Allotetraploidization in Brachypodium May Have Led to the Dominance of One Parent’s Metabolome in Germinating Seeds
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Seed Germination
2.2. Sample Preparation and Metabolite Extraction
2.3. Metabolomic Data Analysis
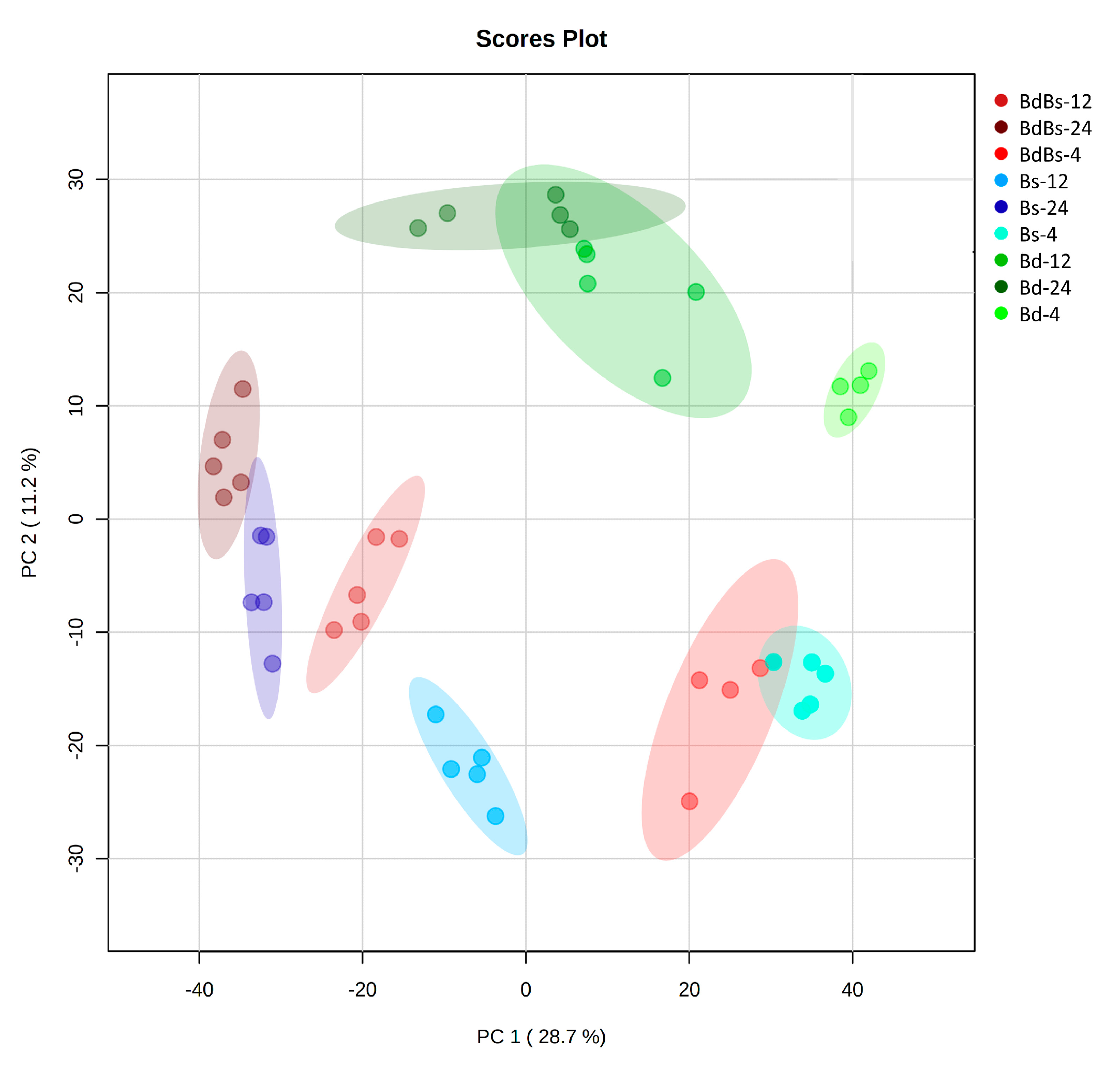
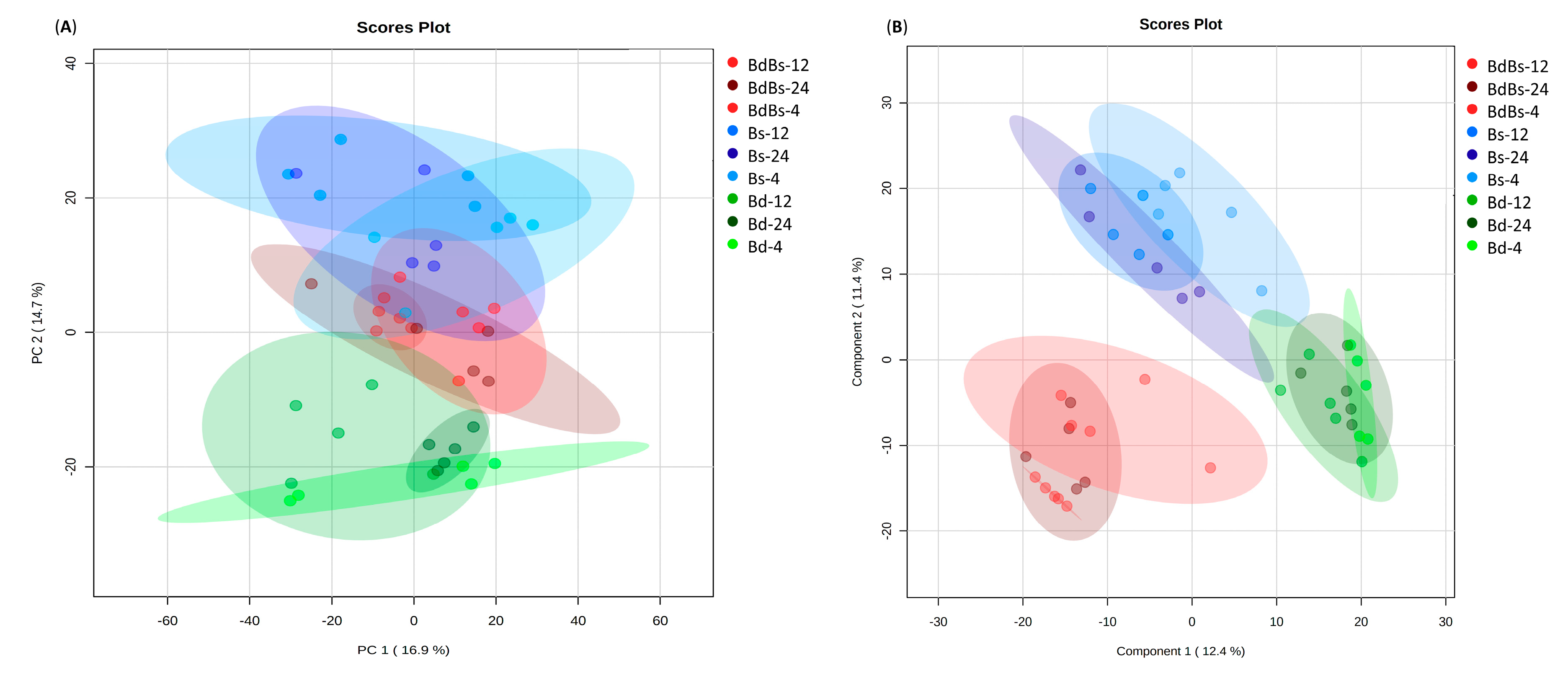
3. Results
3.1. Metabolic Profiling of the Embryo and Endosperm
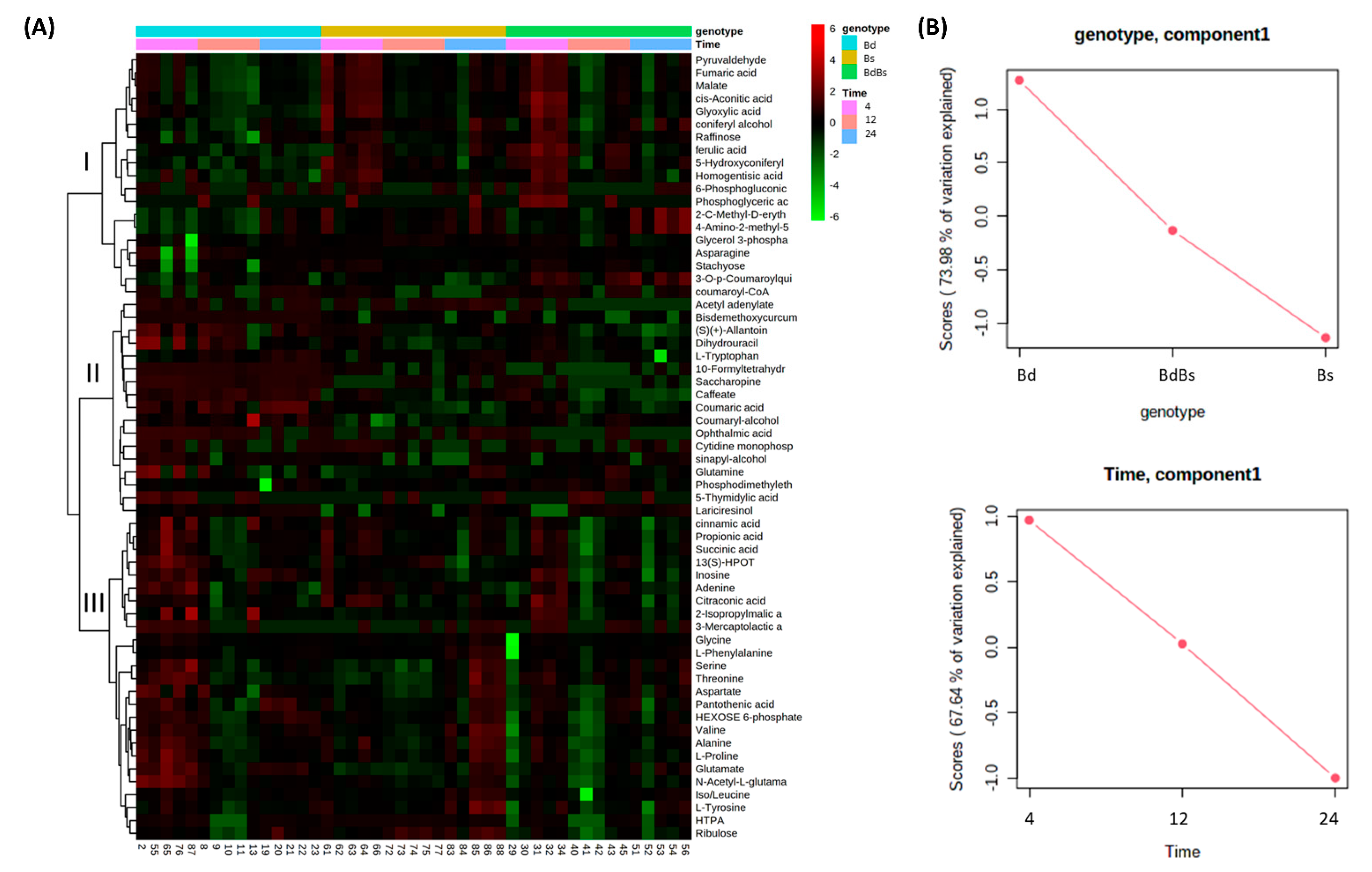
3.2. Assessments of Embryo and Endosperm Metabolomes
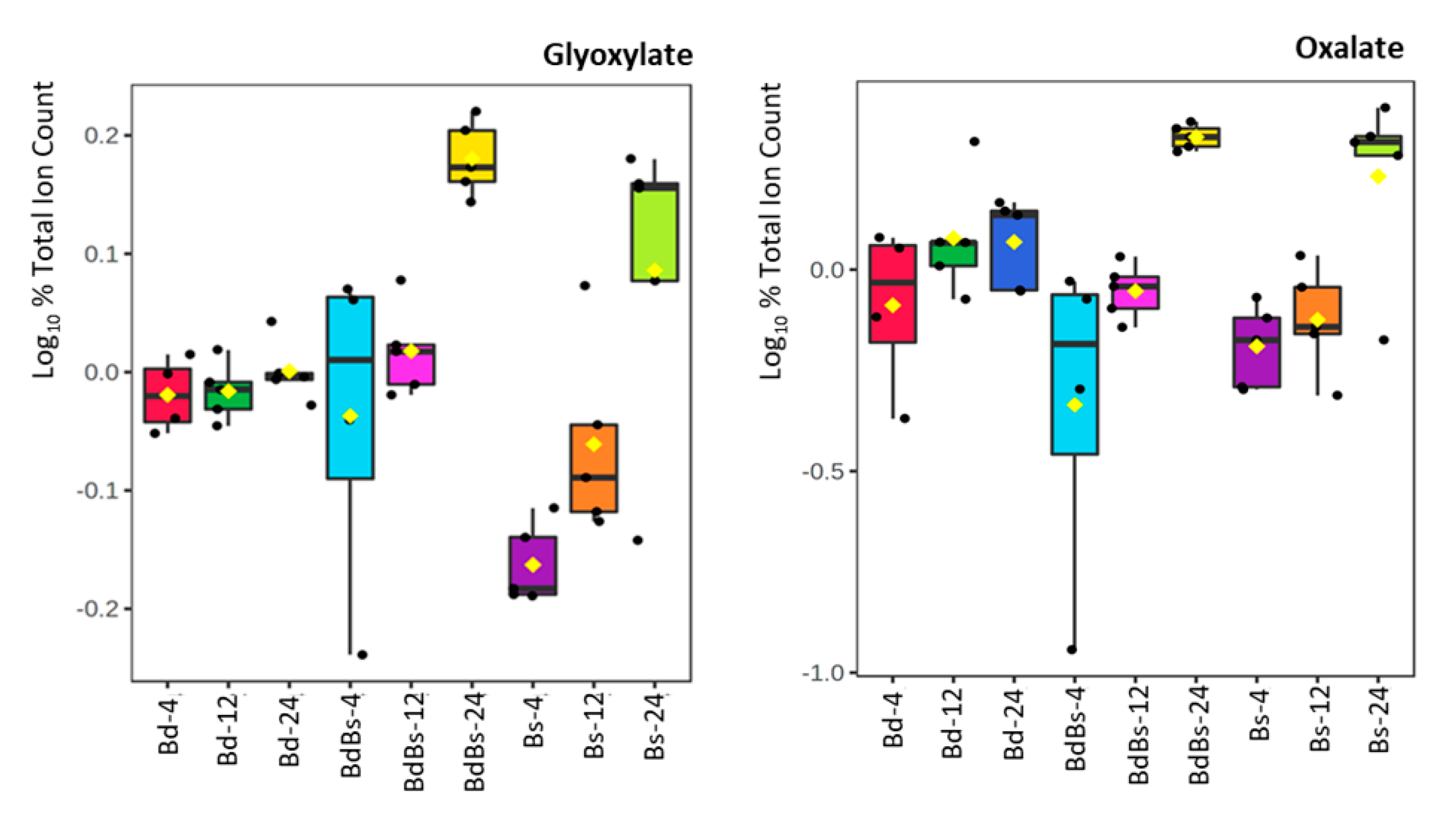
3.3. Species Specific Metabolite Variation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cheng, S.; Zong, Y.; Wang, X. Sub-genome polyploidization Effects on metabolomic signatures in triploid hybrids of Populus. Forests 2019, 10, 1091. [Google Scholar] [CrossRef]
- Liu, Z.; Wang, R.; Wang, J. Comprehensive transcriptomic analysis for developing seeds of a synthetic Brassica hexaploid. Plants 2020, 9, 1141. [Google Scholar] [CrossRef]
- Vergara, F.; Rymen, B.; Kuwahara, A.; Sawada, Y.; Sato, M.; Hirai, M.Y. Autopolyploidization, geographic origin, and metabolome evolution in Arabidopsis thaliana. Am. J. Bot 2017, 104, 905–914. [Google Scholar] [CrossRef]
- Cohen, H.; Fait, A.; Tel-Zur, N. Morphological, cytological and metabolic consequences of autopolyploidization in Hylocereus (Cactaceae) species. BMC Plant Biol. 2013, 13, 173. [Google Scholar] [CrossRef]
- Tan, F.Q.; Tu, H.; Liang, W.J.; Long, J.M.; Wu, X.M.; Zhang, H.Y.; Guo, W.W. Comparative metabolic and transcriptional analysis of a doubled diploid and its diploid citrus rootstock (C. junos cv. Ziyang xiangcheng) suggests its potential value for stress resistance improvement. BMC Plant Biol. 2015, 15, 89. [Google Scholar] [CrossRef]
