Figure 1.
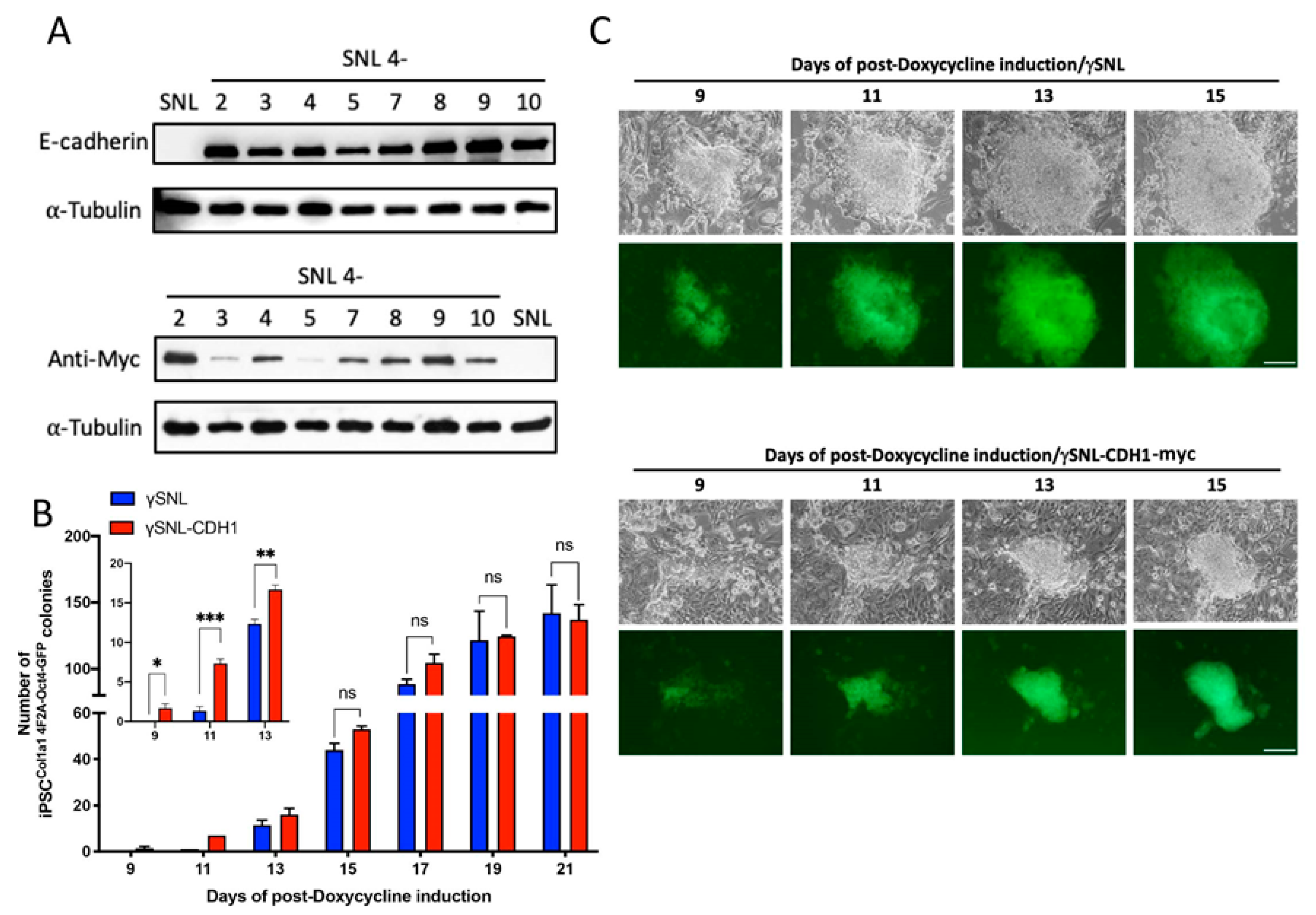
E-cadherin produces in trans facilitation of early-stage somatic reprogramming. (A) The expression of E-cadherin in different engineered SNL-CDH1-MYC cell lines. Each SNL-CDH1-MYC line was subjected to western blot analysis with anti-E-cadherin and anti-MYC antibodies. (B) Quantitative analysis of the Oct4-GFP-positive colonies during iPSC induction (n = 3). (C) Time-lapse photographs of iPSC colony formation from either γSNL or γSNL-CDH1-MYC feeder cell-seeded MEFCol1a1 4F2A Oct4-GFP cells in iSF medium. Fluorescence microscopy was performed on the 9th to 15th days. The iPS clones in the upper panel were seeded with the parental SNL feeder cells, whereas the lower panel shows clones with the SNL-CDH1-MYC feeder cells. The scale bar denotes 50 µm. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ns, not significant.
