The Uncovered Function of the Drosophila GBA1a-Encoded Protein
Abstract
1. Introduction
2. Materials and Methods
2.1. Antibodies
2.2. Construction of Plasmids
2.3. Cells and Transfections
2.4. Fly Strains
2.5. MG132 (Carbobenzoxy-L-Leucyl-L-Leucyl-L-Leucinal) Treatment
2.6. RNA Preparation
2.7. RT-PCR
2.8. Quantitative Real Time PCR (qRT-PCR)
2.9. Transcriptomic Sequencing and Analysis
2.10. Labeling with Activity-Based Probes (ABPs)
2.11. Measurement of Body Fluid Volume
2.12. Midgut Morphology and Quantification
2.13. Developmental Regulation
2.14. Climbing Assay
2.15. Survival Assay
2.16. Egg-Laying Experiment
2.17. Statistical Analysis
3. Results
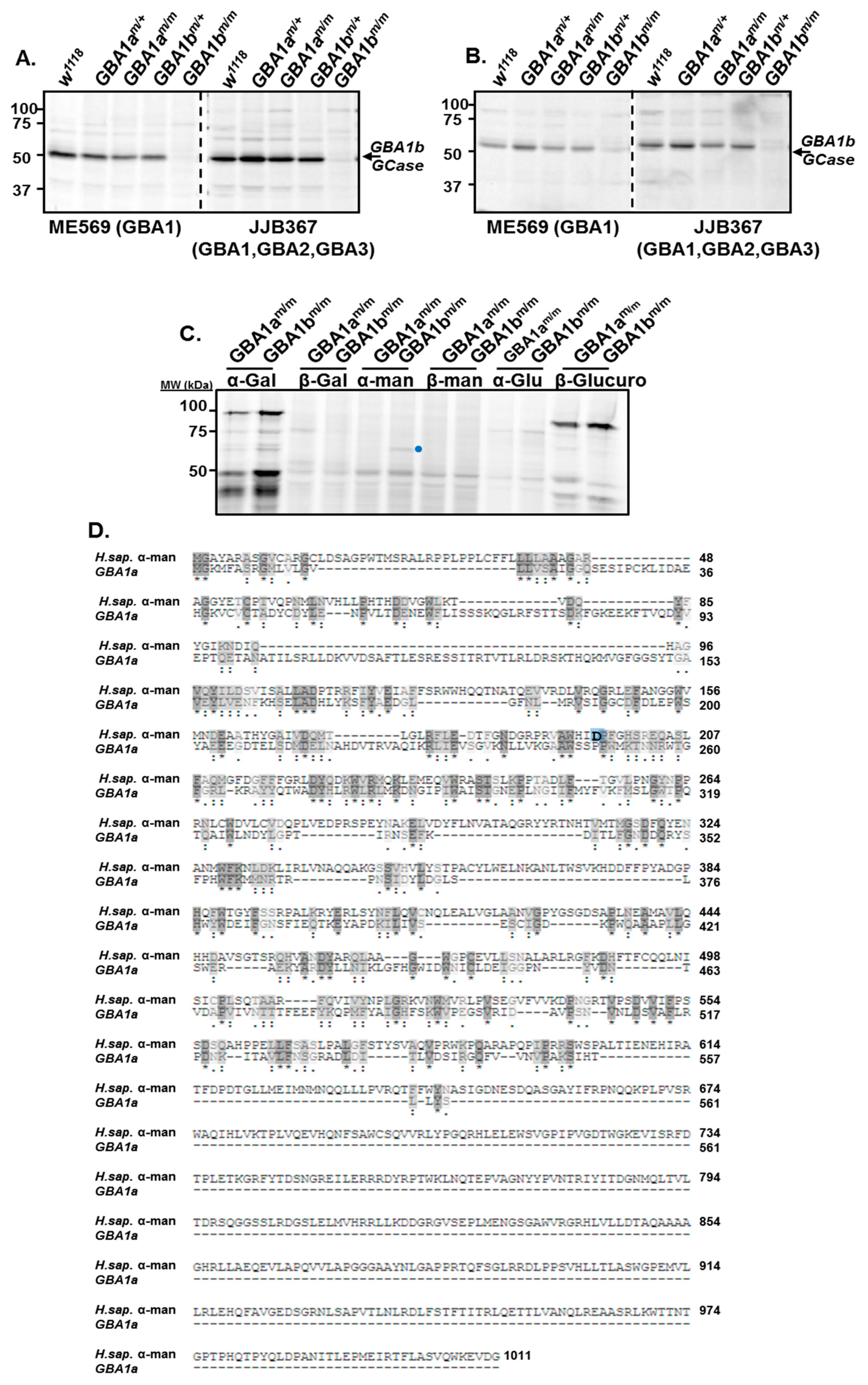
3.1. Activity of GBA1a-Encoded Protein
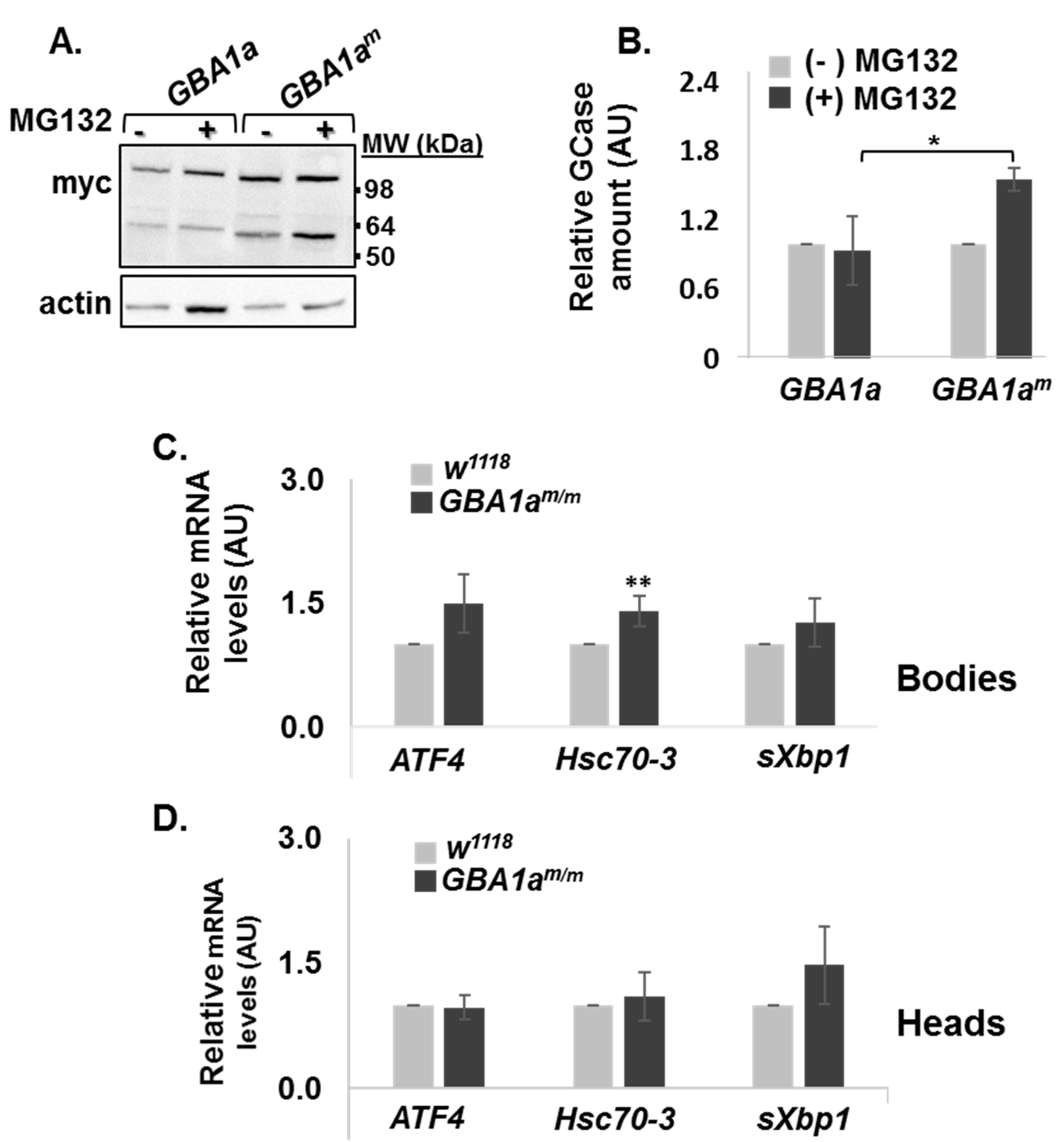
3.2. ERAD and UPR Activation in GBA1am/m Flies
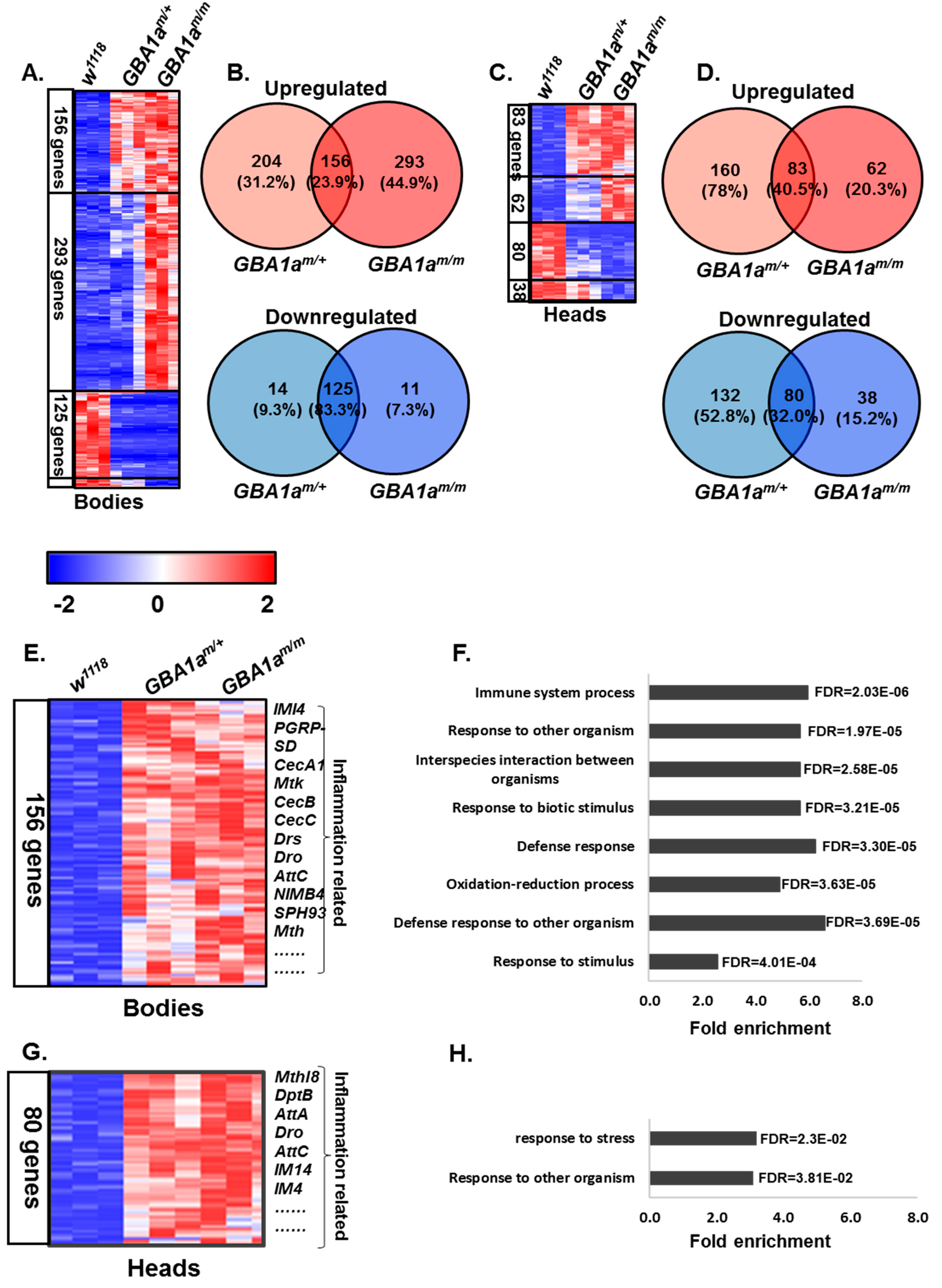
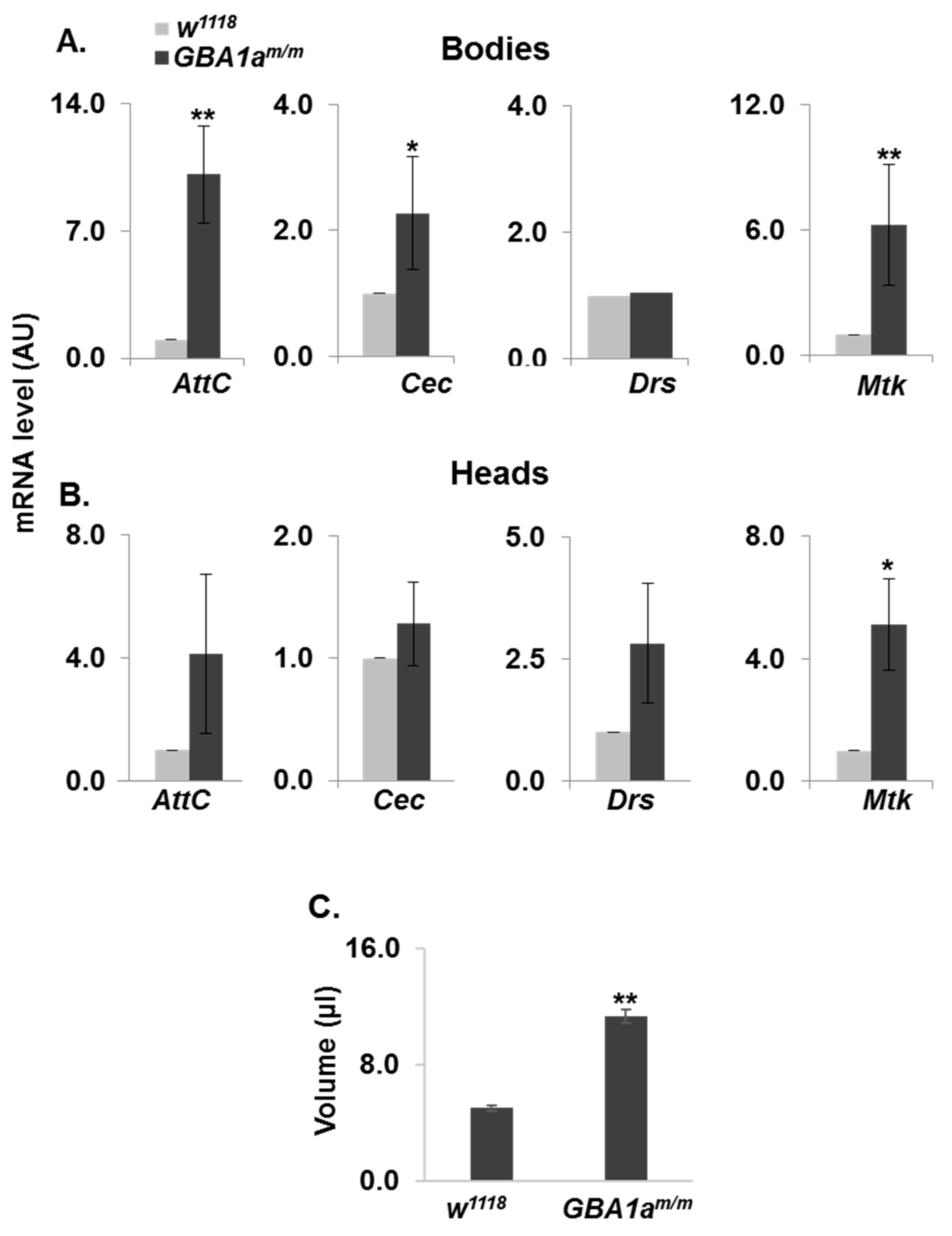
3.3. Inflammation in GBA1am/m Flies
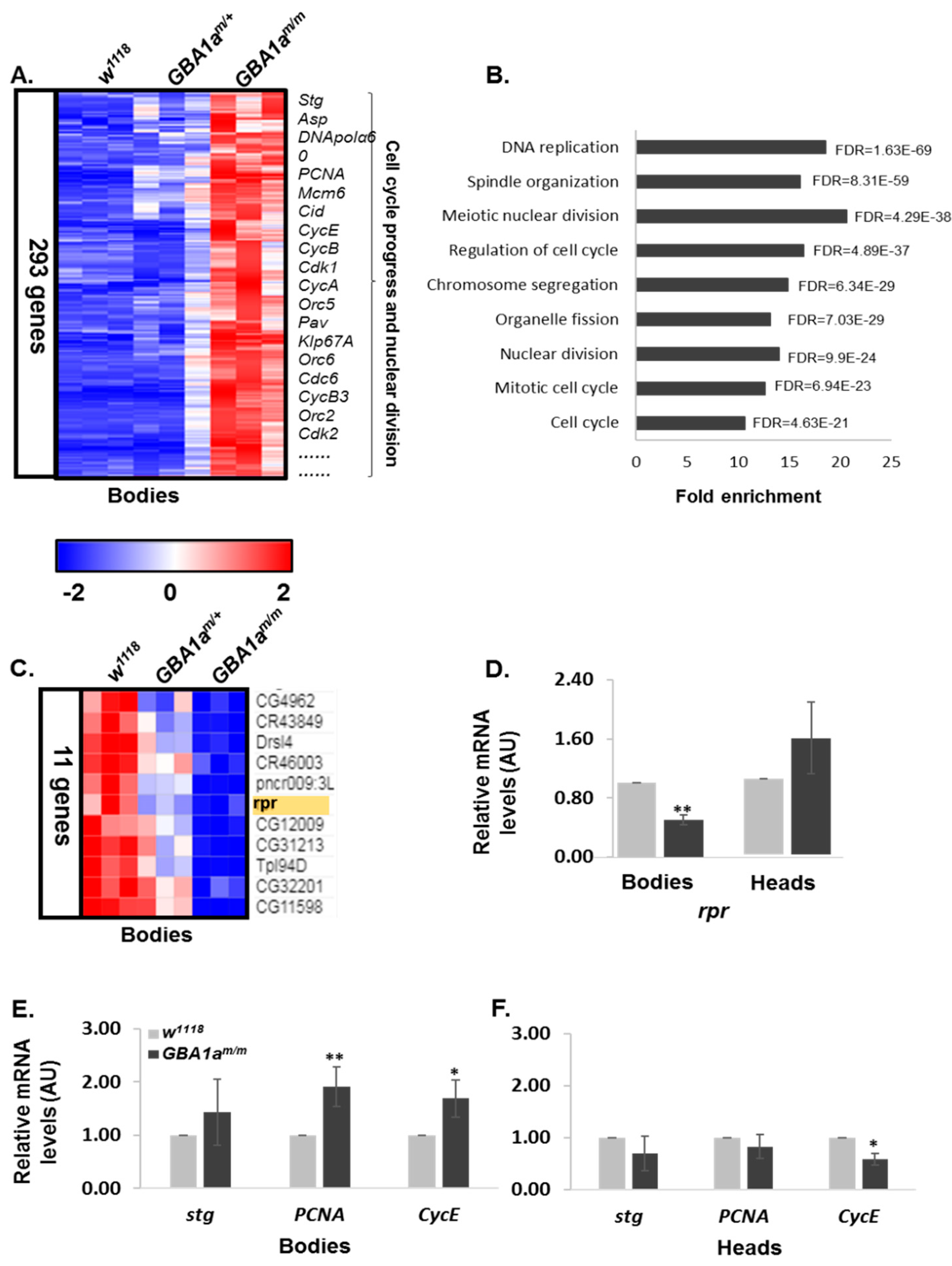
3.4. Enriched Pathways in GBA1am/m Flies
3.5. Delayed Midgut Morphogenesis in GBA1m/m Flies
3.6. Longevity and Locomotion of GBA1am/m Flies
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hruska, K.S.; LaMarca, M.E.; Scott, C.R.; Sidransky, E. Gaucher disease: Mutation and polymorphism spectrum in the gluco-cerebrosidase gene (GBA). Hum. Mutat. 2008, 29, 567–583. [Google Scholar] [CrossRef] [PubMed]
- Dvir, H.; Harel, M.; McCarthy, A.A.; Toker, L.; Silman, I.; Futerman, A.H.; Sussman, J.L. X-ray structure of human ac-id-beta-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003, 4, 704–709. [Google Scholar] [CrossRef] [PubMed]
- Kinghorn, K.J.; Grönke, S.; Castillo-Quan, J.I.; Woodling, N.S.; Li, L.; Sirka, E.; Gegg, M.; Mills, K.; Hardy, J.; Bjedov, I.; et al. A Drosophila Model of Neuronopathic Gaucher Disease Demonstrates Lysosomal-Autophagic Defects and Altered mTOR Signalling and Is Functionally Rescued by Rapamycin. J. Neurosci. 2016, 36, 11654–11670. [Google Scholar] [CrossRef]