- Tan, F.Q.; Tu, H.; Wang, R.; Wu, X.M.; Xie, K.D.; Chen, J.J.; Zhang, H.Y.; Xu, J.; Guo, W.W. Metabolic adaptation following genome doubling in citrus doubled diploids revealed by non-targeted metabolomics. Metabolomics 2017, 13, 1–12. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Bewley, J.D. Seed germination and dormancy. Plant Cell 1997, 9, 1055–1066. [Google Scholar] [CrossRef] [PubMed]
- Feenstra, A.D.; Alexander, L.E.; Song, Z.; Korte, A.R.; Yandeau-Nelson, M.D.; Nikolau, B.J.; Lee, Y.J. Spatial mapping and profiling of metabolite distributions during germination. Plant Physiol. 2017, 174, 2532–2548. [Google Scholar] [CrossRef]
- Shu, X.L.; Frank, T.; Shu, Q.Y.; Engel, K.H. Metabolite profiling of germinating rice seeds. J. Agric. Food Chem. 2008, 56, 11612–11620. [Google Scholar] [CrossRef]
- Han, C.; Zhen, S.; Zhu, G.; Bian, Y.; Yan, Y. Comparative metabolome analysis of wheat embryo and endosperm reveals the dynamic changes of metabolites during seed germination. Plant Physiol. Biochem. 2017, 115, 320–327. [Google Scholar] [CrossRef]
- Fait, A.; Angelovici, R.; Less, H.; Ohad, I.; Urbanczyk-Wochniak, E.; Fernie, A.R.; Galili, G. Arabidopsis seed development and germination is associated with temporally distinct metabolic switches. Plant Physiol. 2006, 142, 839–854. [Google Scholar] [CrossRef]
- Palmiano, E.P.; Juliano, B.O. Biochemical Changes in the Rice Grain during Germination. Plant Physiol. 1972, 49, 751–756. [Google Scholar] [CrossRef] [PubMed]
- Howell, K.A.; Narsai, R.; Carroll, A.; Ivanova, A.; Lohse, M.; Usadel, B.; Millar, A.H.; Whelan, J. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 2009, 149, 961–980. [Google Scholar] [CrossRef] [PubMed]
- Opanowicz, M.; Vain, P.; Draper, J.; Parker, D.; Doonan, J.H. Brachypodium distachyon: Making hay with a wild grass. Trends Plant Sci. 2008, 13, 172–177. [Google Scholar] [CrossRef]
- Betekhtin, A.; Milewska-Hendel, A.; Lusinska, J.; Chajec, L.; Kurczynska, E.; Hasterok, R. Organ and tissue-specific localisation of selected cell wall epitopes in the zygotic embryo of Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19. [Google Scholar] [CrossRef] [PubMed]
