Functional Genomics Approaches to Elucidate Vulnerabilities of Intrinsic and Acquired Chemotherapy Resistance
Abstract
1. Introduction
2. Gene Perturbation Technologies
2.1. RNA Interference (RNAi)
2.2. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)
2.3. Comparison of RNAi and CRISPR Gene Perturbation Technologies
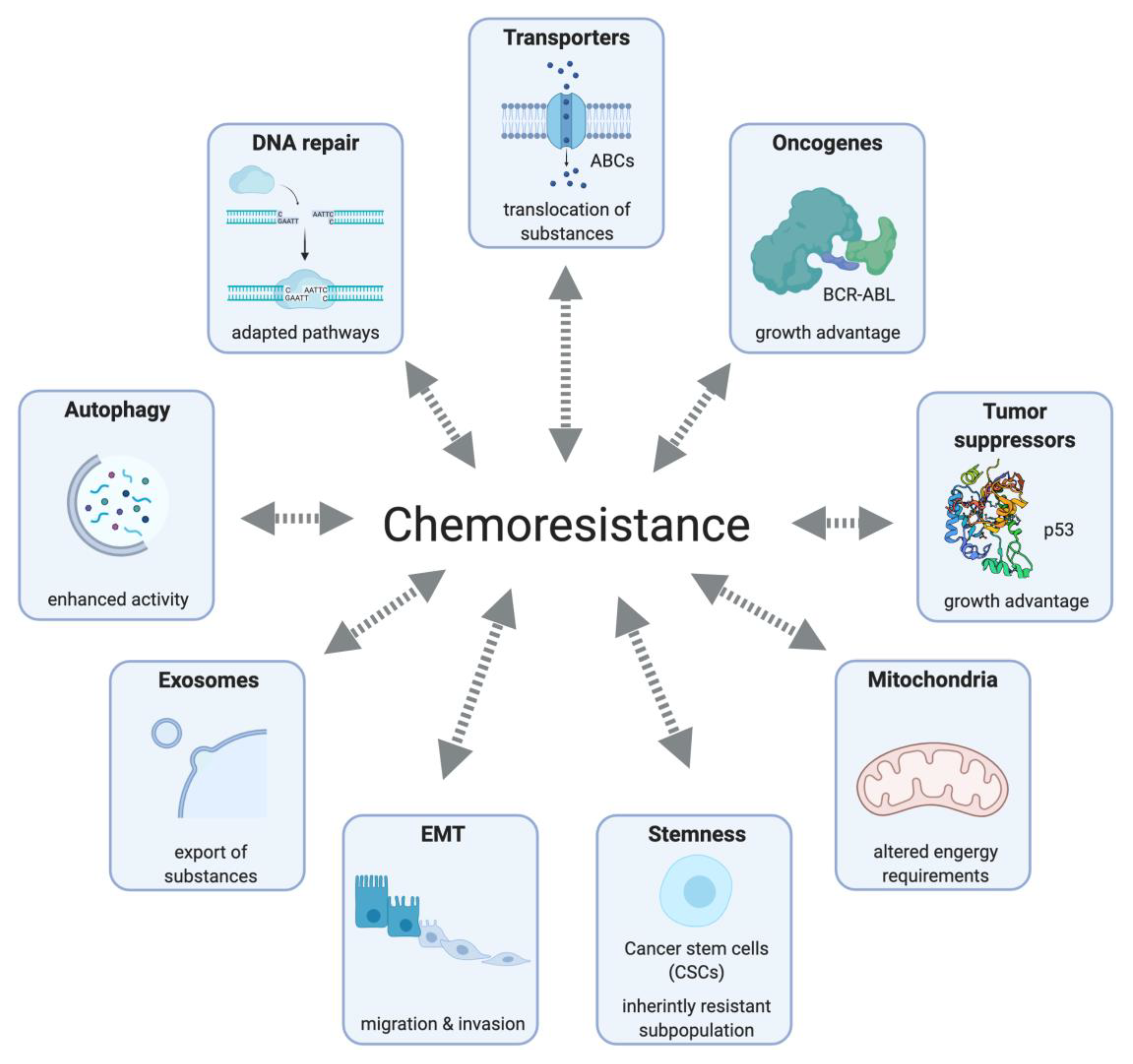
3. Mode of Action, Resistance Mechanisms and Vulnerabilities of Chemotherapeutics
3.1. Alkylating Agents/Platinum-Based Agents
3.2. Topoisomerase Inhibitors
3.3. Mitotic Inhibitors
3.4. Antimetabolites
4. Translating Chemoresistant Vulnerabilities—Conceptual and Technological Strategies
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bertino, J.R.; Göker, E.; Gorlick, R.; Li, W.W.; Banerjee, D. Resistance Mechanisms to Methotrexate in Tumors. Oncologist 1996, 1, 223–226. [Google Scholar] [CrossRef] [PubMed]
- van der Heijden, J.W.; Dijkmans, B.A.C.; Scheper, R.J.; Jansen, G. Drug Insight: Resistance to methotrexate and other disease-modifying antirheumatic drugs--from bench to bedside. Nat. Clin. Pract. Rheumatol. 2007, 3, 26–34. [Google Scholar] [CrossRef]
- Wang, L.; Leite de Oliveira, R.; Wang, C.; Fernandes Neto, J.M.; Mainardi, S.; Evers, B.; Lieftink, C.; Morris, B.; Jochems, F.; Willemsen, L.; et al. High-Throughput Functional Genetic and Compound Screens Identify Targets for Senescence Induction in Cancer. Cell Rep. 2017, 21, 773–783. [Google Scholar] [CrossRef] [PubMed]
- Steinhart, Z.; Pavlovic, Z.; Chandrashekhar, M.; Hart, T.; Wang, X.; Zhang, X.; Robitaille, M.; Brown, K.R.; Jaksani, S.; Overmeer, R.; et al. Genome-wide CRISPR screens reveal a Wnt–FZD5 signaling circuit as a druggable vulnerability of RNF43-mutant pancreatic tumors. Nat. Med. 2017, 23, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Leite de Oliveira, R.; Huijberts, S.; Bosdriesz, E.; Pencheva, N.; Brunen, D.; Bosma, A.; Song, J.-Y.; Zevenhoven, J.; Los-de Vries, G.T.; et al. An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell 2018, 173, 1413–1425.e14. [Google Scholar] [CrossRef] [PubMed]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Elbashir, S.M.; Harborth, J.; Lendeckel, W.; Yalcin, A.; Weber, K.; Tuschl, T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 2001, 411, 494–498. [Google Scholar] [CrossRef] [PubMed]
- Brummelkamp, T.R.; Bernards, R.; Agami, R. A system for stable expression of short interfering RNAs in mammalian cells. Science 2002, 296, 550–553. [Google Scholar] [CrossRef]
- Carthew, R.W.; Sontheimer, E.J. Origins and Mechanisms of miRNAs and siRNAs. Cell 2009, 136, 642–655. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, V.N. Processing of intronic microRNAs. EMBO J. 2007, 26, 775–783. [Google Scholar] [CrossRef]
- Han, J.; Lee, Y.; Yeom, K.-H.; Nam, J.-W.; Heo, I.; Rhee, J.-K.; Sohn, S.Y.; Cho, Y.; Zhang, B.-T.; Kim, V.N. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell 2006, 125, 887–901. [Google Scholar] [CrossRef] [PubMed]
