Graves, J.A.; Ward, A.C. RBMX gene is essential for brain development in zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2005, 234, 682–688. [Google Scholar] [CrossRef]
- Shashi, V.; Xie, P.; Schoch, K.; Goldstein, D.B.; Howard, T.D.; Berry, M.N.; Schwartz, C.E.; Cronin, K.; Sliwa, S.; Allen, A.; et al. The RBMX gene as a candidate for the Shashi X-linked intellectual disability syndrome. Clin. Genet. 2015, 88, 386–390. [Google Scholar] [CrossRef]
- Liu, G.; Li, K. CHCHD2 and Parkinson’s disease. Lancet. Neurol. 2015, 14, 679–680. [Google Scholar] [CrossRef]
- Seo, J.; Park, M. Molecular crosstalk between cancer and neurodegenerative diseases. Cell. Mol. Life Sci. CMLS 2020, 77, 2659–2680. [Google Scholar] [CrossRef]
- Driver, J.A.; Beiser, A.; Au, R.; Kreger, B.E.; Splansky, G.L.; Kurth, T.; Kiel, D.P.; Lu, K.P.; Seshadri, S.; Wolf, P.A. Inverse association between cancer and Alzheimer’s disease: Results from the Framingham Heart Study. Br. Med. J. Clin. Res. Ed. 2012, 344, e1442. [Google Scholar] [CrossRef]
- Marciniak, P. Retropozycja Genów w Tkankach Nowotworowych. Master’s Thesis, Adam Mickiewicz University, Poznań, Poland, 2018. [Google Scholar]
- World Healh Organization. Available online: https://http://www.who.int/ (accessed on 15 January 2021).
- Montaigne, D.; Marechal, X.; Lefebvre, P.; Modine, T.; Fayad, G.; Dehondt, H.; Hurt, C.; Coisne, A.; Koussa, M.; Remy-Jouet, I.; et al. Mitochondrial dysfunction as an arrhythmogenic substrate: A translational proof-of-concept study in patients with metabolic syndrome in whom post-operative atrial fibrillation develops. J. Am. Coll. Cardiol. 2013, 62, 1466–1473. [Google Scholar] [CrossRef]
- Jeganathan, J.; Saraf, R.; Mahmood, F.; Pal, A.; Bhasin, M.K.; Huang, T.; Mittel, A.; Knio, Z.; Simons, R.; Khabbaz, K.; et al. Mitochondrial dysfunction in atrial tissue of patients developing postoperative atrial fibrillation. Ann. Thorac. Surg. 2017, 104, 1547–1555. [Google Scholar] [CrossRef]
