Zebrafish Models of Autosomal Dominant Ataxias
Abstract
1. Introduction
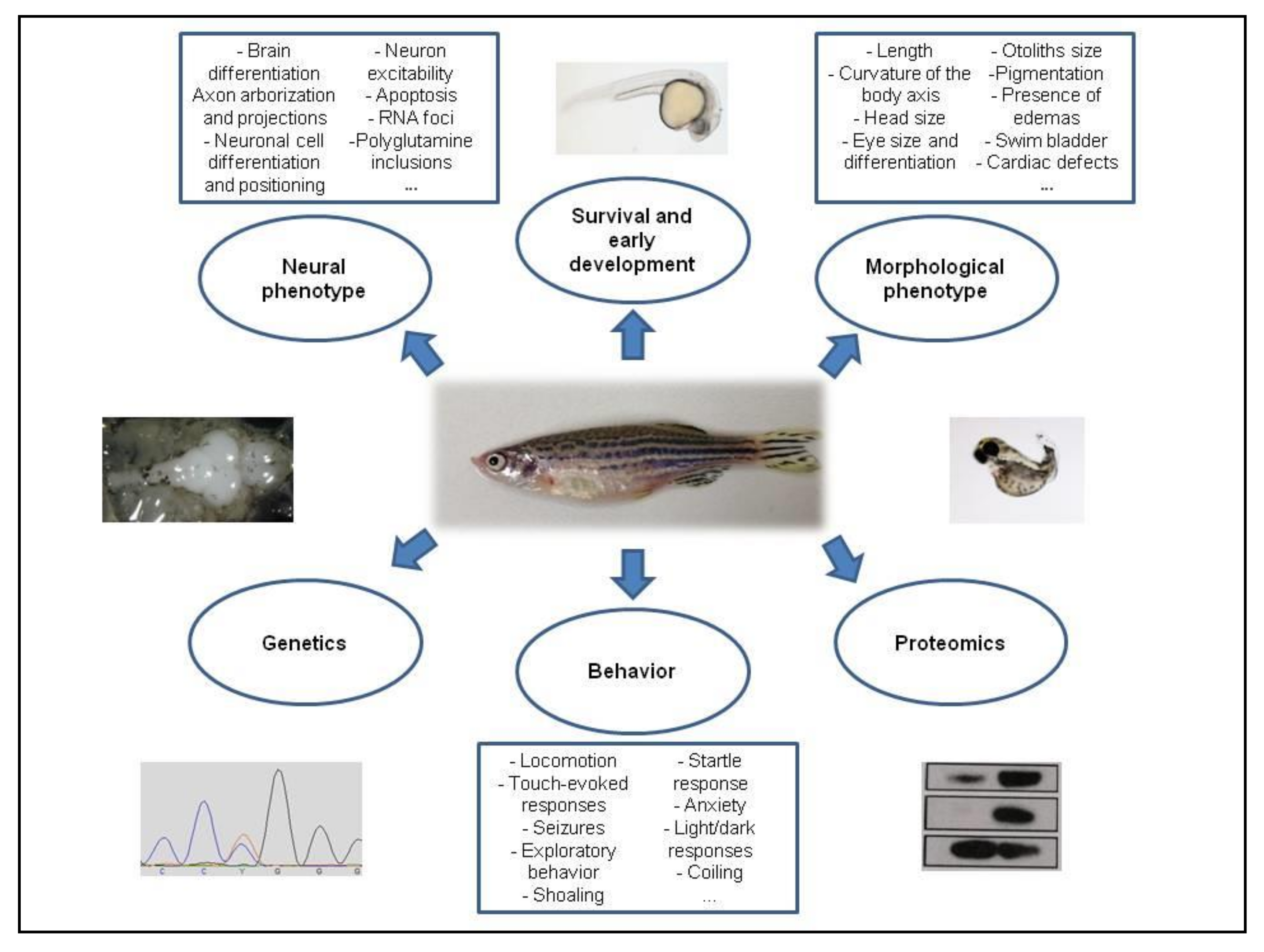
2. Materials and Methods
3. Results
3.1. Spinocerebellar Ataxias
3.1.1. SCA2
3.1.2. SCA3
3.1.3. SCA6 and Episodic Ataxia 2
3.1.4. SCA7
3.1.5. SCA13
3.1.6. SCA14
3.1.7. SCA17
3.1.8. SCA37
3.2. Other Dominant Ataxias
3.2.1. Autosomal Dominant Sensory Ataxia 1
3.2.2. Episodic Ataxia 1
3.2.3. Episodic Ataxia 5
3.3. X-Fragile and FXTAS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hekman, K.E.; Gomez, C.M. The Autosomal Dominant Spinocerebellar Ataxias: Emerging Mechanistic Themes Suggest Pervasive Purkinje Cell Vulnerability. J. Neurol. Neurosurg. Psychiatry 2015, 86, 554–561. [Google Scholar] [CrossRef]
- Jayadev, S.; Bird, T.D. Hereditary Ataxias: Overview. Genet. Med. Off. J. Am. Coll. Med. Genet. 2013, 15, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Matilla-Dueñas, A.; Ashizawa, T.; Brice, A.; Magri, S.; McFarland, K.N.; Pandolfo, M.; Pulst, S.M.; Riess, O.; Rubinsztein, D.C.; Schmidt, J.; et al. Consensus Paper: Pathological Mechanisms Underlying Neurodegeneration in Spinocerebellar Ataxias. Cerebellum Lond. Engl. 2014, 13, 269–302. [Google Scholar] [CrossRef]
- Evidente, V.G.; Gwinn-Hardy, K.A.; Caviness, J.N.; Gilman, S. Hereditary Ataxias. Mayo Clin. Proc. 2000, 75, 475–490. [Google Scholar] [CrossRef]
- Mariotti, C.; Fancellu, R.; Di Donato, S. An Overview of the Patient with Ataxia. J. Neurol. 2005, 252, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Bettencourt, C.; Ryten, M.; Forabosco, P.; Schorge, S.; Hersheson, J.; Hardy, J.; Houlden, H. United Kingdom Brain Expression Consortium Insights from Cerebellar Transcriptomic Analysis into the Pathogenesis of Ataxia. JAMA Neurol. 2014, 71, 831–839. [Google Scholar] [CrossRef]
- Manto, M.; Marmolino, D. Cerebellar Ataxias. Curr. Opin. Neurol. 2009, 22, 419–429. [Google Scholar] [CrossRef]
- Bird, T.D. Hereditary Ataxia Overview. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Mirzaa, G., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Paulson, H. Repeat Expansion Diseases. Handb. Clin. Neurol. 2018, 147, 105–123. [Google Scholar] [CrossRef]
- Chistiakov, D.A.; Hellemans, B.; Volckaert, F.A.M. Microsatellites and Their Genomic Distribution, Evolution, Function and Applications: A Review with Special Reference to Fish Genetics. Aquaculture 2006, 255, 1–29. [Google Scholar] [CrossRef]
- Scoles, D.R.; Meera, P.; Schneider, M.D.; Paul, S.; Dansithong, W.; Figueroa, K.P.; Hung, G.; Rigo, F.; Bennett, C.F.; Otis, T.S.; et al. Antisense Oligonucleotide Therapy for Spinocerebellar Ataxia Type 2. Nature 2017, 544, 362–366. [Google Scholar] [CrossRef] [PubMed]
- Dawson, T.M.; Golde, T.E.; Lagier-Tourenne, C. Animal Models of Neurodegenerative Diseases. Nat. Neurosci. 2018, 21, 1370–1379. [Google Scholar] [CrossRef]
- Lieschke, G.J.; Currie, P.D. Animal Models of Human Disease: Zebrafish Swim into View. Nat. Rev. Genet. 2007, 8, 353–367. [Google Scholar] [CrossRef]
- Howe, K.; Clark, M.D.; Torroja, C.F.; Torrance, J.; Berthelot, C.; Muffato, M.; Collins, J.E.; Humphray, S.; McLaren, K.; Matthews, L.; et al. The Zebrafish Reference Genome Sequence and Its Relationship to the Human Genome. Nature 2013, 496, 498–503. [Google Scholar] [CrossRef]
- Kozol, R.A.; Abrams, A.J.; James, D.M.; Buglo, E.; Yan, Q.; Dallman, J.E. Function Over Form: Modeling Groups of Inherited Neurological Conditions in Zebrafish. Front. Mol. Neurosci. 2016, 9, 55. [Google Scholar] [CrossRef]
- Babin, P.J.; Goizet, C.; Raldúa, D. Zebrafish Models of Human Motor Neuron Diseases: Advantages and Limitations. Prog. Neurobiol. 2014, 118, 36–58. [Google Scholar] [CrossRef]
- Nasevicius, A.; Ekker, S.C. Effective Targeted Gene “knockdown” in Zebrafish. Nat. Genet. 2000, 26, 216–220. [Google Scholar] [CrossRef] [PubMed]
- Heasman, J. Morpholino Oligos: Making Sense of Antisense? Dev. Biol. 2002, 243, 209–214. [Google Scholar] [CrossRef]