- Lund, E.; Dahlberg, J.E. Substrate selectivity of exportin 5 and Dicer in the biogenesis of microRNAs. Cold Spring Harb. Symp. Quant. Biol. 2006, 71, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef] [PubMed]
- Meister, G.; Landthaler, M.; Patkaniowska, A.; Dorsett, Y.; Teng, G.; Tuschl, T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell 2004, 15, 185–197. [Google Scholar] [CrossRef] [PubMed]
- Heigwer, F.; Port, F.; Boutros, M. RNA Interference (RNAi) Screening in Drosophila. Genetics 2018, 208, 853–874. [Google Scholar] [CrossRef]
- Cowley, G.S.; Weir, B.A.; Vazquez, F.; Tamayo, P.; Scott, J.A.; Rusin, S.; East-Seletsky, A.; Ali, L.D.; Gerath, W.F.; Pantel, S.E.; et al. Parallel genome-scale loss of function screens in 216 cancer cell lines for the identification of context-specific genetic dependencies. Sci Data 2014, 1, 140035. [Google Scholar] [CrossRef]
- Marcotte, R.; Brown, K.R.; Suarez, F.; Sayad, A.; Karamboulas, K.; Krzyzanowski, P.M.; Sircoulomb, F.; Medrano, M.; Fedyshyn, Y.; Koh, J.L.Y.; et al. Essential gene profiles in breast, pancreatic, and ovarian cancer cells. Cancer Discov. 2012, 2, 172–189. [Google Scholar] [CrossRef]
- Boutros, M.; Kiger, A.A.; Armknecht, S.; Kerr, K.; Hild, M.; Koch, B.; Haas, S.A.; Paro, R.; Perrimon, N. Heidelberg Fly Array Consortium Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 2004, 303, 832–835. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Iranzo, J.; Shmakov, S.A.; Alkhnbashi, O.S.; Brouns, S.J.J.; Charpentier, E.; Cheng, D.; Haft, D.H.; Horvath, P.; et al. Evolutionary classification of CRISPR-Cas systems: A burst of class 2 and derived variants. Nat. Rev. Microbiol. 2020, 18, 67–83. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 233–239. [Google Scholar] [CrossRef]
- DiCarlo, J.E.; Norville, J.E.; Mali, P.; Rios, X.; Aach, J.; Church, G.M. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013, 41, 4336–4343. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef] [PubMed]
- Nekrasov, V.; Staskawicz, B.; Weigel, D.; Jones, J.D.G.; Kamoun, S. Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 691–693. [Google Scholar] [CrossRef]
- Sander, J.D.; Joung, J.K. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat. Biotechnol. 2014, 32, 347–355. [Google Scholar] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. Elife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, Y.; Kweon, J.; Kim, H.S.; Bae, S.; Kim, J.-S. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014, 24, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar]
- Chakrabarti, A.M.; Henser-Brownhill, T.; Monserrat, J.; Poetsch, A.R.; Luscombe, N.M.; Scaffidi, P. Target-Specific Precision of CRISPR-Mediated Genome Editing. Mol. Cell 2019, 73, 699–713.e6. [Google Scholar] [CrossRef] [PubMed]