- Davis, M.Y.; Trinh, K.; Thomas, R.E.; Yu, S.; Germanos, A.A.; Whitley, B.N.; Sardi, S.P.; Montine, T.J.; Pallanck, L.J. Gluco-cerebrosidase deficiency in w results in alpha-synuclein-independent protein aggregation and neurodegeneration. PLoS Genet. 2016, 12, e1005944. [Google Scholar] [CrossRef]
- Cabasso, O.; Paul, S.; Dorot, O.; Maor, G.; Krivoruk, O.; Pasmanik-Chor, M.; Mirzaian, M.; Ferraz, M.; Aerts, J.; Horowitz, M. Drosophila melanogaster Mutated in its GBA1b Ortholog Recapitulates Neuronopathic Gaucher Disease. J. Clin. Med. 2019, 8, 1420. [Google Scholar] [CrossRef] [PubMed]
- Kawasaki, H.; Suzuki, T.; Ito, K.; Takahara, T.; Goto-Inoue, N.; Setou, M.; Sakata, K.; Ishida, N. Minos-insertion mutant of the Drosophila GBA gene homologue showed abnormal phenotypes of climbing ability, sleep and life span with accumulation of hydroxy-glucocerebroside. Gene 2017, 614, 49–55. [Google Scholar] [CrossRef]
- Suzuki, T.; Shimoda, M.; Ito, K.; Hanai, S.M.; Aizawa, H.; Kato, T.; Kawasaki, K.; Yamaguchi, T.; Ryoo, H.D.; Goto-Inoue, N.; et al. Expression of Human Gaucher Disease Gene GBA Generates Neurodevelopmental Defects and ER Stress in Drosophila Eye. PLoS ONE 2013, 8, e69147. [Google Scholar] [CrossRef]
- Dasari, S.K.; Schejter, E.; Bialik, S.; Shkedy, A.; Levin-Salomon, V.; Levin-Zaidman, S.; Kimchi, A. Death by over-eating: The Gaucher disease associated gene GBA1, identified in a screen for mediators of autophagic cell death, is necessary for devel-opmental cell death in Drosophila midgut. Cell Cycle 2017, 16, 2003–2010. [Google Scholar] [CrossRef]
- Maor, G.; Rencus-Lazar, S.; Filocamo, M.; Steller, H.; Segal, D.; Horowitz, M. Unfolded protein response in Gaucher disease: From human to Drosophila. Orphanet J. Rare Dis. 2013, 8, 140. [Google Scholar] [CrossRef] [PubMed]
