Abstract
In Arabidopsis thaliana there are more than 600 C-to-U RNA editing events in the mitochondria and at least 44 in the chloroplasts. Pentatricopeptide repeat (PPR) proteins provide the specificity for these reactions. They recognize RNA sequences in a partially predictable fashion via key amino acids at the fifth and last position in each PPR motif that bind to individual ribonucleotides. A combined approach of RNA-Seq, mutant complementation, electrophoresis of mitochondrial protein complexes and Western blotting allowed us to show that MEF100, a PPR protein identified in a genetic screen for mutants resistant to an inhibitor of γ -glutamylcysteine synthetase, is required for the editing of nad1-493, nad4-403, nad7-698 and ccmFN2-356 sites in Arabidopsis mitochondria. The absence of editing in mef100 leads to a decrease in mitochondrial Complex I activity, which probably explains the physiological phenotype. Some plants have lost the requirement for MEF100 at one or more of these sites through mutations in the mitochondrial genome. We show that loss of the requirement for MEF100 editing leads to divergence in the MEF100 binding site.
1. Introduction
RNA editing is a post-transcriptional process that alters the nucleotide sequences of RNA molecules such that the information contained in the mature RNA differs from that defined in the genome [1]. In the mitochondria and chloroplasts of angiosperms, RNA editing occurs as singular cytidine (C) to uridine (U) changes by deamination [2]. Editing is catalysed by editosomes, multifactor protein-RNA complexes in which RNA recognition is mediated by pentatricopeptide repeat (PPR) proteins [3]. The majority of the PPR proteins involved in editing belong to the PLS subfamily whose PPR array is composed of three variants of PPR motifs named P, L and S [4]. The PLS proteins usually carry additional PPR-like motifs (E1 and E2) and a cytidine-deaminase-like domain (the DYW domain) at their C-terminus. The zinc-binding DYW domain is required for catalysis of the deamination reaction [5,6,7,8,9]. A PPR-RNA code was developed to describe the observed correlations between amino acids at two key positions of known editing factors and their aligned nucleotides on the transcripts to which they bind [1,10,11]. Subsequently, the amino acids at these positions were shown to bind to the aligned nucleotide via hydrogen bonds [12]. Although this code can be used to predict the targets of some PPR proteins [7,11,13,14,15,16], most PPR proteins required for the editing of mitochondrial sites in Arabidopsis have not been identified. The characterisation of new editing factors remains an important step in understanding how PPR proteins recognise their targets and the role of RNA editing in organelle gene expression.
In wild-type plants, exposure to buthionine sulfoximine (BSO) inhibits the activity of γ-glutamylcysteine synthetase, severely reducing the plant’s ability to produce the tripeptide glutathione (γ-L-glutamyl-L-cysteinylglycine, GSH) [17]. GSH is involved in multiple biochemical processes, including the detoxification of reactive oxygen species [18] and an inability to produce it results in a specific loss of root meristem activity, stopping root growth [19]. In a screen of Col-0 ecotype plants treated with ethane methyl sulfonate (EMS), 10 bir mutants (for BSO-insensitive roots), including two PPR mutants had almost normal root growth in the presence of BSO [20]. This BSO-insensitive-root phenotype observed in the bir6 mutant [20] was also shared by the Arabidopsis Complex I mutants css1 [21] and otp439 [20,22], suggesting that the phenotype is related to the reduced activity of mitochondrial Complex I. In this paper, we show that the mutant named here as mef100 (for mitochondrial editing factor 100) is unable to effectively edit four mitochondrial sites and has moderately reduced levels of assembled Complex I.
2. Materials and Methods
2.1. Plant Growth and Selection
Arabidopsis seeds were surface sterilized in a solution containing 70% (v/v) ethanol and 0.05% Triton X-100 for 5 min, washed in 100% ethanol and then dried in sterile conditions. The seeds were plated (0.5× MS medium), vernalized at 4 °C in the dark for at least 24 h and then grown under a long-day photoperiod (16 h light, 100 μmol photons m−2 s−1). Three weeks after plating, they were transferred to the soil.
The mef100 mutant plants were obtained from Stanislav Kopriva’s lab at the Department of Metabolic Biology at the John Innes Centre (United Kingdom). Plants were confirmed for homozygosity for the point mutation in the coding sequence of the AT3G61170 gene by Sanger sequencing of the PCR product obtained using the primer pair Mef100_859F and Mef100_2211R (Table S1). The mef100 mutants were complemented (see Section 2.2) by floral dip [23] and their seeds were screened for successful transformation on 0.5× MS media containing 25 μg/μL hygromycin. Complementation was confirmed by PCR using primer pair mef100_1865F and pGWB1_C-terminal_R-1 (Table S1).
2.2. Plant Complementation
The region of genomic DNA corresponding to the full mef100 gene and 522 nt upstream of the mef100 start codon was amplified by PCR from Col-0 genomic DNA using mef100_ups_attB_F and mef100-stop_attBR (Table S1), which was cloned into the Gateway donor vector pDONR207 (Thermo Fisher Scientific, Waltham, MA, USA) according to the recommended protocol and used to transform DH5α. The fragment verified by Sanger sequencing was cloned into the pGWB1 binary vector and transferred into plants via Agrobacterium tumefaciens transformation [23].