- Adikusuma, F.; Piltz, S.; Corbett, M.A.; Turvey, M.; McColl, S.R.; Helbig, K.J.; Beard, M.R.; Hughes, J.; Pomerantz, R.T.; Thomas, P.Q. Large deletions induced by Cas9 cleavage. Nature 2018, 560, E8–E9. [Google Scholar] [CrossRef]
- Cullot, G.; Boutin, J.; Toutain, J.; Prat, F.; Pennamen, P.; Rooryck, C.; Teichmann, M.; Rousseau, E.; Lamrissi-Garcia, I.; Guyonnet-Duperat, V.; et al. CRISPR-Cas9 genome editing induces megabase-scale chromosomal truncations. Nat. Commun. 2019, 10, 1136. [Google Scholar] [CrossRef]
- Wang, T.; Wei, J.J.; Sabatini, D.M.; Lander, E.S. Genetic screens in human cells using the CRISPR-Cas9 system. Science 2014, 343, 80–84. [Google Scholar] [CrossRef]
- Kaulich, M.; Dowdy, S.F. Combining CRISPR/Cas9 and rAAV Templates for Efficient Gene Editing. Nucleic Acid Ther. 2015, 25, 287–296. [Google Scholar] [CrossRef]
- Kaulich, M.; Lee, Y.J.; Lönn, P.; Springer, A.D.; Meade, B.R.; Dowdy, S.F. Efficient CRISPR-rAAV engineering of endogenous genes to study protein function by allele-specific RNAi. Nucleic Acids Res. 2015, 43, e45. [Google Scholar]
- Wegner, M.; Diehl, V.; Bittl, V.; de Bruyn, R.; Wiechmann, S.; Matthess, Y.; Hebel, M.; Hayes, M.G.; Schaubeck, S.; Benner, C.; et al. Circular synthesized CRISPR/Cas gRNAs for functional interrogations in the coding and noncoding genome. Elife 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef] [PubMed]
- Chavez, A.; Scheiman, J.; Vora, S.; Pruitt, B.W.; Tuttle, M.; Iyer, E.P.R.; Lin, S.; Kiani, S.; Guzman, C.D.; Wiegand, D.J.; et al. Highly efficient Cas9-mediated transcriptional programming. Nat. Methods 2015, 12, 326–328. [Google Scholar]
- Kuscu, C.; Arslan, S.; Singh, R.; Thorpe, J.; Adli, M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat. Biotechnol. 2014, 32, 677–683. [Google Scholar] [CrossRef]
- Smith, I.; Greenside, P.G.; Natoli, T.; Lahr, D.L.; Wadden, D.; Tirosh, I.; Narayan, R.; Root, D.E.; Golub, T.R.; Subramanian, A.; et al. Evaluation of RNAi and CRISPR technologies by large-scale gene expression profiling in the Connectivity Map. PLoS Biol. 2017, 15, e2003213. [Google Scholar] [CrossRef]
- Jackson, A.L.; Bartz, S.R.; Schelter, J.; Kobayashi, S.V.; Burchard, J.; Mao, M.; Li, B.; Cavet, G.; Linsley, P.S. Expression profiling reveals off-target gene regulation by RNAi. Nat. Biotechnol. 2003, 21, 635–637. [Google Scholar] [CrossRef]
- Evers, B.; Jastrzebski, K.; Heijmans, J.P.M.; Grernrum, W.; Beijersbergen, R.L.; Bernards, R. CRISPR knockout screening outperforms shRNA and CRISPRi in identifying essential genes. Nat. Biotechnol. 2016, 34, 631–633. [Google Scholar] [CrossRef]
- Jiang, W.; Marraffini, L.A. CRISPR-Cas: New Tools for Genetic Manipulations from Bacterial Immunity Systems. Annu. Rev. Microbiol. 2015, 69, 209–228. [Google Scholar] [CrossRef] [PubMed]