- Pekar, O.; Benjamin, S.; Weidberg, H.; Smaldone, S.; Ramirez, F.; Horowitz, M. EHD2 shuttles to the nucleus and represses transcription. Biochem. J. 2012, 444, 383–394. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome. Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. journal of the royal statistical society. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Kallemeijn, W.W.; Witte, M.D.; Voorn-Brouwer, T.M.; Walvoort, M.T.; Li, K.Y.; Codee, J.D.; van der Marel, G.A.; Boot, R.G.; Overkleeft, H.S.; Aerts, J.M. A sensitive gel-based method combining distinct cyclophellitol-based probes for the identification of acid/base residues in human retaining beta-glucosidases. J. Biol. Chem. 2014, 289, 35351–35362. [Google Scholar] [CrossRef] [PubMed]
- Witte, M.D.; Kallemeijn, W.W.; Aten, J.; Li, K.-Y.; Strijland, A.; Donker-Koopman, W.E.; van den Nieuwendijk, A.M.C.H.; Bleijlevens, B.; Kramer, G.; Florea, B.I.; et al. Ultrasensitive in situ visualization of active glucocerebrosidase molecules. Nat. Chem. Biol. 2010, 6, 907–913. [Google Scholar] [CrossRef] [PubMed]
- Willems, L.I.; Beenakker, T.J.M.; Murray, B.; Scheij, S.; Kallemeijn, W.W.; Boot, R.G.; Verhoek, M.; Donker-Koopman, W.E.; Ferraz, M.J.; Van Rijssel, E.R.; et al. Potent and selective activity-based probes for GH27 human retaining α-galactosidases. J. Am. Chem. Soc. 2014, 136, 11622–11625. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, Z.; Kuo, C.-L.; Lahav, D.; Liu, B.; Johnson, R.; Beenakker, T.J.M.; De Boer, C.; Wong, C.-S.; Van Rijssel, E.R.; Debets, M.F.; et al. Manno-epi-cyclophellitols Enable activity-based protein profiling of human α-mannosidases and discovery of new Golgi mannosidase II inhibitors. J. Am. Chem. Soc. 2020, 142, 13021–13029. [Google Scholar] [CrossRef] [PubMed]