2.3. RNA Editing Analyses
RNA from young leaves of mef100, Columbia-0 and three complemented mef100 lines was extracted and DNase treated as previously described [24]. Reverse transcription using random primers and SuperScript III (Thermo Fisher Scientific, Waltham, MA, USA) was done according to their recommended protocol. PCR of the cDNA of mef100, Columbia-0 and complemented lines was done using Taq polymerase (Thermo Fisher Scientific, Waltham, MA, USA) according to their recommended protocol using primers specific to each of the four editing sites. PCR of the nad1-493 site was done using the primer pair nad1-224-F and nad1-861-RV1; nad4-403 using nad4-176-F and nad4-830-RV1; nad7-698 using nad7-382-F and nad7-1004-RV1; ccmFN2-356 using ccmFN2-88-F and ccmFN2-500-RV1 (Table S1).
2.4. Analysis of RNA Editing in RNA-seq Data
Crude mitochondrial pellets were isolated from 8-week-old leaves of mef100 and Col-0 as previously described [25]. Mitochondrial RNA was extracted from these crude mitochondrial fractions using TRIzol (Thermo Fisher Scientific, Waltham, MA, USA). Two μg of RNA were treated with TURBO DNase (Thermo Fisher Scientific, Waltham, MA, USA) for 1 h at 37 °C and tested for DNA contamination as previously described [26]. Three hundred µg of mitochondrial RNA were used for cDNA library preparation using the TruSeq Stranded mRNA Library Prep kit with RIBO zero (Illumina, San Diego, California). The sequencing run (50 nt, single read) was performed on an Illumina Hi-Seq 2500 platform. RNA editing analyses were carried out as described in [27]. Reads were analysed with the BBMap package (sourceforge.net/projects/bbmap/). Clumpify was used to remove optical duplicates (with parameters dedupe optical dist = 40), BBDuk was used to remove adapters (with parameters ktrim = r k = 23 mink = 11 hdist = 1 tpe tbo ftm = 5) and BBMap was used to map trimmed reads to the organelle genomes (with parameters ambiguous = random). Nucleotide counting to identify the extent of editing was carried out with Pyrimid (https://github.com/ian-small/pyrimid, accessed on 21 January 2021). Nucleotide count data was analysed statistically with a Fisher exact test as implemented in the Python scipy.stats package and the p-values were corrected for multiple testing using statsmodels.stats.multitest.multipletests with the Simes-Hochberg procedure. Odds ratios were calculated after adding a pseudocount of 0.5 to all observations to avoid division by zero.
2.5. Analysis of Complex I and Complex III
Crude membrane pellets were prepared from Arabidopsis leaves as previously described [25] and the complexes separated by BN-PAGE using precast NativePAGETM mini-gels (Thermo Fisher Scientific, Waltham, MA, USA). After the run, the gel was rinsed twice in H2O. To detect the NADH oxidase activity of Complex I, the gel was incubated in 50 mL of detection buffer (50 mg of β-Nicotinamide adenine dinucleotide, reduced disodium salt hydrate (NADH) and 50 mg of nitrotetrazolium blue chloride dissolved in 0.1 M Tris, pH 7.4) in the dark for 45 min. The reaction was stopped by incubating the gel in 5% acetic acid for 2 h.
Proteins separated by BN-PAGE were transferred onto a PVDF membrane (Bio-Rad, Hercules, CA, USA) for 1 h in cathode buffer [25] in the XCell tank (100 mA constant current). After transfer, the membrane was destained in 100% ethanol, washed and blocked for 1 h at RT in the Roche blocking reagent (Roche, Mannheim, Germany). Immunodetection was performed using anti-RISP antibody [28] at a 1/3000 dilution, followed by an anti-rabbit secondary antibody (Sigma Aldricht, St Louis, MI, USA) diluted 1/10,000 and subsequently revealed using Clarity ECL reagents (Bio-Rad, Hercules, CA, USA). For SDS PAGE and Western blotting, purified mitochondrial preparations from 4-week-old seedlings were used as previously described [29].
2.6. Prediction of Binding by Editing Factors
Known editing factors and their target sites were aligned (Table S2) and the frequencies with which each 5th/last amino acid combination in the PPR motifs aligned with A, C, G or U were calculated and converted to a scoring table by calculating the natural log of (10 + observed counts)/(10 + counts expected from overall amino acid frequencies). Scoring tables were calculated separately for each motif type (available at https://github.com/ian-small/PPRmatcher (accessed on 21 January 2021) in ‘scoring_tables/Millman’ and Table S3). Scores were then calculated for any PPR-RNA alignment by looking up the score for each motif-nucleotide alignment and summing them.
3. Results
3.1. Mef100 Carries a Mutation in AT3G61170
The natural MEF100 protein consists of 17 PPR motifs and E1-E2-DYW C-terminal domains [4]. The mef100 mutant previously isolated in a screen for BSO-insensitive Arabidopsis mutants [20] contains an EMS-induced point mutation in the gene AT3G61170 that converts the 1497th nucleotide in the coding sequence from a guanine into an adenosine, converting the 499th codon into a stop codon (Figure 1a). This mutation is predicted to terminate the translation of MEF100 in the first helix of the P2 motif (Figure 1a). When grown under standard long-day conditions, the mef100 mutant shows a delayed growth rate compared to wild-type plants (Figure 1b). The wild-type phenotype is completely restored in complemented plants expressing a functional copy of the MEF100 gene including 522 nucleotides upstream of the potential start codon (Figure 1b). Similar development stages are obtained in Col-0 grown for 3.5 weeks and in the mutant grown for 7 weeks (Figure 1c), but the mutant plants display a dark curly foliage typical for Complex I mutants [20,22].