- Bouvier d’Yvoire, M.; Bouchabke-Coussa, O.; Voorend, W.; Antelme, S.; Cezard, L.; Legee, F.; Lebris, P.; Legay, S.; Whitehead, C.; McQueen-Mason, S.J.; et al. Disrupting the cinnamyl alcohol dehydrogenase 1 gene (BdCAD1) leads to altered lignification and improved saccharification in Brachypodium Distachyon. Plant J. 2013, 73, 496–508. [Google Scholar] [CrossRef] [PubMed]
- Christensen, U.; Alonso-Simon, A.; Scheller, H.V.; Willats, W.G.; Harholt, J. Characterization of the primary cell walls of seedlings of Brachypodium distachyon—A potential model plant for temperate grasses. Phytochemistry 2010, 71, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Salazar, A.; Zabotina, O.A.; Hong, M. Structure and dynamics of Brachypodium primary cell wall polysaccharides from two-dimensional (13)C solid-state nuclear magnetic resonance spectroscopy. Biochemistry 2014, 53, 2840–2854. [Google Scholar] [CrossRef]
- Fitzgerald, T.L.; Powell, J.J.; Schneebeli, K.; Hsia, M.M.; Gardiner, D.M.; Bragg, J.N.; McIntyre, C.L.; Manners, J.M.; Ayliffe, M.; Watt, M.; et al. Brachypodium as an emerging model for cereal-pathogen interactions. Ann. Bot. 2015, 115, 717–731. [Google Scholar] [CrossRef]
- Routledge, A.P.; Shelley, G.; Smith, J.V.; Talbot, N.J.; Draper, J.; Mur, L.A. Magnaporthe grisea interactions with the model grass Brachypodium distachyon closely resemble those with rice (Oryza sativa). Mol. Plant Pathol. 2004, 5, 253–265. [Google Scholar] [CrossRef]
- Lenk, I.; Fisher, L.H.C.; Vickers, M.; Akinyemi, A.; Didion, T.; Swain, M.; Jensen, C.S.; Mur, L.A.J.; Bosch, M. Transcriptional and metabolomic analyses indicate that cell wall properties are associated with drought tolerance in Brachypodium distachyon. Int. J. Mol. Sci. 2019, 20, 1758. [Google Scholar] [CrossRef]