- Jiang, J.; Kuo, C.L.; Wu, L.; Franke, C.; Kallemeijn, W.W.; Florea, B.I.; van Meel, E.; van der Marel, G.A.; Codee, J.D.; Boot, R.G.; et al. Detection of active mammalian GH31 alpha-Glucosidases in health and disease using in-class, broad-spectrum ac-tivity-based probes. ACS Cent. Sci. 2016, 2, 351–358. [Google Scholar] [CrossRef]
- Wu, L.; Jiang, J.; Jin, Y.; Kallemeijn, W.W.; Kuo, C.L.; Artola, M.; Dai, W.; van Elk, C.; van Eijk, M.; van der Marel, G.A.; et al. Activity-based probes for functional interrogation of retaining beta-glucuronidases. Nat. Chem. Biol. 2017, 13, 867–873. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Van Meel, E.; Kytidou, K.; Kallemeijn, W.W.; Witte, M.; Overkleeft, H.S.; Artola, M.E.; Aerts, J.M. Activity-Based Probes for Glycosidases: Profiling and Other Applications. Methods Enzymol. 2018, 598, 217–235. [Google Scholar] [CrossRef]
- Inagaki, H.K.; Kamikouchi, A.; Ito, K. Methods for quantifying simple gravity sensing in Drosophila melanogaster. Nat. Protoc. 2010, 5, 20–25. [Google Scholar] [CrossRef]
- Ron, I.; Horowitz, M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum. Mol. Genet. 2005, 14, 2387–2398. [Google Scholar] [CrossRef] [PubMed]
- Bendikov-Bar, I.; Horowitz, M. Gaucher disease paradigm: From ERAD to comorbidity. Hum. Mutat. 2012, 33, 1398–1407. [Google Scholar] [CrossRef] [PubMed]
- Braunstein, H.; Papazian, M.; Maor, G.; Lukas, J.; Rolfs, A.; Horowitz, M. Misfolding of Lysosomal α-Galactosidase a in a Fly Model and its Alleviation by the Pharmacological Chaperone Migalastat. Int. J. Mol. Sci. 2020, 21, 7397. [Google Scholar] [CrossRef]
- Rubin, D.M.; Mehta, A.D.; Zhu, J.; Shoham, S.; Chen, X.; Wells, Q.R.; Palter, K.B. Genomic structure and sequence analysis of Drosophila melanogaster HSC70 genes. Gene 1993, 128, 155–163. [Google Scholar] [CrossRef]
- Chan, S.M.; Zhao, X.; Elfowiris, A.; Ratnam, C.; Herbert, T.P. The role of de novo protein synthesis and SIRT1 in ER stress-induced Atf4 and Chop mRNA expression in mammalian cells. Biochimie 2017, 138, 156–167. [Google Scholar] [CrossRef]