Figure 1.
Characterisation of the mef100 mutant. (a) Schematic representation of the architecture of the MEF100 protein (At3g61170). The EMS mutation creates a STOP codon in the P2 motif; (b) The mef100 mutant shows delayed growth as compared with wild-type (Col-0) and 3 lines complemented with a functional copy of the MEF100 gene (Comp1, 2 and 3); (c) mef100 mutant plants sown 3.5 weeks before Col-0 plants reach comparable development stages.
3.2. MEF100 Is the Specificity Factor for Four Mitochondrial RNA Editing Sites
Targeting prediction software predicts that MEF100 is targeted to mitochondria (TargetP: 0.935, Predotar: 0.48) [30,31]. The fact that mef100 was isolated in a screen for mutants insensitive to BSO [20] suggests that MEF100, a DYW-PPR protein, could be acting in the mitochondrial compartment as an RNA-editing factor. Therefore, we searched for mitochondrial RNA editing events that showed severely reduced levels of editing in the mef100 mutant by sequencing RNA from isolated mitochondria. Approximately 10 million reads were obtained by RNA-seq from each sample and mapped to the Arabidopsis organelle genomes. Of these, 1.01 M reads mapped to the mitochondrial genome in the wild-type sample and 8.97 M reads mapped to the mitochondrial genome in the mef100 sample. The RNA nucleotides mapping to each position were counted to analyse RNA editing. Using a Fisher exact test, we identified seven sites that were differentially edited in the mef100 mutant as compared with Col-0. Four sites showed dramatic decreases of editing in mef100: nad1-493 (genome position 59,314), nad4-403 (215,160), nad7-698 (241,553) and ccmFN2-356 (291,935), from 85–99% editing in Col-0 to 0% editing in the mef100 mutant, except for the nad1 site, which showed residual editing in mef100 (Figure 2a,b and Table 1). The lack of editing at these sites in the mef100 mutant strongly suggests that the MEF100 protein acts as the specificity factor for the editing reactions at these sites. Small but significant decreases were observed at three additional positions (nad1-500 (59,321), nad4-95 (215,468) and nad4-84 (215,479)), but we do not believe these sites are direct targets of MEF100 (see Section 4).
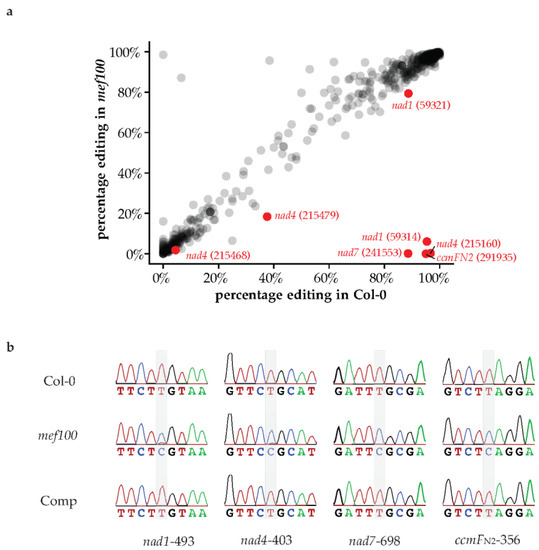
Figure 2.
Analysis of RNA editing in the mef100 mutant, the wild-type Col-0 and complemented line. (a) Dot plot of the difference in percentage of editing in the mef100 mutant as compared with Col-0 at all positions found to show C/U variation in the mitochondrial transcriptome. The seven editing sites recognised by MEF100 are labelled in red. (b) Sanger sequencing of the RT-PCR products of the four major mitochondrial editing sites in mef100, Col-0 and complemented plants. The location of the MEF100 editing sites are highlighted in grey. Some of the sequences contain several editing sites and the sequences shown for Col-0 correspond to the fully edited versions.

Table 1.
Number of edited and unedited reads in the mitochondrial genome at the seven positions impacted in mef100 with percentage editing in mef100 and Col-0. The p-values for the Fisher exact test are given.
To confirm that the lack of a functional MEF100 protein was the cause of these editing deficiencies, the mef100 mutant plants were complemented with a functional copy of the MEF100 gene together with its native promoter. Sequencing of RT-PCR products from the complemented plants along with mef100 mutant and Col-0 plants showed that wild-type editing levels were restored in the complemented plants at all four major editing sites. These results confirm that the MEF100 protein is required for these four editing events in wild-type Arabidopsis plants (Figure 2b).
3.3. Effect of MEF100-Mediated Editing Events at the Protein Level
All four of the main editing events that require MEF100 as the editing factor result in a codon that specifies a different amino acid from that of the unedited codon. Editing of nad1-493 and nad4-403 result in arginine (R) to cysteine (C) transitions while editing of nad7-698 and ccmFN2-356 result in serine (S) to leucine (L) transitions. The nad1, nad4 and nad7 transcripts all encode proteins that are subunits of mitochondrial Complex I (NADH-ubiquinone oxidoreductase), while the protein encoded by the ccmFN2 transcript is a subunit of the cytochrome c maturation system [32] that attaches haem to apocytochromes. A comparison of the amino acid sequences of these four proteins revealed that the amino acids specified by these edited codons are conserved in other land plant species which lack these editing sites as well as in Marchantia polymorpha, a species devoid of RNA editing (Figure S1).