- De Bruijn, E.; Cuppen, E.; Feitsma, H. Highly Efficient ENU Mutagenesis in Zebrafish. Methods Mol. Biol. Clifton NJ 2009, 546, 3–12. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome Editing. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Albadri, S.; Del Bene, F.; Revenu, C. Genome Editing Using CRISPR/Cas9-Based Knock-in Approaches in Zebrafish. Methods San Diego Calif. 2017, 121–122, 77–85. [Google Scholar] [CrossRef]
- Villefranc, J.A.; Amigo, J.; Lawson, N.D. Gateway Compatible Vectors for Analysis of Gene Function in the Zebrafish. Dev. Dyn. Off. Publ. Am. Assoc. Anat. 2007, 236, 3077–3087. [Google Scholar] [CrossRef] [PubMed]
- Choudhry, S.; Mukerji, M.; Srivastava, A.K.; Jain, S.; Brahmachari, S.K. CAG Repeat Instability at SCA2 Locus: Anchoring CAA Interruptions and Linked Single Nucleotide Polymorphisms. Hum. Mol. Genet. 2001, 10, 2437–2446. [Google Scholar] [CrossRef] [PubMed]
- Kozlov, G.; Trempe, J.F.; Khaleghpour, K.; Kahvejian, A.; Ekiel, I.; Gehring, K. Structure and Function of the C-Terminal PABC Domain of Human Poly(A)-Binding Protein. Proc. Natl. Acad. Sci. USA 2001, 98, 4409–4413. [Google Scholar] [CrossRef] [PubMed]
- Lastres-Becker, I.; Rüb, U.; Auburger, G. Spinocerebellar Ataxia 2 (SCA2). Cerebellum Lond. Engl. 2008, 7, 115–124. [Google Scholar] [CrossRef]
- Yokoshi, M.; Li, Q.; Yamamoto, M.; Okada, H.; Suzuki, Y.; Kawahara, Y. Direct Binding of Ataxin-2 to Distinct Elements in 3’ UTRs Promotes MRNA Stability and Protein Expression. Mol. Cell 2014, 55, 186–198. [Google Scholar] [CrossRef]
- Riess, O.; Rüb, U.; Pastore, A.; Bauer, P.; Schöls, L. SCA3: Neurological Features, Pathogenesis and Animal Models. Cerebellum Lond. Engl. 2008, 7, 125–137. [Google Scholar] [CrossRef]
- Do Carmo Costa, M.; Paulson, H.L. Toward Understanding Machado-Joseph Disease. Prog. Neurobiol. 2012, 97, 239–257. [Google Scholar] [CrossRef]
- Zhuchenko, O.; Bailey, J.; Bonnen, P.; Ashizawa, T.; Stockton, D.W.; Amos, C.; Dobyns, W.B.; Subramony, S.H.; Zoghbi, H.Y.; Lee, C.C. Autosomal Dominant Cerebellar Ataxia (SCA6) Associated with Small Polyglutamine Expansions in the Alpha 1A-Voltage-Dependent Calcium Channel. Nat. Genet. 1997, 15, 62–69. [Google Scholar] [CrossRef] [PubMed]
- David, G.; Abbas, N.; Stevanin, G.; Dürr, A.; Yvert, G.; Cancel, G.; Weber, C.; Imbert, G.; Saudou, F.; Antoniou, E.; et al. Cloning of the SCA7 Gene Reveals a Highly Unstable CAG Repeat Expansion. Nat. Genet. 1997, 17, 65–70. [Google Scholar] [CrossRef]
- Holmberg, M.; Duyckaerts, C.; Dürr, A.; Cancel, G.; Gourfinkel-An, I.; Damier, P.; Faucheux, B.; Trottier, Y.; Hirsch, E.C.; Agid, Y.; et al. Spinocerebellar Ataxia Type 7 (SCA7): A Neurodegenerative Disorder with Neuronal Intranuclear Inclusions. Hum. Mol. Genet. 1998, 7, 913–918. [Google Scholar] [CrossRef]
- Cancel, G.; Duyckaerts, C.; Holmberg, M.; Zander, C.; Yvert, G.; Lebre, A.S.; Ruberg, M.; Faucheux, B.; Agid, Y.; Hirsch, E.; et al. Distribution of Ataxin-7 in Normal Human Brain and Retina. Brain J. Neurol. 2000, 123, 2519–2530. [Google Scholar] [CrossRef]
- Aleman, T.S.; Cideciyan, A.V.; Volpe, N.J.; Stevanin, G.; Brice, A.; Jacobson, S.G. Spinocerebellar Ataxia Type 7 (SCA7) Shows a Cone-Rod Dystrophy Phenotype. Exp. Eye Res. 2002, 74, 737–745. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.F.; Minassian, N.A.; Stevanin, G.; Figueroa, K.P.; Bannister, J.P.A.; Nolte, D.; Mock, A.F.; Evidente, V.G.H.; Fee, D.B.; Müller, U.; et al. Mutations in Voltage-Gated Potassium Channel KCNC3 Cause Degenerative and Developmental Central Nervous System Phenotypes. Nat. Genet. 2006, 38, 447–451. [Google Scholar] [CrossRef] [PubMed]
- Waters, M.F.; Pulst, S.M. Sca13. Cerebellum Lond. Engl. 2008, 7, 165–169. [Google Scholar] [CrossRef]
- Vlak, M.H.M.; Sinke, R.J.; Rabelink, G.M.; Kremer, B.P.H.; van de Warrenburg, B.P.C. Novel PRKCG/SCA14 Mutation in a Dutch Spinocerebellar Ataxia Family: Expanding the Phenotype. Mov. Disord. Off. J. Mov. Disord. Soc. 2006, 21, 1025–1028. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, K.; Jeong, S.Y.; Uchihara, T.; Anno, M.; Nagashima, K.; Nagashima, T.; Ikeda, S.; Tsuji, S.; Kanazawa, I. SCA17, a Novel Autosomal Dominant Cerebellar Ataxia Caused by an Expanded Polyglutamine in TATA-Binding Protein. Hum. Mol. Genet. 2001, 10, 1441–1448. [Google Scholar] [CrossRef] [PubMed]