- Ryoo, H.D.; Domingos, P.M.; Kang, M.-J.; Steller, H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2006, 26, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Rigante, D.; Cipolla, C.; Basile, U.; Gulli, F.; Savastano, M.C. Overview of immune abnormalities in lysosomal storage dis-orders. Immunol. Lett. 2017, 188, 79–85. [Google Scholar] [CrossRef]
- Boven, L.A.; van Meurs, M.; Boot, R.G.; Mehta, A.; Boon, L.; Aerts, J.M.; Laman, J.D. Gaucher cells demonstrate a distinct macrophage phenotype and resemble alternatively activated macrophages. Am. J. Clin. Pathol. 2004, 122, 359–369. [Google Scholar] [CrossRef]
- Boot, R.G.; Verhoek, M.; De Fost, M.; Hollak, C.E.M.; Maas, M.; Bleijlevens, B.; Van Breemen, M.J.; Van Meurs, M.; Boven, L.A.; Laman, J.D.; et al. Marked elevation of the chemokine CCL18/PARC in Gaucher disease: A novel surrogate marker for assessing therapeutic intervention. Blood 2004, 103, 33–39. [Google Scholar] [CrossRef]
- De Francesco, P.N.; Mucci, J.M.; Ceci, R.; Fossati, C.A.; Rozenfeld, P.A. Fabry disease peripheral blood immune cells release inflammatory cytokines: Role of globotriaosylceramide. Mol. Genet. Metab. 2013, 109, 93–99. [Google Scholar] [CrossRef]
- Lerner, A.G.; Upton, J.-P.; Praveen, P.; Ghosh, R.; Nakagawa, Y.; Igbaria, A.; Shen, S.; Nguyen, V.; Backes, B.J.; Heiman, M.; et al. IRE1α Induces thioredoxin-interacting protein to activate the NLRP3 inflammasome and promote programmed cell death under irremediable ER stress. Cell Metab. 2012, 16, 250–264. [Google Scholar] [CrossRef] [PubMed]
- Gargalovic, P.S.; Gharavi, N.M.; Clark, M.J.; Pagnon, J.; Yang, W.-P.; He, A.; Truong, A.; Baruch-Oren, T.; Berliner, J.A.; Kirchgessner, T.G.; et al. The unfolded protein response is an important regulator of inflammatory genes in endothelial cells. Arter. Thromb. Vasc. Biol. 2006, 26, 2490–2496. [Google Scholar] [CrossRef] [PubMed]