- Priest, H.D.; Fox, S.E.; Rowley, E.R.; Murray, J.R.; Michael, T.P.; Mockler, T.C. Analysis of global gene expression in Brachypodium distachyon reveals extensive network plasticity in response to abiotic stress. PLoS ONE 2014, 9, e87499. [Google Scholar] [CrossRef]
- Ruíz, M.; Quemada, M.; García, R.M.; Carrillo, J.M.; Benavente, E. Use of thermographic imaging to screen for drought-tolerant genotypes in Brachypodium distachyon. Crop. Pasture Sci. 2016, 67, 99–108. [Google Scholar] [CrossRef][Green Version]
- Barrero, J.M.; Jacobsen, J.V.; Talbot, M.J.; White, R.G.; Swain, S.M.; Garvin, D.F.; Gubler, F. Grain dormancy and light quality effects on germination in the model grass Brachypodium distachyon. New Phytol. 2012, 193, 376–386. [Google Scholar] [CrossRef]
- Wolny, E.; Betekhtin, A.; Rojek, M.; Braszewska-Zalewska, A.; Lusinska, J.; Hasterok, R. Germination and the early stages of seedling development in Brachypodium distachyon. Int. J. Mol. Sci. 2018, 19, 2916. [Google Scholar] [CrossRef] [PubMed]
- Wolny, E.; Braszewska-Zalewska, A.; Kroczek, D.; Hasterok, R. Histone H3 and H4 acetylation patterns are more dynamic than those of DNA methylation in Brachypodium distachyon embryos during seed maturation and germination. Protoplasma 2017, 254, 2045–2052. [Google Scholar] [CrossRef] [PubMed]
- Catalan, P.; Muller, J.; Hasterok, R.; Jenkins, G.; Mur, L.A.; Langdon, T.; Betekhtin, A.; Siwinska, D.; Pimentel, M.; Lopez-Alvarez, D. Evolution and taxonomic split of the model grass Brachypodium distachyon. Ann. Bot. 2012, 109, 385–405. [Google Scholar] [CrossRef]
- Gordon, S.P.; Contreras-Moreira, B.; Levy, J.J.; Djamei, A.; Czedik-Eysenberg, A.; Tartaglio, V.S.; Session, A.; Martin, J.; Cartwright, A.; Katz, A.; et al. Gradual polyploid genome evolution revealed by pan-genomic analysis of Brachypodium hybridum and its diploid progenitors. Nat. Commun. 2020, 11, 3670. [Google Scholar] [CrossRef]