- Seixas, A.I.; Loureiro, J.R.; Costa, C.; Ordóñez-Ugalde, A.; Marcelino, H.; Oliveira, C.L.; Loureiro, J.L.; Dhingra, A.; Brandão, E.; Cruz, V.T.; et al. A Pentanucleotide ATTTC Repeat Insertion in the Non-Coding Region of DAB1, Mapping to SCA37, Causes Spinocerebellar Ataxia. Am. J. Hum. Genet. 2017, 101, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Valdmanis, P.N.; Dupré, N.; Lachance, M.; Stochmanski, S.J.; Belzil, V.V.; Dion, P.A.; Thiffault, I.; Brais, B.; Weston, L.; Saint-Amant, L.; et al. A Mutation in the RNF170 Gene Causes Autosomal Dominant Sensory Ataxia. Brain J. Neurol. 2011, 134, 602–607. [Google Scholar] [CrossRef]
- Hasan, S.M.; D’Adamo, M.C. Episodic Ataxia Type 1. In GeneReviews®; Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J., Mirzaa, G., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Rajakulendran, S.; Schorge, S.; Kullmann, D.M.; Hanna, M.G. Episodic Ataxia Type 1: A Neuronal Potassium Channelopathy. Neurother. J. Am. Soc. Exp. Neurother. 2007, 4, 258–266. [Google Scholar] [CrossRef]
- Escayg, A.; De Waard, M.; Lee, D.D.; Bichet, D.; Wolf, P.; Mayer, T.; Johnston, J.; Baloh, R.; Sander, T.; Meisler, M.H. Coding and Noncoding Variation of the Human Calcium-Channel Beta4-Subunit Gene CACNB4 in Patients with Idiopathic Generalized Epilepsy and Episodic Ataxia. Am. J. Hum. Genet. 2000, 66, 1531–1539. [Google Scholar] [CrossRef]
- Jodice, C.; Mantuano, E.; Veneziano, L.; Trettel, F.; Sabbadini, G.; Calandriello, L.; Francia, A.; Spadaro, M.; Pierelli, F.; Salvi, F.; et al. Episodic Ataxia Type 2 (EA2) and Spinocerebellar Ataxia Type 6 (SCA6) Due to CAG Repeat Expansion in the CACNA1A Gene on Chromosome 19p. Hum. Mol. Genet. 1997, 6, 1973–1978. [Google Scholar] [CrossRef] [PubMed]
- Strupp, M.; Zwergal, A.; Brandt, T. Episodic Ataxia Type 2. Neurother. J. Am. Soc. Exp. Neurother. 2007, 4, 267–273. [Google Scholar] [CrossRef]
- Rajakulendran, S.; Graves, T.D.; Labrum, R.W.; Kotzadimitriou, D.; Eunson, L.; Davis, M.B.; Davies, R.; Wood, N.W.; Kullmann, D.M.; Hanna, M.G.; et al. Genetic and Functional Characterisation of the P/Q Calcium Channel in Episodic Ataxia with Epilepsy. J. Physiol. 2010, 588, 1905–1913. [Google Scholar] [CrossRef]
- Crawford, D.C.; Acuña, J.M.; Sherman, S.L. FMR1 and the Fragile X Syndrome: Human Genome Epidemiology Review. Genet. Med. Off. J. Am. Coll. Med. Genet. 2001, 3, 359–371. [Google Scholar] [CrossRef]
- Hagerman, R.J.; Hagerman, P. Fragile X-Associated Tremor/Ataxia Syndrome−Features, Mechanisms and Management. Nat. Rev. Neurol. 2016, 12, 403–412. [Google Scholar] [CrossRef] [PubMed]
- Sellier, C.; Campanari, M.-L.; Julie Corbier, C.; Gaucherot, A.; Kolb-Cheynel, I.; Oulad-Abdelghani, M.; Ruffenach, F.; Page, A.; Ciura, S.; Kabashi, E.; et al. Loss of C9ORF72 Impairs Autophagy and Synergizes with PolyQ Ataxin-2 to Induce Motor Neuron Dysfunction and Cell Death. EMBO J. 2016, 35, 1276–1297. [Google Scholar] [CrossRef] [PubMed]
- Ciura, S.; Sellier, C.; Campanari, M.-L.; Charlet-Berguerand, N.; Kabashi, E. The Most Prevalent Genetic Cause of ALS-FTD, C9orf72 Synergizes the Toxicity of ATXN2 Intermediate Polyglutamine Repeats through the Autophagy Pathway. Autophagy 2016, 12, 1406–1408. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Ning, G.; Zhu, S.; Ma, X.; Liu, X.; Liu, C.; Huang, M.; Schmitt, I.; Wüllner, U.; et al. The Machado-Joseph Disease Deubiquitinase Ataxin-3 Regulates the Stability and Apoptotic Function of P53. PLoS Biol. 2016, 14, e2000733. [Google Scholar] [CrossRef][Green Version]
- Watchon, M.; Yuan, K.C.; Mackovski, N.; Svahn, A.J.; Cole, N.J.; Goldsbury, C.; Rinkwitz, S.; Becker, T.S.; Nicholson, G.A.; Laird, A.S. Calpain Inhibition Is Protective in Machado-Joseph Disease Zebrafish Due to Induction of Autophagy. J. Neurosci. Off. J. Soc. Neurosci. 2017, 37, 7782–7794. [Google Scholar] [CrossRef]