- Rowe, V.L.; Stevens, S.L.; Reddick, T.T.; Freeman, M.B.; Donnell, R.; Carroll, R.C.; Goldman, M.H. Vascular smooth muscle cell apoptosis in aneurysmal, occlusive, and normal human aortas. J. Vasc. Surg. 2000, 31, 567–576. [Google Scholar] [CrossRef]
- Javitt, D.C. Glutamate as a therapeutic target in psychiatric disorders. Mol. Psychiatry 2004, 9, 984–997. [Google Scholar] [CrossRef]
- Konradi, C.; Öngür, D. Role of mitochondria and energy metabolism in schizophrenia and psychotic disorders. Schizophr. Res. 2017, 187, 1–2. [Google Scholar] [CrossRef]
- Scola, G.; Kim, H.K.; Young, L.T.; Andreazza, A.C. A fresh look at complex I in microarray data: Clues to understanding disease-specific mitochondrial alterations in bipolar disorder. Biol. Psychiatry 2013, 73, e4–e5. [Google Scholar] [CrossRef]
- Keeney, P.M.; Xie, J.; Capaldi, R.A.; Bennett, J.P. Parkinson’s disease brain mitochondrial complex I has oxidatively damaged subunits and is functionally impaired and misassembled. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 5256–5264. [Google Scholar] [CrossRef]
- Tang, J.; Yu, Y.; Yang, W. Long noncoding RNA and its contribution to autism spectrum disorders. CNS Neurosci. Ther. 2017, 23, 645–656. [Google Scholar] [CrossRef]
- Kerin, T.; Ramanathan, A.; Rivas, K.; Grepo, N.; Coetzee, G.A.; Campbell, D.B. A noncoding RNA antisense to moesin at 5p14.1 in autism. Sci. Transl. Med. 2012, 4, 128ra140. [Google Scholar] [CrossRef]
- DeWitt, J.J.; Grepo, N.; Wilkinson, B.; Evgrafov, O.V.; Knowles, J.A.; Campbell, D.B. Impact of the autism-associated long noncoding RNA MSNP1AS on neuronal architecture and gene expression in human neural progenitor cells. Genes 2016, 7, 76. [Google Scholar] [CrossRef]
- Amberger, J.S.; Bocchini, C.A.; Scott, A.F.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019, 47, D1038–D1043. [Google Scholar] [CrossRef] [PubMed]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Kinsella, R.J.; Kähäri, A.; Haider, S.; Zamora, J.; Proctor, G.; Spudich, G.; Almeida-King, J.; Staines, D.; Derwent, P.; Kerhornou, A.; et al. Ensembl BioMarts: A hub for data retrieval across taxonomic space. Database J. Biol. Databases Curation 2011, 2011, bar030. [Google Scholar] [CrossRef] [PubMed]