- Brkljacic, J.; Grotewold, E.; Scholl, R.; Mockler, T.; Garvin, D.F.; Vain, P.; Brutnell, T.; Sibout, R.; Bevan, M.; Budak, H.; et al. Brachypodium as a model for the grasses: Today and the future. Plant Physiol. 2011, 157, 3–13. [Google Scholar] [CrossRef]
- Dinh Thi, V.H.; Coriton, O.; Le Clainche, I.; Arnaud, D.; Gordon, S.P.; Linc, G.; Catalan, P.; Hasterok, R.; Vogel, J.P.; Jahier, J.; et al. Recreating stable Brachypodium hybridum allotetraploids by uniting the divergent genomes of B. distachyon and B. stacei. PLoS ONE 2016, 11, e0167171. [Google Scholar] [CrossRef] [PubMed]
- Fisher, L.H.; Han, J.; Corke, F.M.; Akinyemi, A.; Didion, T.; Nielsen, K.K.; Doonan, J.H.; Mur, L.A.; Bosch, M. Linking dynamic phenotyping with metabolite analysis to study natural variation in drought responses of Brachypodium distachyon. Front. Plant Sci. 2016, 7, 1751. [Google Scholar] [CrossRef] [PubMed]
- Onda, Y.; Hashimoto, K.; Yoshida, T.; Sakurai, T.; Sawada, Y.; Hirai, M.Y.; Toyooka, K.; Mochida, K.; Shinozaki, K. Determination of growth stages and metabolic profiles in Brachypodium distachyon for comparison of developmental context with Triticeae crops. Proc. Biol. Sci. 2015, 282, 20150964. [Google Scholar] [CrossRef]
- Onda, Y.; Inoue, K.; Sawada, Y.; Shimizu, M.; Takahagi, K.; Uehara-Yamaguchi, Y.; Hirai, M.Y.; Garvin, D.F.; Mochida, K. Genetic variation for seed metabolite levels in Brachypodium distachyon. Int. J. Mol. Sci. 2019, 20, 2348. [Google Scholar] [CrossRef]
- Opanowicz, M.; Hands, P.; Betts, D.; Parker, M.L.; Toole, G.A.; Mills, E.N.; Doonan, J.H.; Drea, S. Endosperm development in Brachypodium distachyon. J. Exp. Bot. 2011, 62, 735–748. [Google Scholar] [CrossRef]
- Guillon, F.; Bouchet, B.; Jamme, F.; Robert, P.; Quemener, B.; Barron, C.; Larre, C.; Dumas, P.; Saulnier, L. Brachypodium distachyon grain: Characterization of endosperm cell walls. J. Exp. Bot. 2011, 62, 1001–1015. [Google Scholar] [CrossRef]