Figure 1.
E-cadherin produces in trans facilitation of early-stage somatic reprogramming. (A) The expression of E-cadherin in different engineered SNL-CDH1-MYC cell lines. Each SNL-CDH1-MYC line was subjected to western blot analysis with anti-E-cadherin and anti-MYC antibodies. (B) Quantitative analysis of the Oct4-GFP-positive colonies during iPSC induction (n = 3). (C) Time-lapse photographs of iPSC colony formation from either γSNL or γSNL-CDH1-MYC feeder cell-seeded MEFCol1a1 4F2A Oct4-GFP cells in iSF medium. Fluorescence microscopy was performed on the 9th to 15th days. The iPS clones in the upper panel were seeded with the parental SNL feeder cells, whereas the lower panel shows clones with the SNL-CDH1-MYC feeder cells. The scale bar denotes 50 µm. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ns, not significant.
Figure 2.
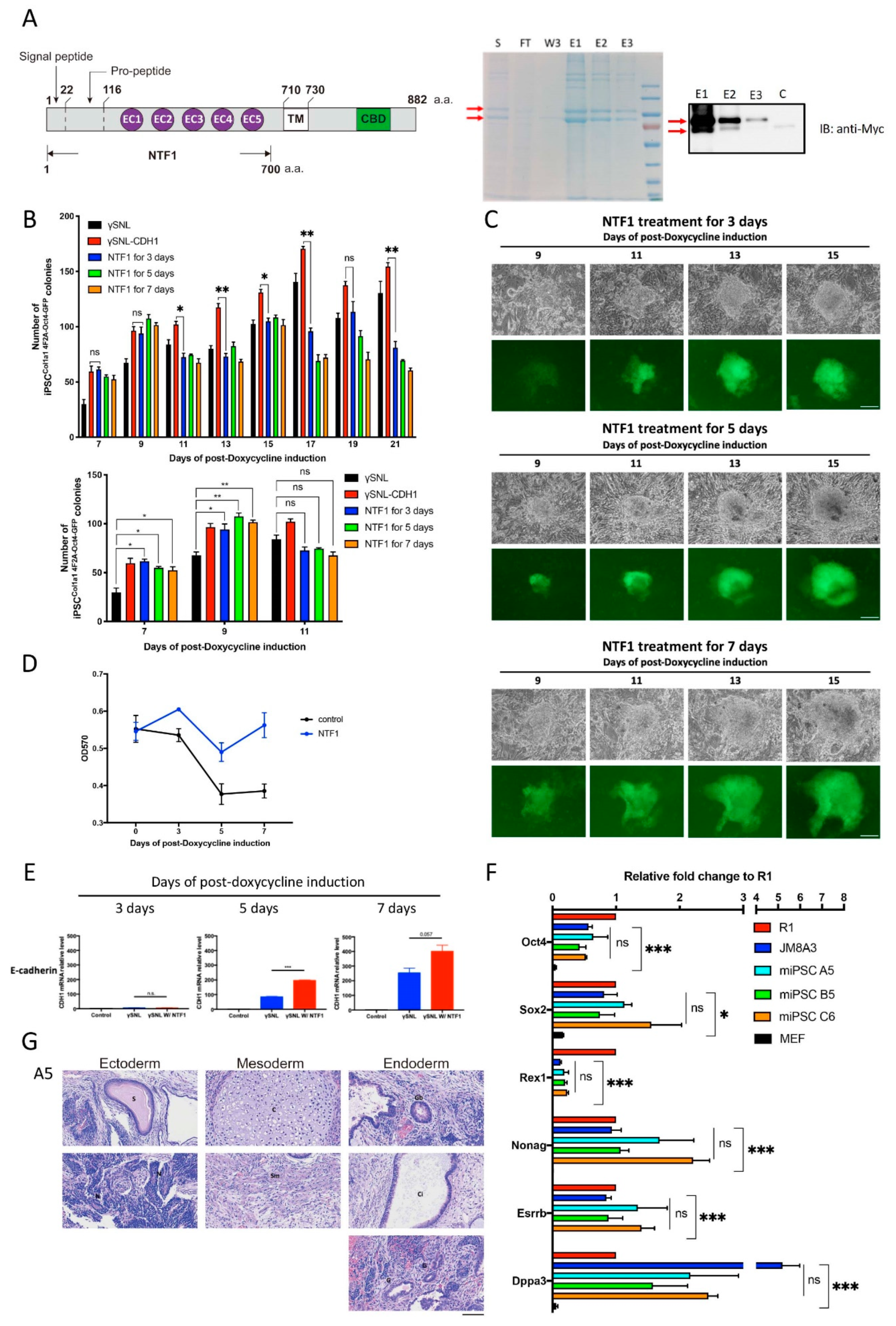
The effect of NTF1 recapitulates the SNL::CHD1-myc feeder adoption at the early stage of reprogramming. (A) Schematic display of the structure of NTF1 protein and purification of the recombinant NTF1 protein. The culture medium collected from the Freestyle 293 cells transfected with pCMV-CDH-NTF1-MYC-His was subjected to NTF1 protein purification. SDS PAGE and western blots were used to analyze the purified product. The red arrows indicate recombinant proteins with different degrees of posttranslational modification. (B) Quantitative analysis of Oct4-GFP-positive clones under different NTF1 treatments during reprogramming. The lower panel shows the data derived from the results of the 7th, 9th, and 11th days in the upper graph. Student’s t-test was applied for statistical evaluation (n = 3). (C) Time-lapse photographs of the Oct4-GFP-positive clones treated with various NTF1 exposure times. The scale bar denotes 50 µm. (D) The growth curve of the induced MEFCol1a1 4F2A Oct4-GFP cells during the first seven days of iPSC induction. An MTT assay was used to quantify cell viability on days 0, 3, 5, and 7 after initiating the induction process. The data points represent the mean of triplicate results. (E) Comparative analysis of endogenous E-cadherin expression in NTF1-treated and untreated MEF cells at the early stage of reprogramming. Harvested cells were subjected to total RNA isolation for the RT-PCR-based gene expression analysis. Cells that did not undergo somatic reprogramming served as controls. (F) Quantitative RT-PCR of pluripotent markers in the above mentioned three iPSC clones compared with two standard ESCs, namely, R1 and JM8A3. (G) Teratoma formation analysis on the A5 iPSC clone to demonstrate its competency for formation of three germ layers. The scale bar denotes 100 µm. S: supernatant; FT: flow-through; W3: 3rd wash; E1: 1st elution; E2: 2nd elution; E3: 3rd elution. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ns, not significant.
Figure 2.
The effect of NTF1 recapitulates the SNL::CHD1-myc feeder adoption at the early stage of reprogramming. (A) Schematic display of the structure of NTF1 protein and purification of the recombinant NTF1 protein. The culture medium collected from the Freestyle 293 cells transfected with pCMV-CDH-NTF1-MYC-His was subjected to NTF1 protein purification. SDS PAGE and western blots were used to analyze the purified product. The red arrows indicate recombinant proteins with different degrees of posttranslational modification. (B) Quantitative analysis of Oct4-GFP-positive clones under different NTF1 treatments during reprogramming. The lower panel shows the data derived from the results of the 7th, 9th, and 11th days in the upper graph. Student’s t-test was applied for statistical evaluation (n = 3). (C) Time-lapse photographs of the Oct4-GFP-positive clones treated with various NTF1 exposure times. The scale bar denotes 50 µm. (D) The growth curve of the induced MEFCol1a1 4F2A Oct4-GFP cells during the first seven days of iPSC induction. An MTT assay was used to quantify cell viability on days 0, 3, 5, and 7 after initiating the induction process. The data points represent the mean of triplicate results. (E) Comparative analysis of endogenous E-cadherin expression in NTF1-treated and untreated MEF cells at the early stage of reprogramming. Harvested cells were subjected to total RNA isolation for the RT-PCR-based gene expression analysis. Cells that did not undergo somatic reprogramming served as controls. (F) Quantitative RT-PCR of pluripotent markers in the above mentioned three iPSC clones compared with two standard ESCs, namely, R1 and JM8A3. (G) Teratoma formation analysis on the A5 iPSC clone to demonstrate its competency for formation of three germ layers. The scale bar denotes 100 µm. S: supernatant; FT: flow-through; W3: 3rd wash; E1: 1st elution; E2: 2nd elution; E3: 3rd elution. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; ns, not significant.
![Cells 10 00284 g002 Cells 10 00284 g002]()
Figure 3.