- Rappaport, N.; Twik, M.; Plaschkes, I.; Nudel, R.; Iny Stein, T.; Levitt, J.; Gershoni, M.; Morrey, C.P.; Safran, M.; Lancet, D. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res. 2017, 45, D877–D887. [Google Scholar] [CrossRef] [PubMed]
- Meng, H.; Yamashita, C.; Shiba-Fukushima, K.; Inoshita, T.; Funayama, M.; Sato, S.; Hatta, T.; Natsume, T.; Umitsu, M.; Takagi, J.; et al. Loss of parkinson’s disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun. 2017, 8, 15500. [Google Scholar] [CrossRef]
- Uwatoko, H.; Hama, Y.; Iwata, I.T.; Shirai, S.; Matsushima, M.; Yabe, I.; Utsumi, J.; Sasaki, H. Identification of plasma microRNA expression changes in multiple system atrophy and Parkinson’s disease. Mol. Brain 2019, 12, 49. [Google Scholar] [CrossRef]
- Zhang, H.; Chen, X.; Wang, J.; Guang, W.; Han, W.; Zhang, H.; Tan, X.; Gu, Y. EGR1 decreases the malignancy of human non-small cell lung carcinoma by regulating KRT18 expression. Sci. Rep. 2014, 4, 5416. [Google Scholar] [CrossRef]
- Mayeda, A.; Munroe, S.H.; Cáceres, J.F.; Krainer, A.R. Function of conserved domains of hnRNP A1 and other hnRNP A/B proteins. Embo J. 1994, 13, 5483–5495. [Google Scholar] [CrossRef]
- Yıldırım, Y.; Orhan, E.K.; Iseri, S.A.U.; Serdaroglu-Oflazer, P.; Kara, B.; Solakoğlu, S.; Tolun, A. A frameshift mutation of ERLIN2 in recessive intellectual disability, motor dysfunction and multiple joint contractures. Hum. Mol. Genet. 2011, 20, 1886–1892. [Google Scholar] [CrossRef]
- Al-Saif, A.; Bohlega, S.; Al-Mohanna, F. Loss of ERLIN2 function leads to juvenile primary lateral sclerosis. Ann. Neurol. 2012, 72, 510–516. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Zhang, B.; Bie, Q.; Qian, H.; Xu, W. Transcriptome Analysis Reveals Key Genes and Pathways Associated with Metastasis in Breast Cancer. Oncotargets Ther. 2020, 13, 323–335. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Liu, Q.; Xu, G.; Mao, F.; Qin, T.; Teng, H.; Cai, W.; Yu, P.; Cai, T.; et al. Comparative RNA-seq analysis reveals potential mechanisms mediating the conversion to androgen independence in an LNCaP progression cell model. Cancer Lett. 2014, 342, 130–138. [Google Scholar] [CrossRef]
- Kim, P.; Zhou, X. FusionGDB: Fusion gene annotation DataBase. Nucleic Acids Res. 2019, 47, D994–D1004. [Google Scholar] [CrossRef]
- Mauri, M.; Elli, T.; Caviglia, G.; Uboldi, G.; Azzi, M. RAWGraphs: A visualisation platform to create open outputs. In Proceedings of the Chitaly ‘17: 12th Biannual Conference of the Italian SIGCHI Chapter, Cagliari, Italy, 18 September 2017; pp. 1–5, Copyright (c), 2013-2021 DensityDesign Lab, Calibro, INMAGIK <hello@rawgraphs.io>. [Google Scholar]
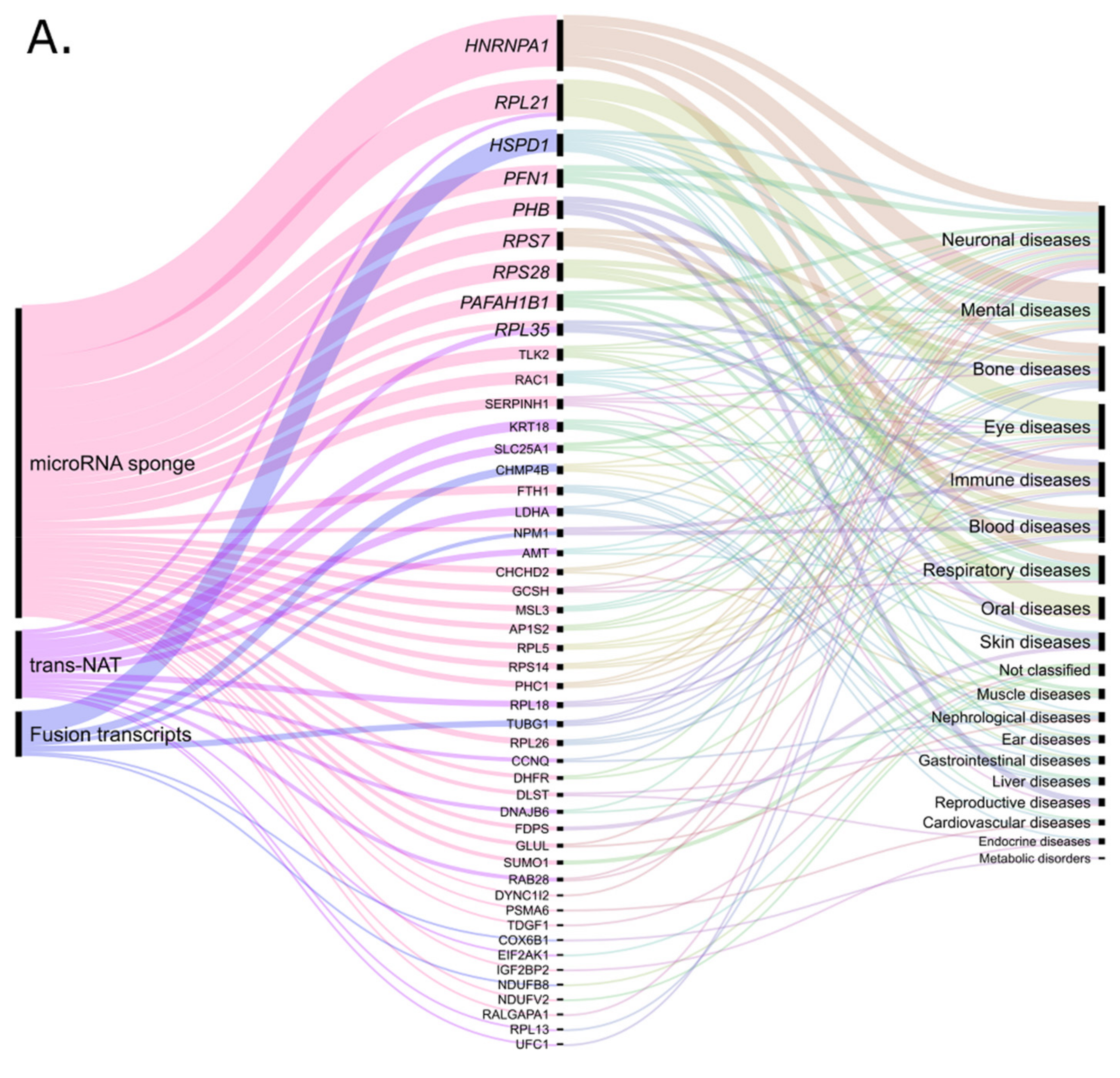
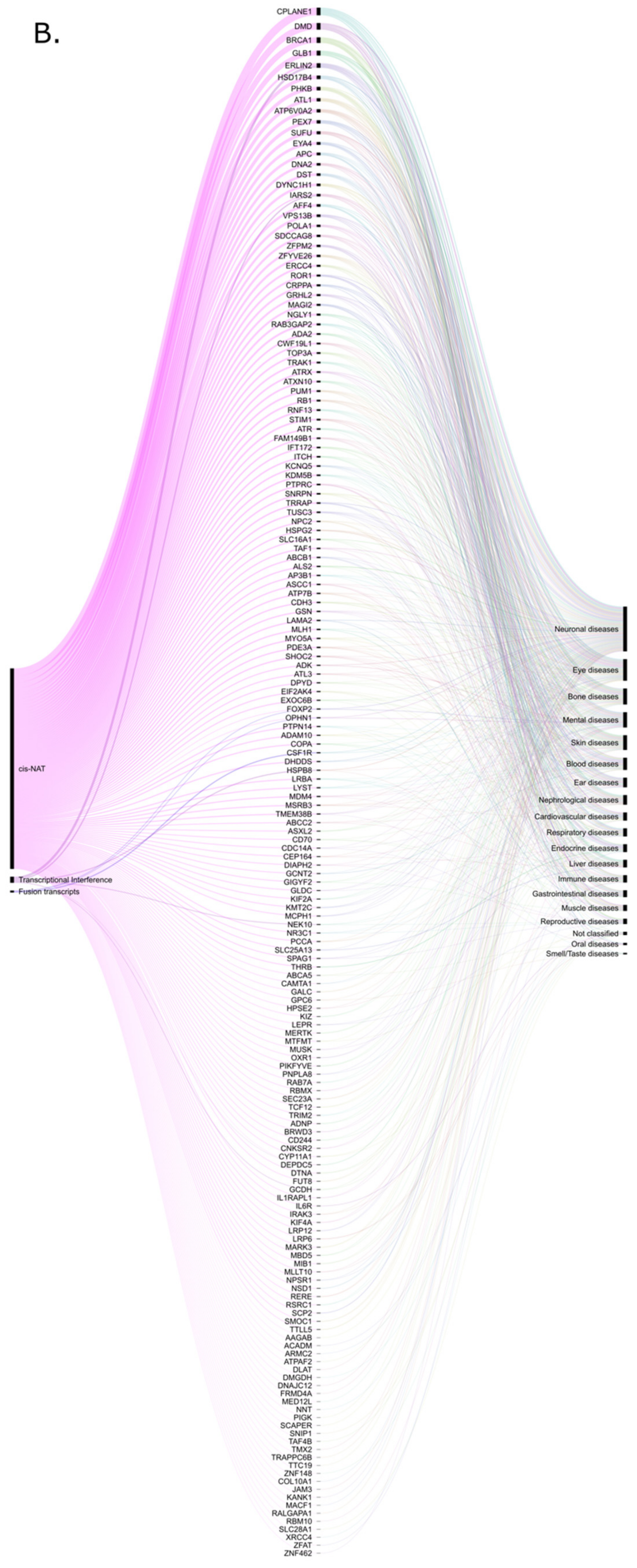
| Retro-lncRNA | Verified or Putative Mechanism | Diseases | Parental Gene |
|---|---|---|---|
| Cancer | |||
| HMGA1P6 | miRNA sponge [43] | endometrial cancer [42], ovarian cancer, thyroid cancer [43] | HMGA1 |
| HMGA1P7 | miRNA sponge [43] | HMGA1 | |
| KRASP1 | miRNA sponge [15] | prostate cancer [15] | KRAS |
| POU5F1P4 (Oct4-pg4) | miRNA sponge [48] | hepatocellular carcinoma [48] | POU5F1 (OCT4) |
| POU5F1P5 (Oct4-pg5) | miRNA sponge [49], epigenetic regulation [50] | endometrial carcinoma [49] | POU5F1 (OCT4) |
| SUMO1P3 | miRNA sponge [3,65]; cis-NAT for host gene [3] | gastric cancer [52], hepatocellular carcinoma [65] | SUMO1 |
| RACGAP1P | miRNA sponge [54,55] | hepatocellular carcinoma [54], breast cancer [55] | RACGAP1 |
| ANXA2P2 | - | hepatocellular carcinoma [59] | ANXA2 |
| UBE2CP3 | - | hepatocellular carcinoma [60] | UBE2CP3 |
| INTS6P1 | miRNA sponge [61] | hepatocellular carcinoma [61] | INTS6 |
| PTENP1 | miRNA sponge [66], epigenetic regulation [30] | hepatocellular carcinoma [66], glioma [62] | PTEN |