- Toulis, V.; García-Monclús, S.; de la Peña-Ramírez, C.; Arenas-Galnares, R.; Abril, J.F.; Todi, S.V.; Khan, N.; Garanto, A.; do Carmo Costa, M.; Marfany, G. The Deubiquitinating Enzyme Ataxin-3 Regulates Ciliogenesis and Phagocytosis in the Retina. Cell Rep. 2020, 33, 108360. [Google Scholar] [CrossRef]
- Pula, J.H.; Towle, V.L.; Staszak, V.M.; Cao, D.; Bernard, J.T.; Gomez, C.M. Retinal Nerve Fibre Layer and Macular Thinning in Spinocerebellar Ataxia and Cerebellar Multisystem Atrophy. Neuro Ophthalmol. Aeolus Press 2011, 35, 108–114. [Google Scholar] [CrossRef]
- Alvarez, G.; Rey, A.; Sanchez-Dalmau, F.B.; Muñoz, E.; Ríos, J.; Adán, A. Optical Coherence Tomography Findings in Spinocerebellar Ataxia-3. Eye Lond. Engl. 2013, 27, 1376–1381. [Google Scholar] [CrossRef] [PubMed]
- Spina Tensini, F.; Sato, M.T.; Shiokawa, N.; Ashizawa, T.; Teive, H.A.G. A Comparative Optical Coherence Tomography Study of Spinocerebellar Ataxia Types 3 and 10. Cerebellum Lond. Engl. 2017, 16, 797–801. [Google Scholar] [CrossRef] [PubMed]
- Low, S.E.; Woods, I.G.; Lachance, M.; Ryan, J.; Schier, A.F.; Saint-Amant, L. Touch Responsiveness in Zebrafish Requires Voltage-Gated Calcium Channel 2.1b. J. Neurophysiol. 2012, 108, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Wen, H.; Linhoff, M.W.; Hubbard, J.M.; Nelson, N.R.; Stensland, D.; Dallman, J.; Mandel, G.; Brehm, P. Zebrafish Calls for Reinterpretation for the Roles of P/Q Calcium Channels in Neuromuscular Transmission. J. Neurosci. Off. J. Soc. Neurosci. 2013, 33, 7384–7392. [Google Scholar] [CrossRef]
- Zhang, Y.; Kecskés, A.; Copmans, D.; Langlois, M.; Crawford, A.D.; Ceulemans, B.; Lagae, L.; de Witte, P.A.M.; Esguerra, C.V. Pharmacological Characterization of an Antisense Knockdown Zebrafish Model of Dravet Syndrome: Inhibition of Epileptic Seizures by the Serotonin Agonist Fenfluramine. PLoS ONE 2015, 10, e0125898. [Google Scholar] [CrossRef]
- Grone, B.P.; Marchese, M.; Hamling, K.R.; Kumar, M.G.; Krasniak, C.S.; Sicca, F.; Santorelli, F.M.; Patel, M.; Baraban, S.C. Epilepsy, Behavioral Abnormalities, and Physiological Comorbidities in Syntaxin-Binding Protein 1 (STXBP1) Mutant Zebrafish. PLoS ONE 2016, 11, e0151148. [Google Scholar] [CrossRef]
- Gawel, K.; Turski, W.A.; van der Ent, W.; Mathai, B.J.; Kirstein-Smardzewska, K.J.; Simonsen, A.; Esguerra, C.V. Phenotypic Characterization of Larval Zebrafish (Danio Rerio) with Partial Knockdown of the Cacna1a Gene. Mol. Neurobiol. 2020, 57, 1904–1916. [Google Scholar] [CrossRef]
- Damaj, L.; Lupien-Meilleur, A.; Lortie, A.; Riou, É.; Ospina, L.H.; Gagnon, L.; Vanasse, C.; Rossignol, E. CACNA1A Haploinsufficiency Causes Cognitive Impairment, Autism and Epileptic Encephalopathy with Mild Cerebellar Symptoms. Eur. J. Hum. Genet. Ejhg 2015, 23, 1505–1512. [Google Scholar] [CrossRef]
- McMahon, S.J.; Pray-Grant, M.G.; Schieltz, D.; Yates, J.R.; Grant, P.A. Polyglutamine-Expanded Spinocerebellar Ataxia-7 Protein Disrupts Normal SAGA and SLIK Histone Acetyltransferase Activity. Proc. Natl. Acad. Sci. USA 2005, 102, 8478–8482. [Google Scholar] [CrossRef]
- Yanicostas, C.; Barbieri, E.; Hibi, M.; Brice, A.; Stevanin, G.; Soussi-Yanicostas, N. Requirement for Zebrafish Ataxin-7 in Differentiation of Photoreceptors and Cerebellar Neurons. PLoS ONE 2012, 7, e50705. [Google Scholar] [CrossRef]
- Carrillo-Rosas, S.; Weber, C.; Fievet, L.; Messaddeq, N.; Karam, A.; Trottier, Y. Loss of Zebrafish Ataxin-7, a SAGA Subunit Responsible for SCA7 Retinopathy, Causes Ocular Coloboma and Malformation of Photoreceptors. Hum. Mol. Genet. 2019, 28, 912–927. [Google Scholar] [CrossRef] [PubMed]
- Bollmann, J.H. The Zebrafish Visual System: From Circuits to Behavior. Annu. Rev. Vis. Sci. 2019, 5, 269–293. [Google Scholar] [CrossRef] [PubMed]
- Seritrakul, P.; Gross, J.M. Genetic and Epigenetic Control of Retinal Development in Zebrafish. Curr. Opin. Neurobiol. 2019, 59, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Akemann, W.; Knöpfel, T. Interaction of Kv3 Potassium Channels and Resurgent Sodium Current Influences the Rate of Spontaneous Firing of Purkinje Neurons. J. Neurosci. Off. J. Soc. Neurosci. 2006, 26, 4602–4612. [Google Scholar] [CrossRef]