The fact that these amino acid changes are conserved in other species suggests that a lack of editing may impact the function of Complex I and/or the cytochrome c maturation complex in Arabidopsis mitochondria. Cytochrome c1 within the cytochrome bc1 complex (Complex III) requires haem attachment by the cytochrome c maturation complex for correct assembly and function [25], so to analyse the effect of the inactivation of MEF100 on the accumulation of complexes I and III, BN-PAGE was performed on leaf membrane proteins isolated from mef100, Col-0 and complemented mef100 plants as well as the Complex I-deficient mutants bir6-1, bir6-2 [20] and the cytochrome c maturation (CCM)-deficient mutant wtf9 [25]. In-gel NADH oxidase activity staining showed that the reduction in Complex I activity in mef100, less marked than that of bir6-1 and bir6-2, was restored in mef100 plants complemented with a functional MEF100 gene (Figure 3a). This suggests that Complex I might be inefficiently assembled or unstable when it includes the three Nad subunits translated from unedited transcripts. This result is supported by the reduced levels of the two Complex I subunits Nad9 and NDUFS4 observed by Western blot (Figure 3b). In contrast, levels of Cytochrome c1 and CcmFN1 were not affected in the mef100 mutant and a Western blot of the BN gel probed with anti-RISP antibody confirmed that Complex III assembly was not affected in mef100 (Figure 3a).
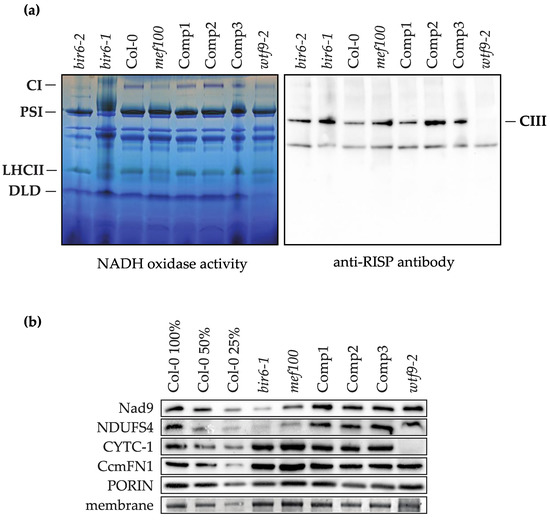
Figure 3.
Analysis of mitochondrial respiratory complexes in mef100 as compared with the bir6-1 and wtf9-2 mutants impaired in Complex I and III respectively, Col-0 and complemented plants (Comp1, 2, 3). (a) BN-PAGE analysis of total membrane extracts: Complex I (CI) and dihydrolipoamide dehydrogenase (DLD) activity bands are indicated on the BN gel (left panel) as well as photosystem I (PSI) and light harvesting complex II (LHCII). Complex III (CIII) is shown on a Western blot probed with an anti-RISP antibody. (b) Western blot analysis using antibodies against two subunits of Complex I (Nad9 and NDUFS4), CYTOCHROME C1 (CYTC-1) and CcmFN1, a subunit of the CCM complex. The anti-porin antibody and a stained membrane were used as loading controls. The full images of the membranes are shown in Figure S2.
3.4. MEF100 Is a Good Match for Its Target According to the PPR Code
PPR editing factors recognize their target sites via interactions between the 5th and last amino acids in each motif and the aligned RNA base [10,13]. These base preferences are to some extent predictable [7,11,13,14,15,16]. To assess whether we could have predicted MEF100 as the specificity factor for these four editing sites, we aligned them with the 205 potential PLS-PPR editing factors encoded by the Arabidopsis nuclear genome. The combined interactions of MEF100 PPR motifs with these target sequences produce very good scores according to the PPR-RNA binding code [10,13], with MEF100 being the top-ranked PLS-PPR editing factor for nad1-493, nad4-403 and nad7-698 (Figure 4b). In contrast, MEF100 is not the top predicted editing factor for the ccmFN2-356 site (Figure 4b), with 17 PLS-PPRs that score better predictions for the sequence.
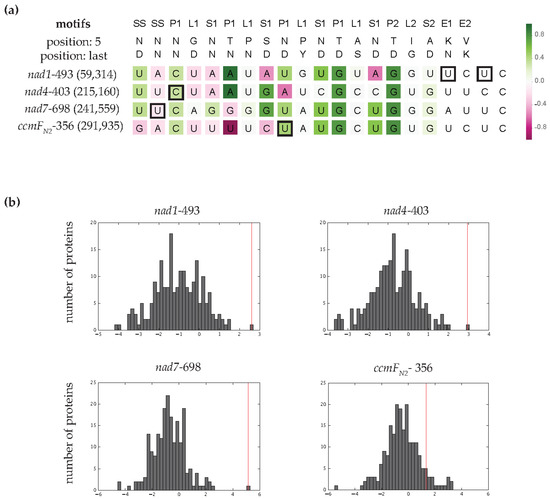
Figure 4.