- Bosher, J.M.; Labouesse, M. RNA interference: Genetic wand and genetic watchdog. Nat. Cell Biol. 2000, 2, E31–E36. [Google Scholar] [CrossRef] [PubMed]
- Hornung, V.; Guenthner-Biller, M.; Bourquin, C.; Ablasser, A.; Schlee, M.; Uematsu, S.; Noronha, A.; Manoharan, M.; Akira, S.; de Fougerolles, A.; et al. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat. Med. 2005, 11, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Bridge, A.J.; Pebernard, S.; Ducraux, A.; Nicoulaz, A.-L.; Iggo, R. Induction of an interferon response by RNAi vectors in mammalian cells. Nat. Genet. 2003, 34, 263–264. [Google Scholar] [CrossRef]
- Morgens, D.W.; Deans, R.M.; Li, A.; Bassik, M.C. Systematic comparison of CRISPR/Cas9 and RNAi screens for essential genes. Nat. Biotechnol. 2016, 34, 634–636. [Google Scholar] [CrossRef]
- Kleinstiver, B.P.; Sousa, A.A.; Walton, R.T.; Esther Tak, Y.; Hsu, J.Y.; Clement, K.; Welch, M.M.; Horng, J.E.; Malagon-Lopez, J.; Scarfò, I.; et al. Engineered CRISPR–Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 2019, 37, 276–282. [Google Scholar]
- Sternberg, S.H.; Redding, S.; Jinek, M.; Greene, E.C.; Doudna, J.A. DNA interrogation by the CRISPR RNA-guided endonuclease Cas9. Nature 2014, 507, 62–67. [Google Scholar]
- Munoz, D.M.; Cassiani, P.J.; Li, L.; Billy, E.; Korn, J.M.; Jones, M.D.; Golji, J.; Ruddy, D.A.; Yu, K.; McAllister, G.; et al. CRISPR Screens Provide a Comprehensive Assessment of Cancer Vulnerabilities but Generate False-Positive Hits for Highly Amplified Genomic Regions. Cancer Discov. 2016, 6, 900–913. [Google Scholar] [CrossRef]
- Aguirre, A.J.; Meyers, R.M.; Weir, B.A.; Vazquez, F.; Zhang, C.-Z.; Ben-David, U.; Cook, A.; Ha, G.; Harrington, W.F.; Doshi, M.B.; et al. Genomic Copy Number Dictates a Gene-Independent Cell Response to CRISPR/Cas9 Targeting. Cancer Discov. 2016, 6, 914–929. [Google Scholar]
- Klann, T.S.; Black, J.B.; Chellappan, M.; Safi, A.; Song, L.; Hilton, I.B.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. CRISPR-Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat. Biotechnol. 2017, 35, 561–568. [Google Scholar]
- Liu, S.J.; Horlbeck, M.A.; Cho, S.W.; Birk, H.S.; Malatesta, M.; He, D.; Attenello, F.J.; Villalta, J.E.; Cho, M.Y.; Chen, Y.; et al. CRISPRi-based genome-scale identification of functional long noncoding RNA loci in human cells. Science 2017, 355. [Google Scholar] [CrossRef] [PubMed]
- Joung, J.; Engreitz, J.M.; Konermann, S.; Abudayyeh, O.O.; Verdine, V.K.; Aguet, F.; Gootenberg, J.S.; Sanjana, N.E.; Wright, J.B.; Fulco, C.P.; et al. Genome-scale activation screen identifies a lncRNA locus regulating a gene neighbourhood. Nature 2017, 548, 343–346. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Zhang, J.; Chen, L.; Shen, B.; Zhou, J.; Hu, B.; Du, Y.; Tate, P.H.; Huang, X.; Zhang, W. Efficient in vivo deletion of a large imprinted lncRNA by CRISPR/Cas9. RNA Biol. 2014, 11, 829–835. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Morita, S.; Noguchi, H.; Horii, T.; Nakabayashi, K.; Kimura, M.; Okamura, K.; Sakai, A.; Nakashima, H.; Hata, K.; Nakashima, K.; et al. Targeted DNA demethylation in vivo using dCas9–peptide repeat and scFv–TET1 catalytic domain fusions. Nat. Biotechnol. 2016, 34, 1060–1065. [Google Scholar] [CrossRef]