- Mock, A.F.; Richardson, J.L.; Hsieh, J.-Y.; Rinetti, G.; Papazian, D.M. Functional Effects of Spinocerebellar Ataxia Type 13 Mutations Are Conserved in Zebrafish Kv3.3 Channels. BMC Neurosci. 2010, 11, 99. [Google Scholar] [CrossRef]
- Issa, F.A.; Mazzochi, C.; Mock, A.F.; Papazian, D.M. Spinocerebellar Ataxia Type 13 Mutant Potassium Channel Alters Neuronal Excitability and Causes Locomotor Deficits in Zebrafish. J. Neurosci. Off. J. Soc. Neurosci. 2011, 31, 6831–6841. [Google Scholar] [CrossRef] [PubMed]
- Issa, F.A.; Mock, A.F.; Sagasti, A.; Papazian, D.M. Spinocerebellar Ataxia Type 13 Mutation That Is Associated with Disease Onset in Infancy Disrupts Axonal Pathfinding during Neuronal Development. Dis. Model. Mech. 2012, 5, 921–929. [Google Scholar] [CrossRef]
- Namikawa, K.; Dorigo, A.; Zagrebelsky, M.; Russo, G.; Kirmann, T.; Fahr, W.; Dübel, S.; Korte, M.; Köster, R.W. Modeling Neurodegenerative Spinocerebellar Ataxia Type 13 in Zebrafish Using a Purkinje Neuron Specific Tunable Coexpression System. J. Neurosci. Off. J. Soc. Neurosci. 2019, 39, 3948–3969. [Google Scholar] [CrossRef]
- Hsieh, J.-Y.; Ulrich, B.N.; Issa, F.A.; Lin, M.-C.A.; Brown, B.; Papazian, D.M. Infant and Adult SCA13 Mutations Differentially Affect Purkinje Cell Excitability, Maturation, and Viability in Vivo. eLife 2020, 9. [Google Scholar] [CrossRef]
- Seki, T.; Adachi, N.; Ono, Y.; Mochizuki, H.; Hiramoto, K.; Amano, T.; Matsubayashi, H.; Matsumoto, M.; Kawakami, H.; Saito, N.; et al. Mutant Protein Kinase Cgamma Found in Spinocerebellar Ataxia Type 14 Is Susceptible to Aggregation and Causes Cell Death. J. Biol. Chem. 2005, 280, 29096–29106. [Google Scholar] [CrossRef]
- Yamamoto, K.; Seki, T.; Adachi, N.; Takahashi, T.; Tanaka, S.; Hide, I.; Saito, N.; Sakai, N. Mutant Protein Kinase C Gamma That Causes Spinocerebellar Ataxia Type 14 (SCA14) Is Selectively Degraded by Autophagy. Genes Cells Devoted Mol. Cell. Mech. 2010, 15, 425–438. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, H.; Adachi, N.; Shirafuji, T.; Danno, S.; Ueyama, T.; Vendruscolo, M.; Shuvaev, A.N.; Sugimoto, T.; Seki, T.; Hamada, D.; et al. Identification and Characterization of PKCγ, a Kinase Associated with SCA14, as an Amyloidogenic Protein. Hum. Mol. Genet. 2015, 24, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Patten, S.A.; Roy, B.; Cunningham, M.E.; Stafford, J.L.; Ali, D.W. Protein Kinase Cgamma Is a Signaling Molecule Required for the Developmental Speeding of Alpha-Amino-3-Hydroxyl-5-Methyl-4-Isoxazole-Propionate Receptor Kinetics. Eur. J. Neurosci. 2010, 31, 1561–1573. [Google Scholar] [CrossRef]
- Patten, S.A.; Ali, D.W. PKCgamma-Induced Trafficking of AMPA Receptors in Embryonic Zebrafish Depends on NSF and PICK1. Proc. Natl. Acad. Sci. USA 2009, 106, 6796–6801. [Google Scholar] [CrossRef]
- Müller, F.; Lakatos, L.; Dantonel, J.; Strähle, U.; Tora, L. TBP Is Not Universally Required for Zygotic RNA Polymerase II Transcription in Zebrafish. Curr. Biol. CB 2001, 11, 282–287. [Google Scholar] [CrossRef][Green Version]
- Ferg, M.; Sanges, R.; Gehrig, J.; Kiss, J.; Bauer, M.; Lovas, A.; Szabo, M.; Yang, L.; Straehle, U.; Pankratz, M.J.; et al. The TATA-Binding Protein Regulates Maternal MRNA Degradation and Differential Zygotic Transcription in Zebrafish. EMBO J. 2007, 26, 3945–3956. [Google Scholar] [CrossRef]
- Bártfai, R.; Balduf, C.; Hilton, T.; Rathmann, Y.; Hadzhiev, Y.; Tora, L.; Orbán, L.; Müller, F. TBP2, a Vertebrate-Specific Member of the TBP Family, Is Required in Embryonic Development of Zebrafish. Curr. Biol. Cb 2004, 14, 593–598. [Google Scholar] [CrossRef] [PubMed]
- Di Donato, V.; De Santis, F.; Albadri, S.; Auer, T.O.; Duroure, K.; Charpentier, M.; Concordet, J.-P.; Gebhardt, C.; Del Bene, F. An Attractive Reelin Gradient Establishes Synaptic Lamination in the Vertebrate Visual System. Neuron 2018, 97, 1049–1062. [Google Scholar] [CrossRef] [PubMed]