RNA target predictions for MEF100 according to the PPR code. (a) Scores of the MEF100 PPR motifs with the ribonucleotide sequences upstream of its target sites. The edited C is in last position in all four sequences. Motifs were aligned such that the E2 motif was aligned with the nucleotide two positions upstream of the edited C. Matches and mismatches of a PPR motif with the aligned nucleotide according to the PPR-RNA binding code are shown in green and magenta, respectively. The opacity is proportional to the strength of the correlation. The editing sites found upstream of the sites edited by MEF100 are indicated by a square. (b) Distribution of prediction scores for the MEF100 target sites aligned with 205 PLS-PPR proteins. Red lines indicate the prediction for the target sites aligned with MEF100.
We aligned MEF100 PPR motifs with the RNA sequences upstream of each edited cytidine (Figure 4a). Many of the predicted PPR-RNA interactions are in accordance with expectations from other editing factor interactions. In particular, five C-terminal motifs (P1–9, S1–11, P1–12, P2–15 and S2–17) match well to the nad7-698, ccmFN2-356, nad1-493 and/or nad4-403 sites. In these motifs, the amino acids likely to be involved in RNA recognition are highly conserved in putative MEF100 orthologues (Figure S1) with a few exceptions when one of the four editing sites is lost. A high proportion of the target nucleotides are conserved: seven out of eight nucleotides are identical between nad1-493 and nad4-403 editing sites at the 5′ end, and six out of seven nucleotides are identical between nad7-698 and ccmFN2-356 at the 3′ end of the putative MEF100 binding region. Within the Poaceae, the loss of the editing sites on nad1 and nad4 correlates with divergence or truncation of the N-terminus of the protein, e.g., eight N-terminal motifs are missing in Zea mays (Table S4). These observations suggest that the N-terminal PPR motifs are more important for nad1 and nad4 binding.
We scored how well different putative MEF100 orthologues were predicted to bind the four potential targets in their respective mitochondrial transcriptomes (Figure 5). For two sites (nad1-493 and nad4-403), the sequences scored significantly higher in species where the site is editable than in species where the site is not editable.
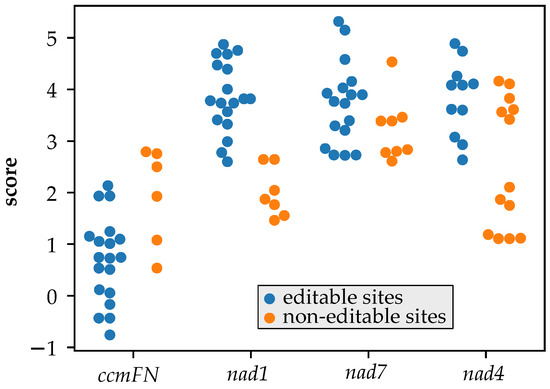
Figure 5.
Variation in predicted recognition by MEF100 orthologues in different species. Putative MEF100 orthologues from a range of species where the mitochondrial genome has been sequenced were aligned to each of the 4 editing sites and the alignment scored using a scoring table developed from alignments of known editing factors and their targets. Higher scores indicate a better match between the protein and the RNA. For each site, species are assorted into two categories, those where the site is editable (C in the genome sequence; blue markers in the plot) and those where the site is not editable (T or rarely A or G in the genome sequence; orange markers in the plot). The non-editable sites score significantly lower for nad1-493 (p-value = 6 × 10−7) and nad4-403 (p-value = 0.003) but not for nad7-698 (p-value = 0.06) or ccmFN2-356 (p-value = 0.99) (one-sided t-test).
4. Discussion
We identified the PPR DYW protein MEF100 as involved in the editing of four mitochondrial sites. Three additional sites were flagged as significantly differentially edited between wild-type and the mef100 mutant, but we think these are secondary effects and that these additional sites are not direct targets of MEF100. Firstly, these three sites are still edited at significant rates in mef100, which is not consistent with their editing being directly affected by MEF100. Secondly, all three sites are on the nad1 or nad4 transcripts and thus susceptible to be influenced by MEF100 binding or editing at the nad1-493 and nad4-403 sites that clearly are direct targets of MEF100. Editing at nad1-493 is seven nucleotides upstream of one of these 3 additional sites and thus presumably alters the cis-element required by the unknown factor that edits nad1-500. The two additional sites in nad4 are ~300 nt upstream of the site at nad4-403, so the mechanism by which binding or editing of nad4-403 by MEF100 affects their editing is not so clear, but it may be by influencing RNA folding or turnover.
Complex I has an important function in cellular redox homeostasis, and when its activity is reduced, the alternative NADH oxidizing enzymes are induced. We previously showed that mutants with a reduced Complex I activity (bir6, css1 and otp439 mutants) had a reduced sensitivity to BSO-induced inhibition of root growth, because they retained more GSH and had a better ability to detoxify reactive oxygen species than wild-type plants [20]. RNA editing of the nad1-493, nad4-403 and nad7-698 sites has effects on the amino acid sequences of Complex I subunits. The accumulation of Complex I is disrupted in mef100, although not to the same extent as in the bir6 mutant. The amino acid differences due to editing may play an important role in the interactions between Complex I subunits in the wild-type (WT) and thus the assembly or stability of the complex. We used the TMHMM software [33] to predict the transmembrane domains of wild-type Nad1 and Nad4 (components of the membrane arm of the L-shaped Complex I) and their counterparts in the mef100 mutant. Although the predicted topology of the mef100 form of Nad1 did not seem to be altered apart from minor changes in the fourth transmembrane domain (Figure S3), the modelling suggested quite major changes in the fourth transmembrane domain of Nad4 (Figure S4). These modifications could be sufficient to alter interactions between subunits and therefore the assembly of the complex.