- Amen, O.M.; Sarker, S.D.; Ghildyal, R.; Arya, A. Endoplasmic reticulum stress activates unfolded protein response signaling and mediates inflammation, Obesity, and Cardiac Dysfunction: Therapeutic and Molecular Approach. Front. Pharmacol. 2019, 10, 977. [Google Scholar] [CrossRef]
- Cao, Y.; Chtarbanova, S.; Petersen, A.J.; Ganetzky, B. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. USA 2013, 110, E1752–E1760. [Google Scholar] [CrossRef] [PubMed]
- Arnold, P.A.; Johnson, K.N.; White, C.R. Physiological and metabolic consequences of viral infection in Drosophila mela-nogaster. J. Exp. Biol. 2013, 216, 3350–3357. [Google Scholar] [CrossRef] [PubMed]
- Goyal, L.; McCall, K.; Agapite, J.; Hartwieg, E.; Steller, H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000, 19, 589–597. [Google Scholar] [CrossRef]
- Lanzotti, D.J.; Kupsco, J.M.; Marzluff, W.F.; Duronio, R.J. string(cdc25) and cyclin E are required for patterned histone expression at different stages of Drosophila embryonic development. Dev. Biol. 2004, 274, 82–93. [Google Scholar] [CrossRef]
- Duronio, R.J.; O’Farrell, P.H. Developmental control of a G1-S transcriptional program in Drosophila. Development 1994, 120, 1503–1515. [Google Scholar]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. ER stress activates immunosuppressive network: Implications for aging and Alzheimer’s disease. J. Mol. Med. 2020, 98, 633–650. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.X.; Sanders, E.; Wang, J.J. Endoplasmic reticulum stress and inflammation: Mechanisms and implications in diabetic retinopathy. J. Ocul. Biol. Dis. Inform. 2011, 4, 51–61. [Google Scholar] [CrossRef] [PubMed]
- Thorp, E.B. Methods and models for monitoring UPR-associated macrophage death during advanced atherosclerosis. Methods Enzymol. 2011, 489, 277–296. [Google Scholar]