- Dalla Vecchia, E.; Di Donato, V.; Young, A.M.J.; Del Bene, F.; Norton, W.H.J. Reelin Signaling Controls the Preference for Social Novelty in Zebrafish. Front. Behav. Neurosci. 2019, 13, 214. [Google Scholar] [CrossRef] [PubMed]
- Nimura, T.; Itoh, T.; Hagio, H.; Hayashi, T.; Di Donato, V.; Takeuchi, M.; Itoh, T.; Inoguchi, F.; Sato, Y.; Yamamoto, N.; et al. Role of Reelin in Cell Positioning in the Cerebellum and the Cerebellum-like Structure in Zebrafish. Dev. Biol. 2019, 455, 393–408. [Google Scholar] [CrossRef]
- Boycott, K.M.; Bonnemann, C.; Herz, J.; Neuert, S.; Beaulieu, C.; Scott, J.N.; Venkatasubramanian, A.; Parboosingh, J.S. Mutations in VLDLR as a Cause for Autosomal Recessive Cerebellar Ataxia with Mental Retardation (Dysequilibrium Syndrome). J. Child Neurol. 2009, 24, 1310–1315. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, Y.; Zou, L.; Zhong, R.; Zhu, B.; Shen, N.; Chen, W.; Lou, J.; Ke, J.; Zhang, T.; et al. Reelin Gene Variants and Risk of Autism Spectrum Disorders: An Integrated Meta-Analysis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet. Off. Publ. Int. Soc. Psychiatr. Genet. 2014, 165B, 192–200. [Google Scholar] [CrossRef] [PubMed]
- Ovadia, G.; Shifman, S. The Genetic Variation of RELN Expression in Schizophrenia and Bipolar Disorder. PLoS ONE 2011, 6, e19955. [Google Scholar] [CrossRef]
- Li, W.; Guo, X.; Xiao, S. Evaluating the Relationship between Reelin Gene Variants (Rs7341475 and Rs262355) and Schizophrenia: A Meta-Analysis. Neurosci. Lett. 2015, 609, 42–47. [Google Scholar] [CrossRef] [PubMed]
- Bufill, E.; Roura-Poch, P.; Sala-Matavera, I.; Antón, S.; Lleó, A.; Sánchez-Saudinós, B.; Tomàs-Abadal, L.; Puig, T.; Abós, J.; Bernades, S.; et al. Reelin Signaling Pathway Genotypes and Alzheimer Disease in a Spanish Population. Alzheimer Dis. Assoc. Disord. 2015, 29, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.; Osborn, D.P.S.; Gehweiler, I.; Nagel, M.; Ulmer, U.; Bakhtiari, S.; Amouri, R.; Boostani, R.; Hentati, F.; Hockley, M.M.; et al. Bi-Allelic Variants in RNF170 Are Associated with Hereditary Spastic Paraplegia. Nat. Commun. 2019, 10, 4790. [Google Scholar] [CrossRef] [PubMed]
- Ibhazehiebo, K.; Gavrilovici, C.; de la Hoz, C.L.; Ma, S.-C.; Rehak, R.; Kaushik, G.; Meza Santoscoy, P.L.; Scott, L.; Nath, N.; Kim, D.-Y.; et al. A Novel Metabolism-Based Phenotypic Drug Discovery Platform in Zebrafish Uncovers HDACs 1 and 3 as a Potential Combined Anti-Seizure Drug Target. Brain J. Neurol. 2018, 141, 744–761. [Google Scholar] [CrossRef]
- Ebert, A.M.; McAnelly, C.A.; Srinivasan, A.; Linker, J.L.; Horne, W.A.; Garrity, D.M. Ca2+ Channel-Independent Requirement for MAGUK Family CACNB4 Genes in Initiation of Zebrafish Epiboly. Proc. Natl. Acad. Sci. USA 2008, 105, 198–203. [Google Scholar] [CrossRef]
- Tucker, B.; Richards, R.I.; Lardelli, M. Contribution of MGluR and Fmr1 Functional Pathways to Neurite Morphogenesis, Craniofacial Development and Fragile X Syndrome. Hum. Mol. Genet. 2006, 15, 3446–3458. [Google Scholar] [CrossRef] [PubMed]
- Den Broeder, M.J.; van der Linde, H.; Brouwer, J.R.; Oostra, B.A.; Willemsen, R.; Ketting, R.F. Generation and Characterization of FMR1 Knockout Zebrafish. PLoS ONE 2009, 4, e7910. [Google Scholar] [CrossRef]
- Ng, M.-C.; Yang, Y.-L.; Lu, K.-T. Behavioral and Synaptic Circuit Features in a Zebrafish Model of Fragile X Syndrome. PLoS ONE 2013, 8, e51456. [Google Scholar] [CrossRef] [PubMed]
- Kim, L.; He, L.; Maaswinkel, H.; Zhu, L.; Sirotkin, H.; Weng, W. Anxiety, Hyperactivity and Stereotypy in a Zebrafish Model of Fragile X Syndrome and Autism Spectrum Disorder. Prog. Neuropsychopharmacol. Biol. Psychiatry 2014, 55, 40–49. [Google Scholar] [CrossRef]
- Wu, Y.-J.; Hsu, M.-T.; Ng, M.-C.; Amstislavskaya, T.G.; Tikhonova, M.A.; Yang, Y.-L.; Lu, K.-T. Fragile X Mental Retardation-1 Knockout Zebrafish Shows Precocious Development in Social Behavior. Zebrafish 2017, 14, 438–443. [Google Scholar] [CrossRef] [PubMed]