STAT3 and Erk1/2 regulated Snail2 expression are the potential signal transducers activated by NTF1 (A) Identification of the NTF1-mediated signal transducers through blotting of the phospho-kinase array. After 24-h serum starvation, MEFCol1a1 4F2A Oct4-GFP cells treated with NTF1 for 120 and 150 min were subjected to the phospho-kinase array analysis, and untreated MEFsCol1a1 4F2A Oct4-GFP served as the control. The numbers labeled on the bar graph correspond to the numbers on the blot images. The upper panel shows the 120 min NTF1 treatment, whereas the lower panel displays the 150 min treatment. (B) The time-dependent effect of NTF1 treatment on the phosphorylation of STAT3 and Erk1/2. After 24-h of serum starvation, MEFCol1a1 4F2A Oct4-GFP cells were subjected to a time-course experiment via treatment with 1 µg/mL NTF1 for 5, 15, 30, 60, 120, or 150 min. The NTF1-untreated MEFCol1a1 4F2A Oct4-GFP cells served as the control. In the 120 min NTF1 treatment group, both phosphorylated STAT3 and Erk1/2 signals reached a plateau on the blot. (C) The dose-dependent effect of NTF1 treatment. MEFCol1a1 4F2A Oct4-GFP cells were subjected to different concentrations of NTF1 (0.5, 1, and 2.5 µg/mL). Both the phosphorylated STAT3 and Erk1/2 signals reached a plateau at 1 µg/mL after 120 min of NTF1 treatment. Untreated MEFCol1a1 4F2A Oct4-GFP cells served as the control. (D) The effects of NTF1 treatment in the presence of doxycycline. After 24 h of serum starvation, MEFCol1a1 4F2A Oct4-GFP cells were simultaneously subjected to NTF1 and doxycycline treatment for 120 min. (E) A comparative analysis of the MET-associated genes, including Snail1, Snail2, and Thy1, revealed that Snail2 is the downstream effector of NTF1 treatment targeted by STAT3 and Erk1/2. (n = 3). ***, p < 0.0005; ns, not significant.
Figure 3.
STAT3 and Erk1/2 regulated Snail2 expression are the potential signal transducers activated by NTF1 (A) Identification of the NTF1-mediated signal transducers through blotting of the phospho-kinase array. After 24-h serum starvation, MEFCol1a1 4F2A Oct4-GFP cells treated with NTF1 for 120 and 150 min were subjected to the phospho-kinase array analysis, and untreated MEFsCol1a1 4F2A Oct4-GFP served as the control. The numbers labeled on the bar graph correspond to the numbers on the blot images. The upper panel shows the 120 min NTF1 treatment, whereas the lower panel displays the 150 min treatment. (B) The time-dependent effect of NTF1 treatment on the phosphorylation of STAT3 and Erk1/2. After 24-h of serum starvation, MEFCol1a1 4F2A Oct4-GFP cells were subjected to a time-course experiment via treatment with 1 µg/mL NTF1 for 5, 15, 30, 60, 120, or 150 min. The NTF1-untreated MEFCol1a1 4F2A Oct4-GFP cells served as the control. In the 120 min NTF1 treatment group, both phosphorylated STAT3 and Erk1/2 signals reached a plateau on the blot. (C) The dose-dependent effect of NTF1 treatment. MEFCol1a1 4F2A Oct4-GFP cells were subjected to different concentrations of NTF1 (0.5, 1, and 2.5 µg/mL). Both the phosphorylated STAT3 and Erk1/2 signals reached a plateau at 1 µg/mL after 120 min of NTF1 treatment. Untreated MEFCol1a1 4F2A Oct4-GFP cells served as the control. (D) The effects of NTF1 treatment in the presence of doxycycline. After 24 h of serum starvation, MEFCol1a1 4F2A Oct4-GFP cells were simultaneously subjected to NTF1 and doxycycline treatment for 120 min. (E) A comparative analysis of the MET-associated genes, including Snail1, Snail2, and Thy1, revealed that Snail2 is the downstream effector of NTF1 treatment targeted by STAT3 and Erk1/2. (n = 3). ***, p < 0.0005; ns, not significant.
![Cells 10 00284 g003 Cells 10 00284 g003]()
Figure 4.