| TUSC2P1 | miRNA sponge [64] | esophageal squamous cell carcinoma [67] | TUSC2 |
| PDIA3P1 | miRNA sponge, cis-NAT for host gene [3] | hepatocellular carcinoma [68] | PDIA3 |
| CSDAP1 (YBX3P1) | - | lung cancer [69] | CSDA (YBX3) |
| LGMNP1 | - | glioblastoma [70] | LGMN |
| PTTG3P | miRNA sponge [71] | breast cancer [71] | PTTG1 |
| CKS1BP7 | - | breast cancer [72] | CKS1B |
| MSL3P1 | miRNA sponge [3] | renal cell carcinoma [73] | MSL3 |
| CTNNA1P1 | miRNA sponge [74] | colorectal cancer [74] | CTNNA1 |
| PPIAP43 | miRNA sponge [75] | lung cancer [75] | PPIA |
| FTH1P3 | miRNA sponge [76] | breast cancer [76] | FTH1 |
| E2F3P1 | - | hepatocellular carcinoma [77] | E2F3 |
| AC107983.1 | miRNA sponge, cis-NATs for host gene [3] | cancer cell lines [3] | RPS28 |
| SYPL1P2 | - | SYPL1 | |
| NDUFB1P1 | cis-NATs for host gene [3] | NDUFB1 | |
| Neurodegenerative Disorders | |||
| BZW1P2 | miRNA sponge [78] | Huntington’s disease [78] | BZW1 |
| COX7A2P2 | miRNA sponge [78] | COX7A2 | |
| DGKZP1 | miRNA sponge [78] | DGKZ | |
| EEF1A1P5 | miRNA sponge [78], cis-NAT for host gene [3] | EEF1A1 | |
| EIF2S2P4 | miRNA sponge [78], fusion transcript [3] | EIF2S2 | |
| ETF1P1 | miRNA sponge [78] | ETF1 | |
| FABP5P1 | miRNA sponge [78] | FABP5 | |
| HIGD1AP14 | miRNA sponge [78] | HIGD1A | |
| HMGB1P1 | miRNA sponge [78] | HMGB1 | |
| HMGB1P10 | miRNA sponge [3,78]; cis-NAT for host gene [3] | HMGB1 | |
| HMGB1P5 | miRNA sponge [78] | HMGB1 | |
| HMGN1P36 | miRNA sponge [78] | HMGN1 | |
| HMGN2P3 | miRNA sponge [78] | HMGN2 | |
| HNRNPA3P1 | miRNA sponge [78] | HNRNPA3 | |
| HTR7P1 | miRNA sponge [78] | HTR7 | |
| POU5F1P4 (Oct4-pg4) | miRNA sponge [78] | POU5F1(OCT4) | |
| PTENP1 | miRNA sponge [78] | PTEN | |
| RBBP4P4 | miRNA sponge [78] | RBBP4 | |
| RBMS1P1 | miRNA sponge [3,78], cis-NAT for host gene [3] | RBMS1 | |
| RHOQP2 | miRNA sponge [78] | RHOQ | |
| RPLP0P6 | miRNA sponge [78] | RPLP0 | |
| S100A11P1 | miRNA sponge [78] | S100A11 | |
| SKP1P1 | miRNA sponge [78] | SKP1 | |