- Constantin, L.; Poulsen, R.E.; Scholz, L.A.; Favre-Bulle, I.A.; Taylor, M.A.; Sun, B.; Goodhill, G.J.; Vanwalleghem, G.C.; Scott, E.K. Altered Brain-Wide Auditory Networks in a Zebrafish Model of Fragile X Syndrome. BMC Biol. 2020, 18, 125. [Google Scholar] [CrossRef] [PubMed]
- Van der Molen, M.J.W.; Van der Molen, M.W.; Ridderinkhof, K.R.; Hamel, B.C.J.; Curfs, L.M.G.; Ramakers, G.J.A. Auditory Change Detection in Fragile X Syndrome Males: A Brain Potential Study. Clin. Neurophysiol. Off. J. Int. Fed. Clin. Neurophysiol. 2012, 123, 1309–1318. [Google Scholar] [CrossRef] [PubMed]
- Shamay-Ramot, A.; Khermesh, K.; Porath, H.T.; Barak, M.; Pinto, Y.; Wachtel, C.; Zilberberg, A.; Lerer-Goldshtein, T.; Efroni, S.; Levanon, E.Y.; et al. Fmrp Interacts with Adar and Regulates RNA Editing, Synaptic Density and Locomotor Activity in Zebrafish. PLoS Genet. 2015, 11, e1005702. [Google Scholar] [CrossRef]
- Doll, C.A.; Yergert, K.M.; Appel, B.H. The RNA Binding Protein Fragile X Mental Retardation Protein Promotes Myelin Sheath Growth. Glia 2020, 68, 495–508. [Google Scholar] [CrossRef]
- Hu, J.; Chen, L.; Yin, J.; Yin, H.; Huang, Y.; Tian, J. Hyperactivity, Memory Defects, and Craniofacial Abnormalities in Zebrafish Fmr1 Mutant Larvae. Behav. Genet. 2020, 50, 152–160. [Google Scholar] [CrossRef]
- Medishetti, R.; Rani, R.; Kavati, S.; Mahilkar, A.; Akella, V.; Saxena, U.; Kulkarni, P.; Sevilimedu, A. A DNAzyme Based Knockdown Model for Fragile-X Syndrome in Zebrafish Reveals a Critical Window for Therapeutic Intervention. J. Pharm. Toxicol. Methods 2020, 101, 106656. [Google Scholar] [CrossRef]
- Stainier, D.Y.R.; Raz, E.; Lawson, N.D.; Ekker, S.C.; Burdine, R.D.; Eisen, J.S.; Ingham, P.W.; Schulte-Merker, S.; Yelon, D.; Weinstein, B.M.; et al. Guidelines for Morpholino Use in Zebrafish. PLoS Genet. 2017, 13, e1007000. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-Cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed]
- Cascella, R.; Capitini, C.; Fani, G.; Dobson, C.M.; Cecchi, C.; Chiti, F. Quantification of the Relative Contributions of Loss-of-Function and Gain-of-Function Mechanisms in TAR DNA-Binding Protein 43 (TDP-43) Proteinopathies. J. Biol. Chem. 2016, 291, 19437–19448. [Google Scholar] [CrossRef] [PubMed]
- Baskoylu, S.N.; Yersak, J.; O’Hern, P.; Grosser, S.; Simon, J.; Kim, S.; Schuch, K.; Dimitriadi, M.; Yanagi, K.S.; Lins, J.; et al. Single Copy/Knock-in Models of ALS SOD1 in C. Elegans Suggest Loss and Gain of Function Have Different Contributions to Cholinergic and Glutamatergic Neurodegeneration. PLoS Genet. 2018, 14, e1007682. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Fernandez, J.P.; Rouet, R.; Vejnar, C.E.; Lane, M.A.; Mis, E.; Khokha, M.K.; Doudna, J.A.; Giraldez, A.J. CRISPR-Cpf1 Mediates Efficient Homology-Directed Repair and Temperature-Controlled Genome Editing. Nat. Commun. 2017, 8, 2024. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Rajanikant, G.K. Huntington Disease: Can a Zebrafish Trail Leave More than a Ripple? Neurosci. Biobehav. Rev. 2014, 45, 258–261. [Google Scholar] [CrossRef]
- Veldman, M.B.; Rios-Galdamez, Y.; Lu, X.-H.; Gu, X.; Qin, W.; Li, S.; Yang, X.W.; Lin, S. The N17 Domain Mitigates Nuclear Toxicity in a Novel Zebrafish Huntington’s Disease Model. Mol. Neurodegener. 2015, 10, 67. [Google Scholar] [CrossRef] [PubMed]
- Carlson, K.M.; Melcher, L.; Lai, S.; Zoghbi, H.Y.; Clark, H.B.; Orr, H.T. Characterization of the Zebrafish Atxn1/Axh Gene Family. J. Neurogenet. 2009, 23, 313–323. [Google Scholar] [CrossRef] [PubMed]
- Wullimann, M.F.; Mueller, T.; Distel, M.; Babaryka, A.; Grothe, B.; Köster, R.W. The Long Adventurous Journey of Rhombic Lip Cells in Jawed Vertebrates: A Comparative Developmental Analysis. Front. Neuroanat. 2011, 5, 27. [Google Scholar] [CrossRef]
- Iwaniuk, A.N.; Whishaw, I.Q. On the Origin of Skilled Forelimb Movements. Trends Neurosci. 2000, 23, 372–376. [Google Scholar] [CrossRef]
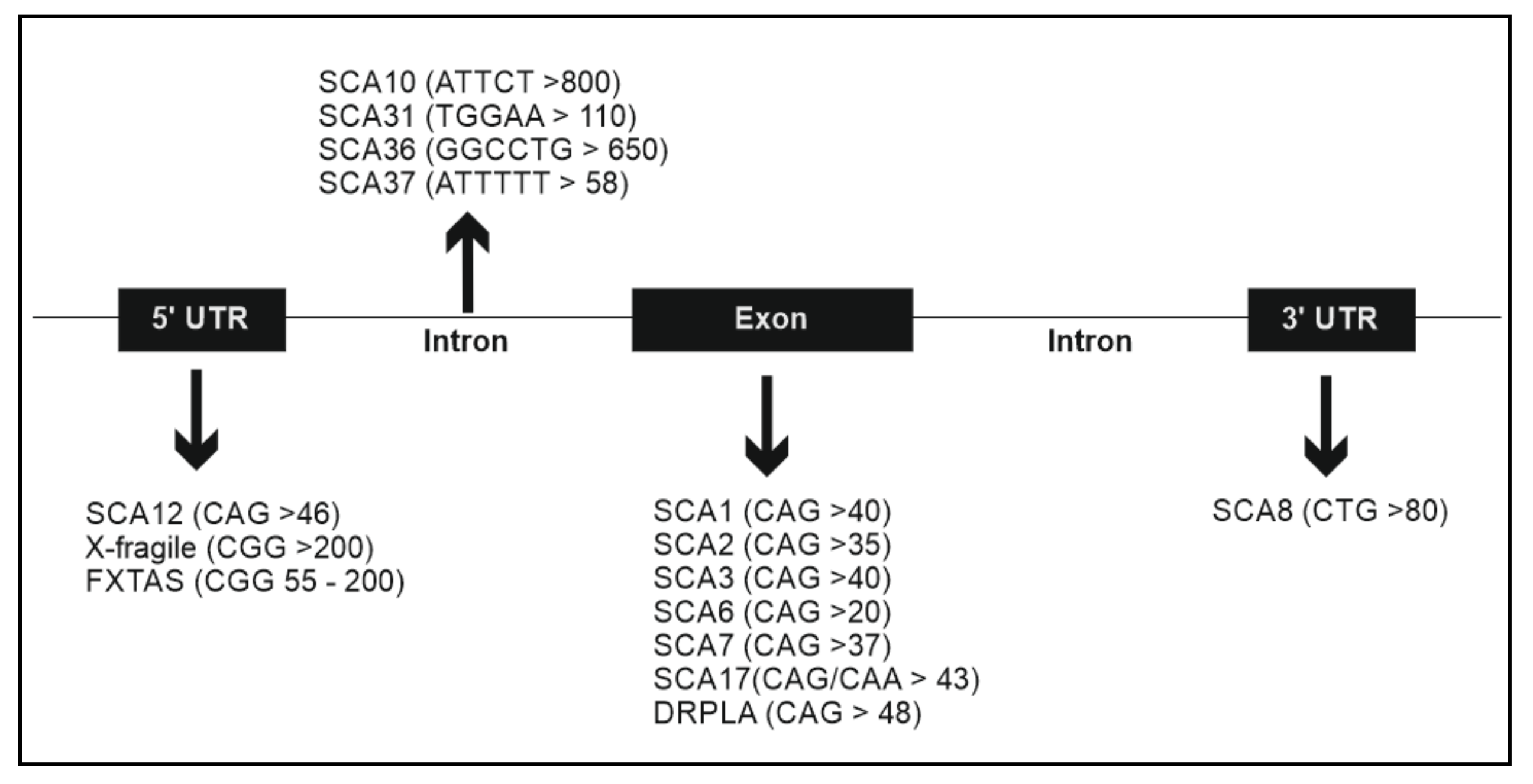