- Baptista, R.; Fazakerley, D.M.; Beckmann, M.; Baillie, L.; Mur, L.A.J. Untargeted metabolomics reveals a new mode of action of pretomanid (PA-824). Sci. Rep. 2018, 8, 5084. [Google Scholar] [CrossRef]
- Chong, J.; Wishart, D.S.; Xia, J. Using MetaboAnalyst 4.0 for Comprehensive and Integrative Metabolomics Data Analysis. Curr. Protoc. Bioinform. 2019, 68, e86. [Google Scholar] [CrossRef]
- Wolny, E.; Braszewska-Zalewska, A.; Hasterok, R. Spatial distribution of epigenetic modifications in Brachypodium distachyon embryos during seed maturation and germination. PLoS ONE 2014, 9, e101246. [Google Scholar] [CrossRef]
- Burke, M.; Small, D.M.; Antolasic, F.; Hughes, J.G.; Spencer, M.J.S.; Blanch, E.W.; Jones, O.A.H. Infrared Spectroscopy-Based Metabolomic Analysis for the Detection of Preharvest Sprouting in Grain. Cereal Chem. 2016, 93, 444–449. [Google Scholar] [CrossRef]
- Das, A.; Kim, D.W.; Khadka, P.; Rakwal, R.; Rohila, J.S. Unraveling Key Metabolomic Alterations in Wheat Embryos Derived from Freshly Harvested and Water-Imbibed Seeds of Two Wheat Cultivars with Contrasting Dormancy Status. Front. Plant Sci. 2017, 8, 1203. [Google Scholar] [CrossRef]
- Kazmi, R.H.; Willems, L.A.J.; Joosen, R.V.L.; Khan, N.; Ligterink, W.; Hilhorst, H.W.M. Metabolomic analysis of tomato seed germination. Metabolomics 2017, 13, 145. [Google Scholar] [CrossRef]
- Jones, O.A. Assessing pre-harvest sprouting in cereals using near-infrared spectroscopy-based metabolomics. NIR News 2017, 28, 15–19. [Google Scholar] [CrossRef]
- Gu, Y.Q.; Wanjugi, H.; Coleman-Derr, D.; Kong, X.; Anderson, O.D. Conserved globulin gene across eight grass genomes identify fundamental units of the loci encoding seed storage proteins. Funct. Integr. Genom. 2010, 10, 111–122. [Google Scholar] [CrossRef]
- Hands, P.; Drea, S. A comparative view of grain development in Brachypodium distachyon. J. Cereal Sci. 2012, 56, 2–8. [Google Scholar] [CrossRef]