- Giusti, C.; Luciani, M.-F.; Golstein, P. A second signal for autophagic cell death? Autophagy 2010, 6, 823–824. [Google Scholar] [CrossRef] [PubMed]
- Dasari, S.K.; Bialik, S.; Levin-Zaidman, S.; Levin-Salomon, V.; Jr, A.H.M.; Futerman, A.H.; Kimchi, A. Signalome-wide RNAi screen identifies GBA1 as a positive mediator of autophagic cell death. Cell Death Differ. 2017, 24, 1288–1302. [Google Scholar] [CrossRef] [PubMed]
- Jeffery, C.J. Moonlighting proteins: Old proteins learning new tricks. Trends Genet. 2003, 19, 415–417. [Google Scholar] [CrossRef]
- Tamhane, T.; Lllukkumbura, R.; Lu, S.; Maelandsmo, G.M.; Haugen, M.H.; Brix, K. Nuclear cathepsin L activity is required for cell cycle progression of colorectal carcinoma cells. Biochimie 2016, 122, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of Apoptotic Program in Cell-Free Extracts: Requirement for dATP and Cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef]
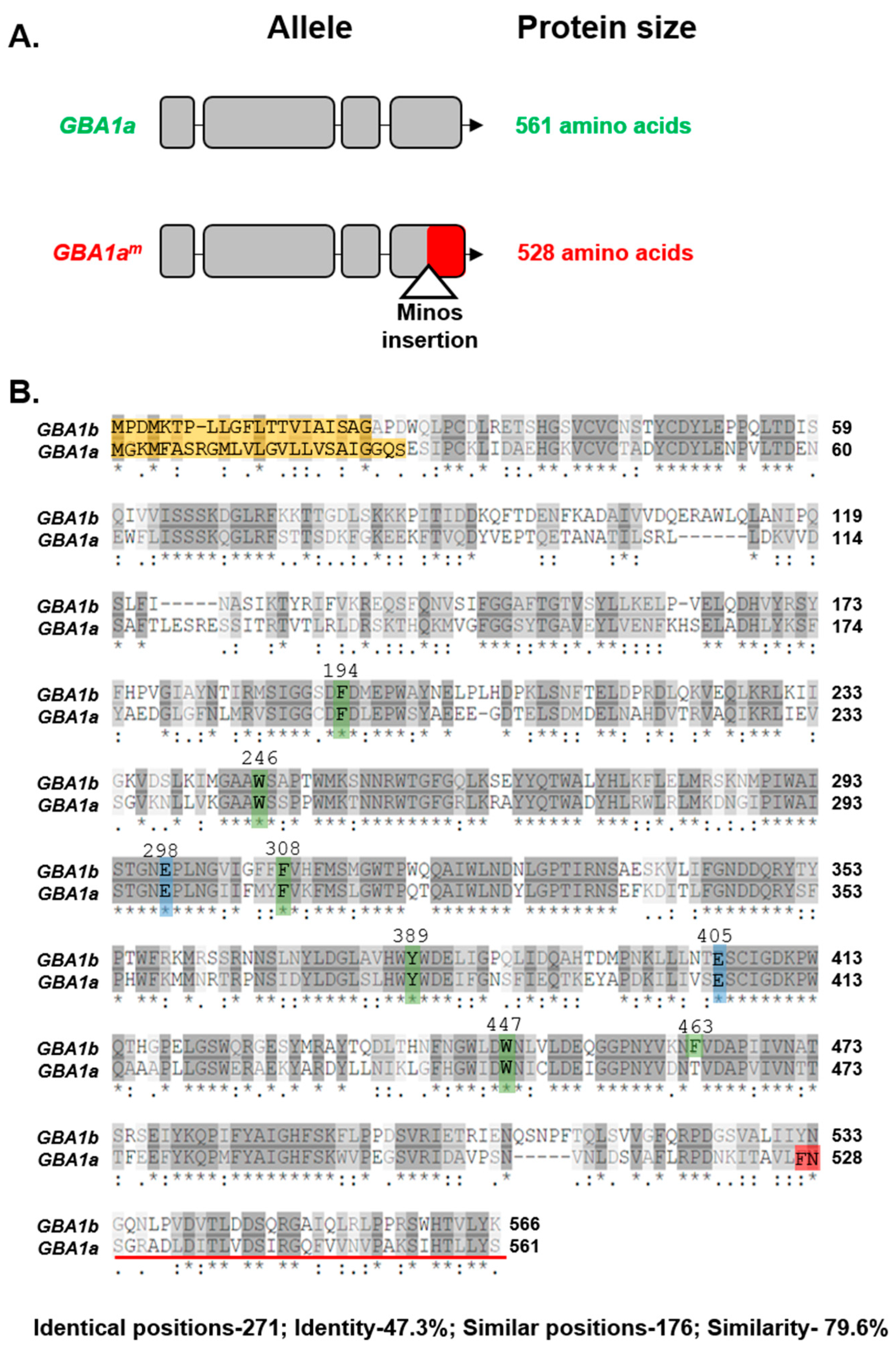


| Role in Active Site | Human GCase | GBA1a Encoded | GBA1b Encoded |
|---|---|---|---|
| Catalytic amino acid | E235 | E298 | E298 |
| Catalytic amino acid | E340 | E405 | E405 |
| Stabilizing the substrate | F128 | F194 | F194 |
| Stabilizing the substrate | W179 | W246 | W246 |
| Stabilizing the substrate | F246 | F308 | F308 |
| Stabilizing the substrate | Y313 | Y389 | Y389 |
| Stabilizing the substrate | W381 | W408 | W408 |
| Stabilizing the substrate | F397 | --- | F463 |
| Name | Primer Sequence |
|---|---|
| ATF4 | F: 5′-AGACGCTGCTTCGCTTCCTTC-3′ |
| R: 5′-GCCCGTAAGTGCGAGTACGCT-3′ | |
| Hsc70-3 | F: 5′-GCTGGTGTTATTGCCGGTCTGC-3′ |
| R: 5′-GATGCCTCGGGATGGTTCCTTGC-3′ | |
| sXBP1 | F: 5′-CCGAACTGAAGCAGCAACAGC-3′ |
| R: 5′-GTATACCCTGCGGCAGATCC-3′ | |
| AttC | F: 5′-CTGCACTGGACTACTCCCACATCA-3′ |
| R: 5′-CGATCCTGCGACTGCCAAAGATTG-3′ | |
| CEC | F: 5′-CATTGGACAATCGGAAGCTGGGTG-3′ |
| R: 5′-TAATCATCGTGGTCAACCTCGGGC-3′ | |
| DRS | F: 5′-AGTACTTGTTCGCCCTCTTCGCTG-3′ |