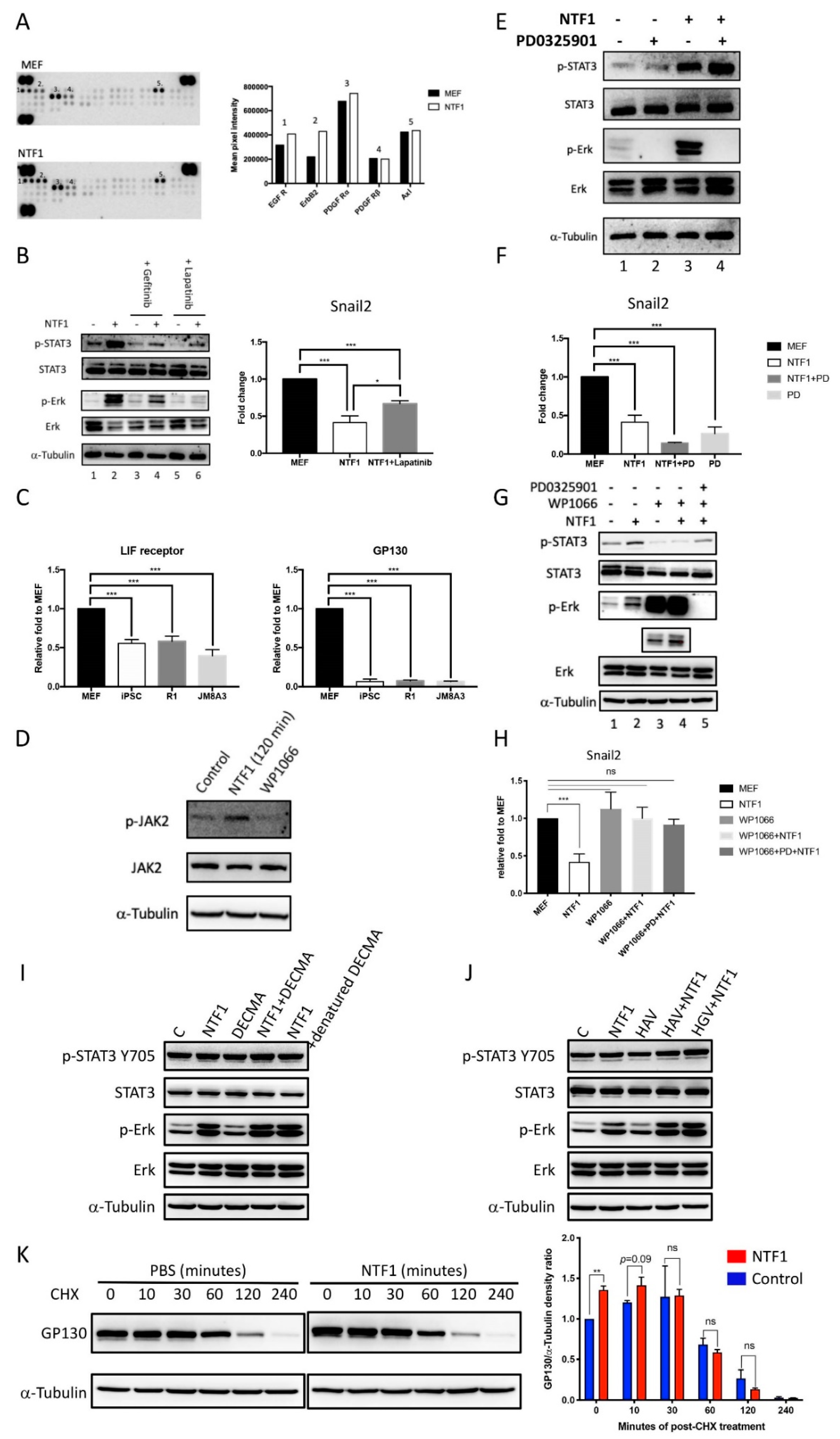
NTF1 may stabilize the LIFR/gp130 complex to transduce the STAT3-mediated signal cascade (A) Use of a receptor tyrosine kinase array revealed the potential NTF1-interacting candidates on the MEFCol1a1 4F2A Oct4-GFP membrane. The numbers on the bar graph correspond to the numbers on the blot images. (B) Examination of the signals involved in the NTF1-mediated signal transduction pathways using Gefitinib and Lapatinib, which are specific inhibitors of EGFR and EGFR/ERbB2, respectively. Using the same treatments, the harvested total RNA of the treated cells was subjected to quantitative RT-PCR analysis with a Snail2-specific primer set. (C) A comparative RT-PCR analysis of LIFR and gp130 in MEFCol1a1 4F2A Oct4-GFP, iPSCCol1a1 4F2A Oct4-GFP, R1 ESC, and JM8A3 ESC cells. (D) Treatment with NTF1 also activates JAK2. MEFCol1a1 4F2A Oct4-GFP cells treated with NTF1 or WP1066 for 120 min were subjected to phosphorylation analysis of Jak2, and untreated MEFs served as the control. (E,F) A synergistic effect on repression of Snail2 expression was observed in the presence of both PD0325901 and NTF1. Using the same treatments, the counterpart experiment was subjected to quantitative RT-PCR analysis with a Snail2-specific primer set. (G,H) Inactivation of STAT3 abolishes the repression of Snail2 expression. MEFCol1a1 4F2A Oct4-GFP cells were subjected to 24-h serum starvation before treatment with NTF1, WP1066, NTF1+WP1066, or NTF1+WP1066+PD0325901. Both cell lysates and total RNA were extracted and subjected to western blotting and RT-PCR analyses, respectively. (I,J) The effect of the DECMA-1 antibody and HAV and HGV peptides in the NTF1-treated experiments. The working concentrations of DECMA-1 (ThermoFisher Scientific, Waltham, MA, USA; 14-3249-82) and the HAV and HGV peptides were 5 µg/mL, 5 mM, and 5 mM, respectively. (K) NTF1 stabilized gp130 in the first 10 min treatment. In the left panel, cycloheximide (CHX) was applied to MEF cells following treatment with NTF1 or PBS. A time-course experiment was performed to observe the change in the gp130 expression level at 0, 10, 30, 60, 120, and 240 min after applying the treatments. The working concentration of CHX was 50 mM. The right panel displays the quantitative result of western blot signals. All groups were normalized to the 0 min treatment in PBS. In each group, the normalized gp130 value was divided by the amount of α-tubulin. Bars represent the mean ratios of each group, and the experiment was performed in duplicate. “+” and “-” denote that the factor was present or absent in the medium, respectively. The working concentrations of NTF1, WP1066, and PD0325901 were 1 µg/mL, 10 µM and 1 µM, respectively. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005; ns, not significant.