- Martin, S.L.; Husband, B.C. Whole genome duplication affects evolvability of flowering time in an autotetraploid plant. PLoS ONE 2012, 7, e44784. [Google Scholar] [CrossRef]
- Yao, H.; Kato, A.; Mooney, B.; Birchler, J.A. Phenotypic and gene expression analyses of a ploidy series of maize inbred Oh43. Plant Mol. Biol. 2011, 75, 237–251. [Google Scholar] [CrossRef]
- Chao, D.Y.; Dilkes, B.; Luo, H.; Douglas, A.; Yakubova, E.; Lahner, B.; Salt, D.E. Polyploids exhibit higher potassium uptake and salinity tolerance in Arabidopsis. Science 2013, 341, 658–659. [Google Scholar] [CrossRef] [PubMed]
- Del Pozo, J.C.; Ramirez-Parra, E. Deciphering the molecular bases for drought tolerance in Arabidopsis autotetraploids. Plant Cell Environ. 2014, 37, 2722–2737. [Google Scholar] [CrossRef]
- Yi, G.; Shin, H.; Park, H.R.; Park, J.E.; Ahn, J.H.; Lim, S.; Lee, J.G.; Lee, E.J.; Huh, J.H. Revealing biomass heterosis in the allodiploid xBrassicoraphanus, a hybrid between Brassica rapa and Raphanus sativus, through integrated transcriptome and metabolites analysis. BMC Plant Biol. 2020, 20, 252. [Google Scholar] [CrossRef] [PubMed]
- Von Well, E.; Fossey, A. A comparative investigation of seed germination, metabolism and seedling growth between two polyploid Triticum species. Euphytica 1998, 101, 83–89. [Google Scholar] [CrossRef]
- Yan, D.; Duermeyer, L.; Leoveanu, C.; Nambara, E. The functions of the endosperm during seed germination. Plant Cell Physiol. 2014, 55, 1521–1533. [Google Scholar] [CrossRef]
- The International Brachypodium Initiative. Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature 2010, 463, 763–768. [Google Scholar] [CrossRef]
- Fujimoto, R.; Taylor, J.M.; Shirasawa, S.; Peacock, W.J.; Dennis, E.S. Heterosis of Arabidopsis hybrids between C24 and Col is associated with increased photosynthesis capacity. Proc. Natl. Acad. Sci. USA 2012, 109, 7109–7114. [Google Scholar] [CrossRef]