| R: 5′-CCTTGTATCTTCCGGACAGGCAGT-3′ | |
| MTK | F: 5′-CATCAATCAATTCCCGCCACCGAG-3′ |
| R: 5′-AAATGGGTCCCTGGTGACGATGAG-3′ | |
| stg | F: 5’-CGTCGTCGAGTCAACAGCTCTTC-3′ |
| R: 5′-GTATTTCGGAGTGTGGTTGTGCG-3′ | |
| PCNA | F: 5’-GCAGCGACTCCGGCATTCAG-3′ |
| R: 5′-CGCAGGGTCAGCGAGACAAG-3′ | |
| R | F: 5’-GCTGGATGGAGCCATTCTTCCG-3′ |
| R: 5′-CCTGGGCCATAAGCACTTCGTC-3′ | |
| RPR | F: 5’CATACCCGATCAGGCGACTC-3′ |
| R: 5’-GCTTGCGATATTTGCCGGAC-3′ | |
| rp49 | F: 5′-TAAGAAGCGCACAAAGCACT-3′ |
| R: 5′-GGGCATCAGATATTGTCCCT-3′ |
| Temperature | Genotype | Number of Laid Eggs | Number of Hatching |
|---|---|---|---|
| 25 °C | w1118 | 105 | 72 |
| GBA1am/m | 103 | 64 | |
| p = 0.61 | p = 0.35 |
| Temperature | Genotype | % Larvae Surviving to Pupae | % Pupae Surviving to Adult |
|---|---|---|---|
| 25 °C | w1118 | 100 | 94 |
| GBA1am/+ | 98.1 | 92 | |
| GBA1am/m | 90 * | 87 | |
| 29 °C | w1118 | 96.9 | 85.6 |
| GBA1am/+ | 95.2 | 90.3 | |
| GBA1am/m | 86.7 * | 80.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cabasso, O.; Paul, S.; Maor, G.; Pasmanik-Chor, M.; Kallemeijn, W.; Aerts, J.; Horowitz, M. The Uncovered Function of the Drosophila GBA1a-Encoded Protein. Cells 2021, 10, 630. https://doi.org/10.3390/cells10030630
Cabasso O, Paul S, Maor G, Pasmanik-Chor M, Kallemeijn W, Aerts J, Horowitz M. The Uncovered Function of the Drosophila GBA1a-Encoded Protein. Cells. 2021; 10(3):630. https://doi.org/10.3390/cells10030630
Chicago/Turabian StyleCabasso, Or, Sumit Paul, Gali Maor, Metsada Pasmanik-Chor, Wouter Kallemeijn, Johannes Aerts, and Mia Horowitz. 2021. "The Uncovered Function of the Drosophila GBA1a-Encoded Protein" Cells 10, no. 3: 630. https://doi.org/10.3390/cells10030630
APA StyleCabasso, O., Paul, S., Maor, G., Pasmanik-Chor, M., Kallemeijn, W., Aerts, J., & Horowitz, M. (2021). The Uncovered Function of the Drosophila GBA1a-Encoded Protein. Cells, 10(3), 630. https://doi.org/10.3390/cells10030630