Figure 4.
NTF1 may stabilize the LIFR/gp130 complex to transduce the STAT3-mediated signal cascade (A) Use of a receptor tyrosine kinase array revealed the potential NTF1-interacting candidates on the MEFCol1a1 4F2A Oct4-GFP membrane. The numbers on the bar graph correspond to the numbers on the blot images. (B) Examination of the signals involved in the NTF1-mediated signal transduction pathways using Gefitinib and Lapatinib, which are specific inhibitors of EGFR and EGFR/ERbB2, respectively. Using the same treatments, the harvested total RNA of the treated cells was subjected to quantitative RT-PCR analysis with a Snail2-specific primer set. (C) A comparative RT-PCR analysis of LIFR and gp130 in MEFCol1a1 4F2A Oct4-GFP, iPSCCol1a1 4F2A Oct4-GFP, R1 ESC, and JM8A3 ESC cells. (D) Treatment with NTF1 also activates JAK2. MEFCol1a1 4F2A Oct4-GFP cells treated with NTF1 or WP1066 for 120 min were subjected to phosphorylation analysis of Jak2, and untreated MEFs served as the control. (E,F) A synergistic effect on repression of Snail2 expression was observed in the presence of both PD0325901 and NTF1. Using the same treatments, the counterpart experiment was subjected to quantitative RT-PCR analysis with a Snail2-specific primer set. (G,H) Inactivation of STAT3 abolishes the repression of Snail2 expression. MEFCol1a1 4F2A Oct4-GFP cells were subjected to 24-h serum starvation before treatment with NTF1, WP1066, NTF1+WP1066, or NTF1+WP1066+PD0325901. Both cell lysates and total RNA were extracted and subjected to western blotting and RT-PCR analyses, respectively. (I,J) The effect of the DECMA-1 antibody and HAV and HGV peptides in the NTF1-treated experiments. The working concentrations of DECMA-1 (ThermoFisher Scientific, Waltham, MA, USA; 14-3249-82) and the HAV and HGV peptides were 5 µg/mL, 5 mM, and 5 mM, respectively. (K) NTF1 stabilized gp130 in the first 10 min treatment. In the left panel, cycloheximide (CHX) was applied to MEF cells following treatment with NTF1 or PBS. A time-course experiment was performed to observe the change in the gp130 expression level at 0, 10, 30, 60, 120, and 240 min after applying the treatments. The working concentration of CHX was 50 mM. The right panel displays the quantitative result of western blot signals. All groups were normalized to the 0 min treatment in PBS. In each group, the normalized gp130 value was divided by the amount of α-tubulin. Bars represent the mean ratios of each group, and the experiment was performed in duplicate. “+” and “-” denote that the factor was present or absent in the medium, respectively. The working concentrations of NTF1, WP1066, and PD0325901 were 1 µg/mL, 10 µM and 1 µM, respectively. *, p ≤ 0.05; **, p ≤ 0.005; ***, p ≤ 0.0005; ns, not significant.
![Cells 10 00284 g004 Cells 10 00284 g004]()
Figure 5.