- Fuchs, A.; Riegler, S.; Ayatollahi, Z.; Cavallari, N.; Giono, L.E.; Nimeth, B.A.; Mutanwad, K.V.; Schweighofer, A.; Lucyshyn, D.; Barta, A.; et al. Targeting alternative splicing by RNAi: From the differential impact on splice variants to triggering artificial pre-mRNA splicing. Nucleic Acids Res. 2021. [Google Scholar] [CrossRef]
- Rosenbluh, J.; Xu, H.; Harrington, W.; Gill, S.; Wang, X.; Vazquez, F.; Root, D.E.; Tsherniak, A.; Hahn, W.C. Complementary information derived from CRISPR Cas9 mediated gene deletion and suppression. Nat. Commun. 2017, 8, 15403. [Google Scholar] [CrossRef]
- Boettcher, M.; McManus, M.T. Choosing the Right Tool for the Job: RNAi, TALEN, or CRISPR. Mol. Cell 2015, 58, 575–585. [Google Scholar] [CrossRef]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-Resolution CRISPR Screens Reveal Fitness Genes and Genotype-Specific Cancer Liabilities. Cell 2015, 163, 1515–1526. [Google Scholar]
- Crudele, J.M.; Chamberlain, J.S. Cas9 immunity creates challenges for CRISPR gene editing therapies. Nat. Commun. 2018, 9, 3497. [Google Scholar] [CrossRef] [PubMed]
- Charlesworth, C.T.; Deshpande, P.S.; Dever, D.P.; Camarena, J.; Lemgart, V.T.; Cromer, M.K.; Vakulskas, C.A.; Collingwood, M.A.; Zhang, L.; Bode, N.M.; et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat. Med. 2019, 25, 249–254. [Google Scholar] [CrossRef] [PubMed]
- Najm, F.J.; Strand, C.; Donovan, K.F.; Hegde, M.; Sanson, K.R.; Vaimberg, E.W.; Sullender, M.E.; Hartenian, E.; Kalani, Z.; Fusi, N.; et al. Orthologous CRISPR-Cas9 enzymes for combinatorial genetic screens. Nat. Biotechnol. 2018, 36, 179–189. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Rossi, J. RNAi mechanisms and applications. Biotechniques 2008, 44, 613–616. [Google Scholar] [CrossRef] [PubMed]
- Tuladhar, R.; Yeu, Y.; Tyler Piazza, J.; Tan, Z.; Rene Clemenceau, J.; Wu, X.; Barrett, Q.; Herbert, J.; Mathews, D.H.; Kim, J.; et al. CRISPR-Cas9-based mutagenesis frequently provokes on-target mRNA misregulation. Nat. Commun. 2019, 10, 4056. [Google Scholar] [CrossRef]
- Sharpe, J.J.; Cooper, T.A. Unexpected consequences: Exon skipping caused by CRISPR-generated mutations. Genome Biol. 2017, 18, 109. [Google Scholar] [CrossRef]
- Bennett, E.P.; Petersen, B.L.; Johansen, I.E.; Niu, Y.; Yang, Z.; Chamberlain, C.A.; Met, Ö.; Wandall, H.H.; Frödin, M. INDEL detection, the “Achilles heel” of precise genome editing: A survey of methods for accurate profiling of gene editing induced indels. Nucleic Acids Res. 2020, 48, 11958–11981. [Google Scholar]