Nevertheless, the residual accumulation of Complex I may explain the mild growth phenotype of mef100 as compared with the tang2 [22] or ndufs4 [34] mutants, which almost entirely lack assembled Complex I. Whilst editing at the ccmFN2-356 site would be expected to be important based on conservation of the amino acid specified when the site is edited, the absence of editing does not seem to have any noticeable impact on the function of the CcmFN2 protein. The accumulation of Complex III is not disrupted in mef100, and the levels of cytochrome c1 are unchanged, suggesting that the CCM complex is functional in mef100. The topology of the E. coli CcmF integral membrane protein [35] shows that the leucine residue resulting from the ccmFN2-356 editing event is located in a trans-membrane alpha helix and the alignment of Arabidopsis thaliana CcmFN1 and CcmFN2 with E. coli and other plant CcmF proteins suggest that it is conserved (after considering RNA editing in plants). Although, unlike leucine, serine is not a hydrophobic amino acid, it is found in several places in trans-membrane helices of the bacterial CcmF and can provide the necessary H bonds to maintain the alpha helix structure. Our assumption is that the presence of serine rather than leucine at this position does not destabilise CcmFN2 or compromise the function of the CCM complex, at least under standard growth conditions. Interestingly, the lack of editing of ccmFN-1553 in Zea mays, which leads to a phenylalanine to serine substitution in a neighbouring region of the same protein, leads to a reduction in complex III, mitochondrial malfunction and an embryo-lethal phenotype [36]. In contrast to the leucine residue inserted as a result of ccmFN2-356 editing, which merely contributes to the secondary structure of a membrane helix, this phenylalanine residue is one of 3 highly conserved amino acids (SVHAF in E. coli) located in a loop where the histidine residue is crucial for haem attachment.
The PPR-RNA interactions of MEF100 with its target sites are mostly consistent with what would be expected with the current understanding of the PPR-RNA binding code. When aligned with the sequences upstream of the nad1-493, nad4-403 and nad7-698 editing sites, the interactions between the majority of the PPR motifs of MEF100 and the corresponding ribonucleotides fit well with expectations from the analysis of other editing factors aligned to their targets [1,10,11,13]. In comparison to the other sites, the ccmFN2-356 binding sequence is a poorer match, consistent with it being a less important event. The loss of one or more of these target sites in different plants (via mutation of the genome sequence, usually to T, thus obviating the need for RNA editing at that site) allowed us to test the hypothesis that the sequence upstream of the editing site is constrained not only by the requirement to encode suitable amino acids but also by the requirement to bind MEF100. In plants that no longer edit a particular site, we thus hypothesised that the sequence would tend to diverge in a way that would not necessarily preserve the MEF100 binding site. For the nad1-493 and nad4-403 sites the results are consistent with this hypothesis, but for the nad7-698 and ccmFN2-356 sites we do not see a significant drop in the match score to MEF100 in species where these sites are no longer edited (Figure 5). This may indicate that the nad1-493 and nad4-403 sites are under the strongest selection for recognition by MEF100, and illustrates the potential complexity of the co-evolution of transcripts and editing factors when multiple sites are involved [37], which is frequently the case for mitochondrial editing factors.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/10/2/468/s1, Figure S1: Conservation of MEF100 binding sites in land plants, Figure S2: Western blot membranes used in Figure 3, Figure S3: Trans-membrane helix predictions obtained by TMHMM for Nad1, Figure S4: Trans-membrane helix predictions obtained by TMHMM for Nad4, Table S1: Primers used in this work, Table S2: Alignment of known editing factors and their target sites, Table S3: Scoring tables, Table S4: Conservation of the amino acid 5 and last in MEF100 orthologs and the editing sites recognised by MEF100 in Arabidopsis.
Author Contributions
Conceptualisation and methodology, C.C.d.F.-S., B.G. and I.S.; validation, C.C.d.F.-S., B.G., M.M. and L.V.P.S.; formal analysis, C.C.d.F.-S., B.G., M.M. and I.S.; writing—original draft preparation, figures, review and editing, C.C.d.F.-S., B.G., M.M. and I.S.; supervision, C.C.d.F.-S., B.G. and I.S.; funding acquisition, I.S. and B.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the Australian Research Council to I.S. (CE140100008 and FL140100179) and to B.G. (Australian Research Council Discovery Early Career Research Award; DE150101484).
Data Availability Statement
The RNA-seq data is available from SRA under the BioProject ID: PRJNA686212.