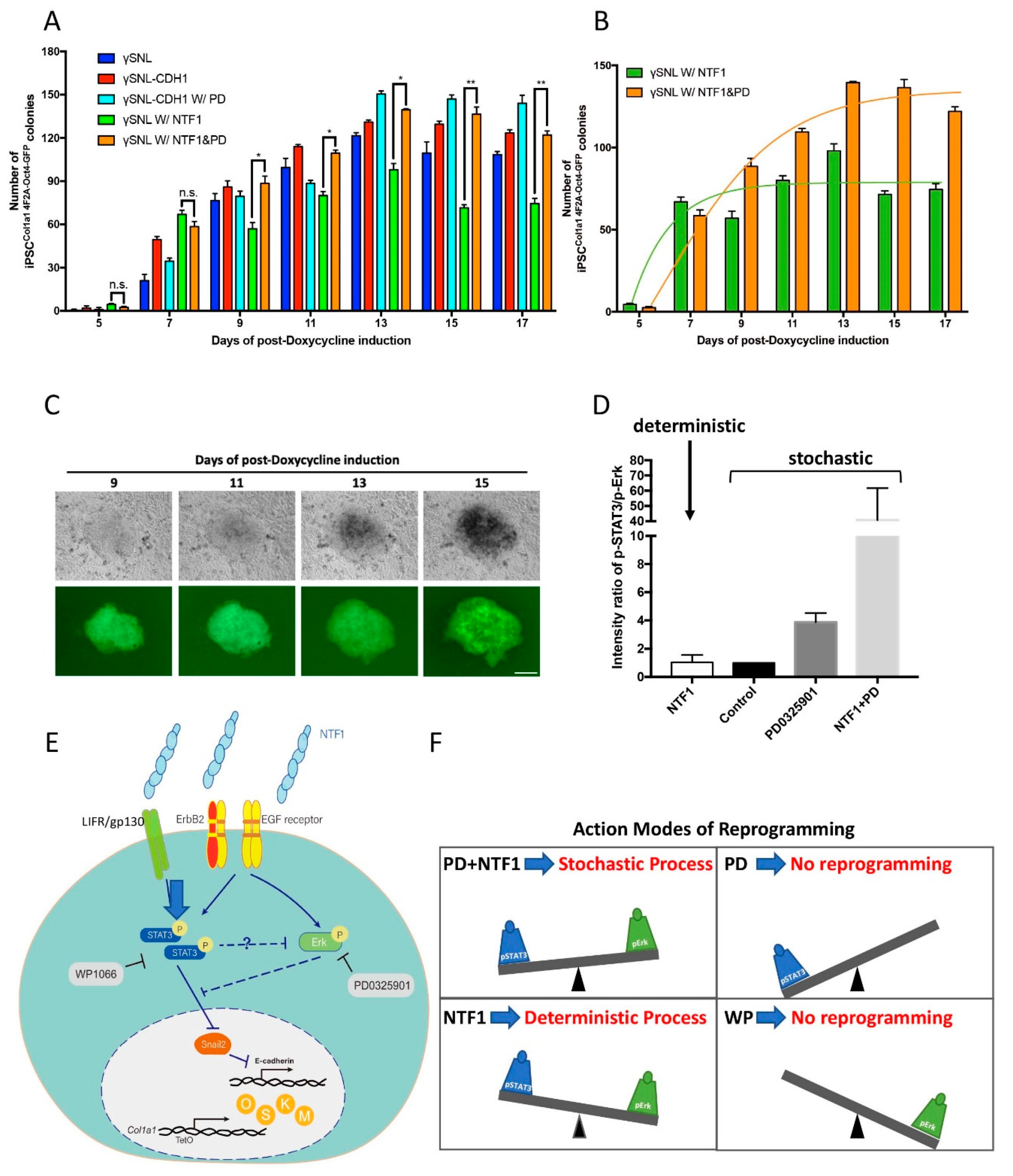
The differential effects of NTF1 and PD0325901 on the choice of programming process at the onset of iPSC induction (A) The quantitative analysis of iPSC colony formation under various treatments. The number of the OCT4-GFP-positive clones is indistinguishable between MEF cells treated with NTF1+PD0325901 and those treated with NTF1 alone on the 7th day postdoxycycline induction. However, the NTF1 alone condition maintained the OCT4-GFP-positive clone number observed on the 7th day throughout the entire course of induction (green bar). In contrast, the NTF1+PD0325901-treated group showed an increase the OCT4-GFP-positive clone number until the end of induction (orange bar). (B) The curve-fitting data of the NTF1 (green) and NTF1+PD (orange) groups extracted from panel A. The NTF1-treated group (green) represents the deterministic process, whereas the NTF1+PD group (orange) shows the stochastic model. (C) Representative time-lapse photographs of iPSCCol1a1 4F2A Oct4-GFP clone formation induced by NTF1+PD0325901. The series of photographs was obtained from the same clone from the 9th to 15th day postdoxycycline induction. The scale bar denotes 50 µm. (D) The selection of a reprogramming process depends on the pSTAT3 vs. pErk1/2 ratio. In the presence of NTF1, the pSTAT3/pErk1/2 ratio creates a unique cell state and a threshold where it engages the deterministic process during reprogramming. If the ratio is higher than the NTF1-established ratio, reprogramming will occur through the stochastic route instead. (E) A proposed mechanism addresses the selection of the somatic reprogramming mode. The current model represents an example in which the surrounding microenvironment imposed upon MEFCol1a1 4F2A Oct4-GFP cells changes the cell fate at the onset of iPSC formation. Extraneous E-cadherin transduces the signal through EGFR/ErbB2 and LIFR/gp130 resulting in activation of the downstream effectors STAT3 and Erk1/2. As the activities of pSTAT3 and pErk1/2 counteract each other, the net output through the respective down- and upregulation of Snail2 and E-cadherin overcomes the MET barrier. In this scenario, pErk acts as a negative regulator on the entry of reprogramming. Accordingly, the balance between STAT3 and Erk1/2 activity is thus essential in setting the stage for cell fate at the entry point of somatic reprogramming. (F) In addition to controlling the entry, the balance between STAT3 and Erk1/2 activity also regulates the choice of entry model, where either the deterministic or stochastic process could be chosen at the onset of reprogramming. The presence of NTF1 in the surrounding environment alters the ratio of pSTAT3 and pErk activity, resulting in adoption of the deterministic model. In contrast, the increment of STAT3 activity, along with the deactivation of Erk, engages the stochastic model in the presence of NTF1+PD0325901 for the first three days of induction. Previous reports have supported that either deterministic or stochastic processes could be adopted during reprogramming depending on the source of the cell type. In the present study, our evidence supports that different modes of operation can act on the same MEF cells and are interchangeable via modulation of the STAT3 and Erk activity. n = 3. PD: PD0325901; WP: WP1066. *, p < 0.05; **, p < 0.005; ns, not significant.
Figure 5.