- Haapaniemi, E.; Botla, S.; Persson, J.; Schmierer, B.; Taipale, J. CRISPR-Cas9 genome editing induces a p53-mediated DNA damage response. Nat. Med. 2018, 24, 927–930. [Google Scholar]
- Enache, O.M.; Rendo, V.; Abdusamad, M.; Lam, D.; Davison, D.; Pal, S.; Currimjee, N.; Hess, J.; Pantel, S.; Nag, A.; et al. Cas9 activates the p53 pathway and selects for p53-inactivating mutations. Nat. Genet. 2020, 52, 662–668. [Google Scholar] [CrossRef]
- Bowden, A.R.; Morales-Juarez, D.A.; Sczaniecka-Clift, M.; Agudo, M.M.; Lukashchuk, N.; Thomas, J.C.; Jackson, S.P. Parallel CRISPR-Cas9 screens clarify impacts of p53 on screen performance. Elife 2020, 9. [Google Scholar] [CrossRef]
- Bast, R.C., Jr.; Croce, C.M.; Hait, W.N.; Hong, W.K.; Kufe, D.W.; Piccart-Gebhart, M.; Pollock, R.E.; Weichselbaum, R.R.; Wang, H.; Holland, J.F. Holland-Frei Cancer Medicine; John Wiley & Sons: Hoboken, NJ, USA, 2017; ISBN 9781119000846. [Google Scholar]
- Pinto, A.L.; Lippard, S.J. Sequence-dependent termination of in vitro DNA synthesis by cis- and trans-diamminedichloroplatinum (II). Proc. Natl. Acad. Sci. USA 1985, 82, 4616–4619. [Google Scholar] [CrossRef] [PubMed]
- Villani, G.; Hübscher, U.; Butour, J.L. Sites of termination of in vitro DNA synthesis on cis-diamminedichloroplatinum(II) treated single-stranded DNA: A comparison between E. coli DNA polymerase I and eucaryotic DNA polymerases alpha. Nucleic Acids Res. 1988, 16, 4407–4418. [Google Scholar] [CrossRef] [PubMed]
- Kartalou, M.; Essigmann, J.M. Mechanisms of resistance to cisplatin. Mutat. Res. 2001, 478, 23–43. [Google Scholar] [CrossRef]
- Chaney, S.G.; Campbell, S.L.; Bassett, E.; Wu, Y. Recognition and processing of cisplatin- and oxaliplatin-DNA adducts. Crit. Rev. Oncol./Hematol. 2005, 53, 3–11. [Google Scholar]
- Jamieson, E.R.; Lippard, S.J. Structure, Recognition, and Processing of Cisplatin−DNA Adducts. Chem. Rev. 1999, 99, 2467–2498. [Google Scholar] [CrossRef]
- Zhou, J.; Kang, Y.; Chen, L.; Wang, H.; Liu, J.; Zeng, S.; Yu, L. The Drug-Resistance Mechanisms of Five Platinum-Based Antitumor Agents. Front. Pharmacol. 2020, 11, 343. [Google Scholar] [CrossRef] [PubMed]
- Dijt, F.J.; Fichtinger-Schepman, A.M.; Berends, F.; Reedijk, J. Formation and repair of cisplatin-induced adducts to DNA in cultured normal and repair-deficient human fibroblasts. Cancer Res. 1988, 48, 6058–6062. [Google Scholar]
- Wynne, P.; Newton, C.; Ledermann, J.A.; Olaitan, A.; Mould, T.A.; Hartley, J.A. Enhanced repair of DNA interstrand crosslinking in ovarian cancer cells from patients following treatment with platinum-based chemotherapy. Br. J. Cancer 2007, 97, 927–933. [Google Scholar]
- Dann, R.B.; DeLoia, J.A.; Timms, K.M.; Zorn, K.K.; Potter, J.; Flake, D.D., 2nd; Lanchbury, J.S.; Krivak, T.C. BRCA1/2 mutations and expression: Response to platinum chemotherapy in patients with advanced stage epithelial ovarian cancer. Gynecol. Oncol. 2012, 125, 677–682. [Google Scholar] [CrossRef]