Acknowledgments
The authors would like to thank Stanislas Kopriva and Anna Koprivova for the gift of the mef100 seeds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Takenaka, M.; Zehrmann, A.; Brennicke, A.; Graichen, K. Improved computational target site prediction for pentatricopeptide repeat RNA editing factors. PLoS ONE 2013, 8, e65343. [Google Scholar] [CrossRef] [PubMed]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef]
- Sun, T.; Bentolila, S.; Hanson, M.R. The Unexpected Diversity of Plant Organelle RNA Editosomes. Trends Plant Sci. 2016, 21, 962–973. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Gutmann, B.; Zhong, X.; Ye, Y.; Fisher, M.F.; Bai, F.; Castleden, I.; Song, Y.; Song, B.; Huang, J.; et al. Redefining the structural motifs that determine RNA binding and RNA editing by pentatricopeptide repeat proteins in land plants. Plant J. 2016, 85, 532–547. [Google Scholar] [CrossRef] [PubMed]
- Boussardon, C.; Avon, A.; Kindgren, P.; Bond, C.S.; Challenor, M.; Lurin, C.; Small, I. The cytidine deaminase signature HxE(x)n CxxC of DYW1 binds zinc and is necessary for RNA editing of ndhD-1. New Phytol. 2014, 203, 1090–1095. [Google Scholar] [CrossRef] [PubMed]
- Hayes, M.L.; Santibanez, P.I. A plant pentatricopeptide repeat protein with a DYW-deaminase domain is sufficient for catalyzing C-to-U RNA editing in vitro. J. Biol. Chem. 2020, 295, 3497–3505. [Google Scholar] [CrossRef]
- Hayes, M.L.; Giang, K.; Berhane, B.; Mulligan, R.M. Identification of two pentatricopeptide repeat genes required for RNA editing and zinc binding by C-terminal cytidine deaminase-like domains. J. Biol. Chem. 2013, 288, 36519–36529. [Google Scholar] [CrossRef]
- Wagoner, J.A.; Sun, T.; Lin, L.; Hanson, M.R. Cytidine deaminase motifs within the DYW domain of two pentatricopeptide repeat-containing proteins are required for site-specific chloroplast RNA editing. J. Biol. Chem. 2015, 290, 2957–2968. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yang, Y.; Lesch, E.; Knoop, V.; Schallenberg-Rüdinger, M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019, 2, 85. [Google Scholar] [CrossRef]
- Barkan, A.; Rojas, M.; Fujii, S.; Yap, A.; Chong, Y.S.; Bond, C.S.; Small, I. A combinatorial amino acid code for RNA recognition by pentatricopeptide repeat proteins. PLoS Genet. 2012, 8, e1002910. [Google Scholar] [CrossRef] [PubMed]
- Yagi, Y.; Hayashi, S.; Kobayashi, K.; Hirayama, T.; Nakamura, T. Elucidation of the RNA recognition code for pentatricopeptide repeat proteins involved in organelle RNA editing in plants. PLoS ONE 2013, 8, e57286. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhang, D.; Guan, Z.; Liu, Y.; Yang, Z.; Yang, Y.; Wang, X.; Wang, Q.; Zhang, Q.; Fan, S.; et al. Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins. Nat. Commun. 2016, 7, 11285. [Google Scholar] [CrossRef]
- Kobayashi, T.; Yagi, Y.; Nakamura, T. Comprehensive prediction of target RNA editing sites for PLS-class PPR proteins in Arabidopsis thaliana. Plant Cell Physiol. 2019. [Google Scholar] [CrossRef] [PubMed]
- Harrison, T.; Ruiz, J.; Sloan, D.B.; Ben-Hur, A.; Boucher, C. aPPRove: An HMM-Based Method for Accurate Prediction of RNA-Pentatricopeptide Repeat Protein Binding Events. PLoS ONE 2016, 11, e0160645. [Google Scholar]
- Sun, Y.K.; Gutmann, B.; Yap, A.; Kindgren, P.; Small, I. Editing of chloroplast rps14 by PPR editing factor EMB2261 is essential for Arabidopsis development. Front. Plant Sci. 2018, 9, 841. [Google Scholar] [CrossRef] [PubMed]
- Yap, A.; Kindgren, P.; Colas des Francs-Small, C.; Kazama, T.; Tanz, S.K.; Toriyama, K.; Small, I. AEF1/MPR25 is implicated in RNA editing of plastid atpF and mitochondrial nad5, and also promotes atpF splicing in Arabidopsis and rice. Plant J. 2015, 81, 661–669. [Google Scholar] [CrossRef]
- Griffith, O.W.; Meister, A. Potent and specific inhibition of glutathione synthesis by buthionine sulfoximine (Sn-butyl homocysteine sulfoximine). J. Biol. Chem. 1979, 254, 7558–7560. [Google Scholar] [CrossRef]
- Rouhier, N.; Lemaire, S.D.; Jacquot, J.-P. The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 2008, 59, 143–166. [Google Scholar] [CrossRef] [PubMed]
- Vernoux, T.; Wilson, R.C.; Seeley, K.A.; Reichheld, J.-P.; Muroy, S.; Brown, S.; Maughan, S.C.; Cobbett, C.S.; Van Montagu, M.; Inzé, D.; et al. The ROOT MERISTEMLESS1/CADMIUM SENSITIVE2 Gene Defines a Glutathione-Dependent Pathway Involved in Initiation and Maintenance of Cell Division during Postembryonic Root Development. Plant Cell 2000, 12, 97–109. [Google Scholar] [CrossRef] [PubMed]