- Korn, M.; Peterek, S.; Mock, H.P.; Heyer, A.G.; Hincha, D.K. Heterosis in the freezing tolerance, and sugar and flavonoid contents of crosses between Arabidopsis thaliana accessions of widely varying freezing tolerance. Plant Cell Environ. 2008, 31, 813–827. [Google Scholar] [CrossRef]
- Song, G.S.; Zhai, H.L.; Peng, Y.G.; Zhang, L.; Wei, G.; Chen, X.Y.; Xiao, Y.G.; Wang, L.; Chen, Y.J.; Wu, B.; et al. Comparative transcriptional profiling and preliminary study on heterosis mechanism of super-hybrid rice. Mol. Plant 2010, 3, 1012–1025. [Google Scholar] [CrossRef]
- Bernard, S.M.; Habash, D.Z. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009, 182, 608–620. [Google Scholar] [CrossRef]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Skalska, A.; Wolny, E.; Beckmann, M.; Doonan, J.H.; Hasterok, R.; Mur, L.A.J. Allotetraploidization in Brachypodium May Have Led to the Dominance of One Parent’s Metabolome in Germinating Seeds. Cells 2021, 10, 828. https://doi.org/10.3390/cells10040828
Skalska A, Wolny E, Beckmann M, Doonan JH, Hasterok R, Mur LAJ. Allotetraploidization in Brachypodium May Have Led to the Dominance of One Parent’s Metabolome in Germinating Seeds. Cells. 2021; 10(4):828. https://doi.org/10.3390/cells10040828
Chicago/Turabian StyleSkalska, Aleksandra, Elzbieta Wolny, Manfred Beckmann, John H. Doonan, Robert Hasterok, and Luis A. J. Mur. 2021. "Allotetraploidization in Brachypodium May Have Led to the Dominance of One Parent’s Metabolome in Germinating Seeds" Cells 10, no. 4: 828. https://doi.org/10.3390/cells10040828
APA StyleSkalska, A., Wolny, E., Beckmann, M., Doonan, J. H., Hasterok, R., & Mur, L. A. J. (2021). Allotetraploidization in Brachypodium May Have Led to the Dominance of One Parent’s Metabolome in Germinating Seeds. Cells, 10(4), 828. https://doi.org/10.3390/cells10040828