| Disease | Gene | Mutation | Protein Function | Clinical Features | References |
|---|---|---|---|---|---|
| SCA2 | ATXN2 | 35 to 59 CAG repeats | RNA binding protein that regulates mRNA stability | Progressive gait ataxia, dysarthria, Parkinsonian rigidity/bradykinesia, slow saccadic eye movements, tremor, muscle cramps, initial hyperreflexia followed by early hyporeflexia and myoclonus or fasciculation-like movements | [23,24,25,26] |
| SCA3 | ATXN3 | >40 CAG repeats | Involved in ubiquitin-proteasome system degradation of proteins | Slowly progressive ataxia accompanied with ophthalmoplegia, dysarthria, dysphagia, pyramidal signs, dystonia, rigidity and distal muscle atrophies | [27,28] |
| SCA6 | CACNA1A | >20 CAG repeats | Control of neurotransmitter release | Slowly progressive cerebellar ataxia, dysarthria, dysarthria and mild vibratory and proprioceptive sensory loss | [29] |
| SCA7 | ATXN7 | 37 to 200 CAG repeats | Involved in histone acetylation and transcription regulation | Cerebellar ataxia, dysarthria and dysphagia and loss of visual acuity | [30, 31, 32, 33] |
| SCA13 | KCNC3 | Missense mutations | Potassium channel activity | Early or late onset cerebellar ataxia with dysarthria often followed by mild intellectual disability and seizures | [34, 35] |
| SCA14 | PRKCG | Missense mutations | Intracellular signaling in the Central Nervous System | Middle age of onset, slowly progressive cerebellar ataxia, dysarthria, nystagmus and myoclonus | [36] |
| SCA17 | TBP | >43 CAG/CAA repeats | Transcription initiation factor binding DNA pol II | Ataxia, dementia and parkinsonism | [37] |
| SCA37 | DAB1 | >58 ATTTT repeats | Reelin adaptor, functions in neuronal development | Pure cerebellar ataxia and, distinctively, onset of dysarthria in late adolescence to adulthood | [38] |
| Sensory dominant ataxia 1 | RNF170 | Missense mutations | Ubiquitination | Severe loss of proprioception causing gait ataxia and a reduced ability to feel pain, temperature and vibration, particularly in the hands and feet | [39] |
| Episodic Ataxia 1 | KCNA1 | Missense, nonsense and splice site variants | Potassium channel | Early onset, spastic contractions of skeletal muscle, recurrent midline cerebellar dysfunction with loss of motor coordination and balance. Sometimes associated with epilepsy | [40, 41] |
| Episodic Ataxia 5 | CACNB4 | Missense mutations | Calcium channel | Early onset, generalized epilepsy and hereditary episodic ataxia | [42] |
| Episodic Ataxia 2 | CACNA1A | Missense and nonsense mutations | Control neurotransmitter release | Ataxia episodic weakness, vertigo, dystonia, epilepsy nystagmus, and cognitive impairment | [43, 44, 45] |
| X-fragile | FMR1 | X-fragile: >200 CGG repeats | RNA binding protein, Development of synapses | Mental retardation, facial dysmorphism, macroorchidism and autism like behavior or other psychiatric symptoms | [46] |
| FXTAS | FMR1 | FXTAS: premutation alleles on FMR1 gene with 55 to 200 CGG repeats | RNA binding protein, Development of synapses | Late onset, cerebellar ataxia, intention tremor, dementia, parkinsonism and in sometimes psychological symptoms | [47] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quelle-Regaldie, A.; Sobrido-Cameán, D.; Barreiro-Iglesias, A.; Sobrido, M.J.; Sánchez, L. Zebrafish Models of Autosomal Dominant Ataxias. Cells 2021, 10, 421. https://doi.org/10.3390/cells10020421
Quelle-Regaldie A, Sobrido-Cameán D, Barreiro-Iglesias A, Sobrido MJ, Sánchez L. Zebrafish Models of Autosomal Dominant Ataxias. Cells. 2021; 10(2):421. https://doi.org/10.3390/cells10020421
Chicago/Turabian StyleQuelle-Regaldie, Ana, Daniel Sobrido-Cameán, Antón Barreiro-Iglesias, María Jesús Sobrido, and Laura Sánchez. 2021. "Zebrafish Models of Autosomal Dominant Ataxias" Cells 10, no. 2: 421. https://doi.org/10.3390/cells10020421
APA StyleQuelle-Regaldie, A., Sobrido-Cameán, D., Barreiro-Iglesias, A., Sobrido, M. J., & Sánchez, L. (2021). Zebrafish Models of Autosomal Dominant Ataxias. Cells, 10(2), 421. https://doi.org/10.3390/cells10020421