- Koprivova, A.; des Francs-Small, C.C.; Calder, G.; Mugford, S.T.; Tanz, S.; Lee, B.-R.; Zechmann, B.; Small, I.; Kopriva, S. Identification of a pentatricopeptide repeat protein implicated in splicing of intron 1 of mitochondrial nad7 transcripts. J. Biol. Chem. 2010, 285, 32192–32199. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, N.; Sakurai, N. A mutation in At-nMat1a, which encodes a nuclear gene having high similarity to group II intron maturase, causes impaired splicing of mitochondrial nad4 transcript and altered carbon metabolism in Arabidopsis thaliana. Plant Cell Physiol. 2006, 47, 772–783. [Google Scholar] [CrossRef] [PubMed]
- Colas des Francs-Small, C.; Falcon de Longevialle, A.; Li, Y.; Lowe, E.; Tanz, S.K.; Smith, C.; Bevan, M.W.; Small, I. The pentatricopeptide repeat proteins TANG2 and ORGANELLE TRANSCRIPT PROCESSING439 are involved in the splicing of the multipartite nad5 transcript encoding a subunit of mitochondrial complex I. Plant Physiol. 2014, 165, 1409–1416. [Google Scholar] [CrossRef] [PubMed]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Colas des Francs-Small, C.; Vincis Pereira Sanglard, L.; Small, I. Targeted cleavage of nad6 mRNA induced by a modified pentatricopeptide repeat protein in plant mitochondria. Commun. Biol. 2018, 1, 166. [Google Scholar] [CrossRef] [PubMed]
- Colas des Francs-Small, C.; Kroeger, T.; Zmudjak, M.; Ostersetzer-Biran, O.; Rahimi, N.; Small, I.; Barkan, A. A PORR domain protein required for rpl2 and ccmFC intron splicing and for the biogenesis of c-type cytochromes in Arabidopsis mitochondria. Plant J. 2012, 69, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Delannoy, E.; Falcon de Longevialle, A.; Colas des Francs-Small, C. Mitochondrial RNA transcript analysis assay of Arabidopsis leaf tissues. Bio-Protocol 2015, 5, 1620e. [Google Scholar] [CrossRef]
- Royan, S.; Gutmann, B.; des Francs-Small, C.C.; Honkanen, S.; Schmidberger, J.; Soet, A.; Sun, Y.; Sanglard, L.V.P.; Bond, C.; Small, I. A synthetic RNA editing factor edits its target site in chloroplasts and bacteria. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Carrie, C.; Giraud, E.; Duncan, O.; Xu, L.; Wang, Y.; Huang, S.; Clifton, R.; Murcha, M.; Filipovska, A.; Rackham, O.; et al. Conserved and novel functions for Arabidopsis thaliana MIA40 in assembly of proteins in mitochondria and peroxisomes. J. Biol. Chem. 2010, 285, 36138–36148. [Google Scholar] [CrossRef]
- Lee, K.; Han, J.H.; Park, Y.-I.; Colas des Francs-Small, C.; Small, I.; Kang, H. The mitochondrial pentatricopeptide repeat protein PPR19 is involved in the stabilization of NADH dehydrogenase 1 transcripts and is crucial for mitochondrial function and Arabidopsis thaliana development. New Phytol. 2017, 215, 202–216. [Google Scholar] [CrossRef] [PubMed]
- Emanuelsson, O.; Nielsen, H.; Brunak, S.; von Heijne, G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 2000, 300, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Small, I.; Peeters, N.; Legeai, F.; Lurin, C. Predotar: A tool for rapidly screening proteomes for N-terminal targeting sequences. Proteomics 2004, 4, 1581–1590. [Google Scholar] [CrossRef]
- Giegé, P.; Grienenberger, J.M.; Bonnard, G. Cytochrome c biogenesis in mitochondria. Mitochondrion 2008, 8, 61–73. [Google Scholar] [CrossRef]
- Krogh, A.; Larsson, B.; von Heijne, G.; Sonnhammer, E.L. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J. Mol. Biol. 2001, 305, 567–580. [Google Scholar] [CrossRef]
- Meyer, E.H.; Tomaz, T.; Carroll, A.J.; Estavillo, G.; Delannoy, E.; Tanz, S.K.; Small, I.D.; Pogson, B.J.; Millar, A.H. Remodeled respiration in ndufs4 with low phosphorylation efficiency suppresses Arabidopsis germination and growth and alters control of metabolism at night. Plant Physiol. 2009, 151, 603–619. [Google Scholar] [CrossRef] [PubMed]
- Kranz, R.G.; Richard-Fogal, C.; Taylor, J.-S.; Frawley, E.R. Cytochrome c biogenesis: Mechanisms for covalent modifications and trafficking of heme and for heme-iron redox control. Microbiol. Mol. Biol. Rev. 2009, 73, 510–528. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Wang, X.; Bonnard, G.; Shen, Y.; Xiu, Z.; Li, X.; Gao, D.; Zhang, Z.; Tan, B.-C. Empty pericarp7 encodes a mitochondrial E-subgroup pentatricopeptide repeat protein that is required for ccmFN editing, mitochondrial function and seed development in maize. Plant J. 2015, 84, 283–295. [Google Scholar] [CrossRef] [PubMed]
- Hein, A.; Knoop, V. Expected and unexpected evolution of plant RNA editing factors CLB19, CRR28 and RARE1: Retention of CLB19 despite a phylogenetically deep loss of its two known editing targets in Poaceae. BMC Evol. Biol. 2018, 18, 85. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).