The differential effects of NTF1 and PD0325901 on the choice of programming process at the onset of iPSC induction (A) The quantitative analysis of iPSC colony formation under various treatments. The number of the OCT4-GFP-positive clones is indistinguishable between MEF cells treated with NTF1+PD0325901 and those treated with NTF1 alone on the 7th day postdoxycycline induction. However, the NTF1 alone condition maintained the OCT4-GFP-positive clone number observed on the 7th day throughout the entire course of induction (green bar). In contrast, the NTF1+PD0325901-treated group showed an increase the OCT4-GFP-positive clone number until the end of induction (orange bar). (B) The curve-fitting data of the NTF1 (green) and NTF1+PD (orange) groups extracted from panel A. The NTF1-treated group (green) represents the deterministic process, whereas the NTF1+PD group (orange) shows the stochastic model. (C) Representative time-lapse photographs of iPSCCol1a1 4F2A Oct4-GFP clone formation induced by NTF1+PD0325901. The series of photographs was obtained from the same clone from the 9th to 15th day postdoxycycline induction. The scale bar denotes 50 µm. (D) The selection of a reprogramming process depends on the pSTAT3 vs. pErk1/2 ratio. In the presence of NTF1, the pSTAT3/pErk1/2 ratio creates a unique cell state and a threshold where it engages the deterministic process during reprogramming. If the ratio is higher than the NTF1-established ratio, reprogramming will occur through the stochastic route instead. (E) A proposed mechanism addresses the selection of the somatic reprogramming mode. The current model represents an example in which the surrounding microenvironment imposed upon MEFCol1a1 4F2A Oct4-GFP cells changes the cell fate at the onset of iPSC formation. Extraneous E-cadherin transduces the signal through EGFR/ErbB2 and LIFR/gp130 resulting in activation of the downstream effectors STAT3 and Erk1/2. As the activities of pSTAT3 and pErk1/2 counteract each other, the net output through the respective down- and upregulation of Snail2 and E-cadherin overcomes the MET barrier. In this scenario, pErk acts as a negative regulator on the entry of reprogramming. Accordingly, the balance between STAT3 and Erk1/2 activity is thus essential in setting the stage for cell fate at the entry point of somatic reprogramming. (F) In addition to controlling the entry, the balance between STAT3 and Erk1/2 activity also regulates the choice of entry model, where either the deterministic or stochastic process could be chosen at the onset of reprogramming. The presence of NTF1 in the surrounding environment alters the ratio of pSTAT3 and pErk activity, resulting in adoption of the deterministic model. In contrast, the increment of STAT3 activity, along with the deactivation of Erk, engages the stochastic model in the presence of NTF1+PD0325901 for the first three days of induction. Previous reports have supported that either deterministic or stochastic processes could be adopted during reprogramming depending on the source of the cell type. In the present study, our evidence supports that different modes of operation can act on the same MEF cells and are interchangeable via modulation of the STAT3 and Erk activity. n = 3. PD: PD0325901; WP: WP1066. *, p < 0.05; **, p < 0.005; ns, not significant.
![Cells 10 00284 g005 Cells 10 00284 g005]()