| TLK2P1 | miRNA sponge [3,78] | TLK2 | |
| VDAC1P1 | miRNA sponge [78] | VDAC1 | |
| VEZF1P1 | miRNA sponge [78] | VEZF1 | |
| YWHAZP3 | miRNA sponge [78] | YWHAZ | |
| ZFAND6P1 | miRNA sponge [78] | ZFAND6 | |
| CHCHD2P2 | miRNA sponge [78] | Parkinson’s disease [78] | CHCHD2 |
| PHC1P1 | miRNA sponge [3,78] | PHC1 | |
| RBMXP2 | miRNA sponge [78] | RBMX | |
| CHCHD2P2 | miRNA sponge [78] | Huntington’s disease, Parkinson’s disease [78] | CHCHD2 |
| Other Diseses | |||
| LOC646616 | miRNA sponge [79] | essential hypertension [79] | TMEM183A |
| LAP3P2 | LAP3 | ||
| VDAC2P2 | - | atrial fibrillation [80] | VDAC2 |
| PTENP1 | miRNA sponge [81] | aortic dissection [81] | PTEN |
| NDUFV2P1 | miRNA sponge [3] | shizophrenia [82] | NDUFV2 |
| MSNP1AS | - | autism spectrum disorder [83] | MSN |
| PGK1P2 | miRNA sponge [84] | (severe) preeclampsia [84] | PGK1 |
| HK2P1 | miRNA sponge [85] | (severe) preeclampsia [85] | HK2 |
| ANXA2P3 | - | biliary atresia [86] | ANXA2 |
| HMGA1P8 | competition for factor [34] | diabetes [34] | HMGA1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciomborowska-Basheer, J.; Staszak, K.; Kubiak, M.R.; Makałowska, I. Not So Dead Genes—Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts. Cells 2021, 10, 912. https://doi.org/10.3390/cells10040912
Ciomborowska-Basheer J, Staszak K, Kubiak MR, Makałowska I. Not So Dead Genes—Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts. Cells. 2021; 10(4):912. https://doi.org/10.3390/cells10040912
Chicago/Turabian StyleCiomborowska-Basheer, Joanna, Klaudia Staszak, Magdalena Regina Kubiak, and Izabela Makałowska. 2021. "Not So Dead Genes—Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts" Cells 10, no. 4: 912. https://doi.org/10.3390/cells10040912
APA StyleCiomborowska-Basheer, J., Staszak, K., Kubiak, M. R., & Makałowska, I. (2021). Not So Dead Genes—Retrocopies as Regulators of Their Disease-Related Progenitors and Hosts. Cells, 10(4), 912. https://doi.org/10.3390/cells10040912