- Muggia, F.; Safra, T. “BRCAness” and its implications for platinum action in gynecologic cancer. Anticancer Res. 2014, 34, 551–556. [Google Scholar]
- Bartz, S.R.; Zhang, Z.; Burchard, J.; Imakura, M.; Martin, M.; Palmieri, A.; Needham, R.; Guo, J.; Gordon, M.; Chung, N.; et al. Small interfering RNA screens reveal enhanced cisplatin cytotoxicity in tumor cells having both BRCA network and TP53 disruptions. Mol. Cell. Biol. 2006, 26, 9377–9386. [Google Scholar] [CrossRef] [PubMed]
- Soyano, A.E.; Baldeo, C.; Kasi, P.M. BRCA Mutation and Its Association With Colorectal Cancer. Clin. Colorectal Cancer 2018, 17, e647–e650. [Google Scholar] [CrossRef] [PubMed]
- Golan, T.; Kanji, Z.S.; Epelbaum, R.; Devaud, N.; Dagan, E.; Holter, S.; Aderka, D.; Paluch-Shimon, S.; Kaufman, B.; Gershoni-Baruch, R.; et al. Overall survival and clinical characteristics of pancreatic cancer in BRCA mutation carriers. Br. J. Cancer 2014, 111, 1132–1138. [Google Scholar]
- Guillemette, S.; Serra, R.W.; Peng, M.; Hayes, J.A.; Konstantinopoulos, P.A.; Green, M.R.; Cantor, S.B. Resistance to therapy in BRCA2 mutant cells due to loss of the nucleosome remodeling factor CHD4. Genes Dev. 2015, 29, 489–494. [Google Scholar] [PubMed]
- Sawant, A.; Kothandapani, A.; Zhitkovich, A.; Sobol, R.W.; Patrick, S.M. Role of mismatch repair proteins in the processing of cisplatin interstrand cross-links. DNA Repair 2015, 35, 126–136. [Google Scholar] [CrossRef]
- Goodspeed, A.; Jean, A.; Costello, J.C. A Whole-genome CRISPR Screen Identifies a Role of MSH2 in Cisplatin-mediated Cell Death in Muscle-invasive Bladder Cancer. Eur. Urol. 2019, 75, 242–250. [Google Scholar] [CrossRef]
- Morandell, S.; Yaffe, M.B. Exploiting synthetic lethal interactions between DNA damage signaling, checkpoint control, and p53 for targeted cancer therapy. Prog. Mol. Biol. Transl. Sci. 2012, 110, 289–314. [Google Scholar] [PubMed]
- Xu, D.; Liang, S.-Q.; Yang, H.; Bruggmann, R.; Berezowska, S.; Yang, Z.; Marti, T.M.; Hall, S.R.R.; Gao, Y.; Kocher, G.J.; et al. CRISPR Screening Identifies WEE1 as a Combination Target for Standard Chemotherapy in Malignant Pleural Mesothelioma. Mol. Cancer Ther. 2020, 19, 661–672. [Google Scholar] [CrossRef]
- MacKeigan, J.P.; Murphy, L.O.; Blenis, J. Sensitized RNAi screen of human kinases and phosphatases identifies new regulators of apoptosis and chemoresistance. Nat. Cell Biol. 2005, 7, 591–600. [Google Scholar] [CrossRef]
- Beuvink, I.; Boulay, A.; Fumagalli, S.; Zilbermann, F.; Ruetz, S.; O’Reilly, T.; Natt, F.; Hall, J.; Lane, H.A.; Thomas, G. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell 2005, 120, 747–759. [Google Scholar] [CrossRef]
- Geoerger, B.; Kerr, K.; Tang, C.B.; Fung, K.M.; Powell, B.; Sutton, L.N.; Phillips, P.C.; Janss, A.J. Antitumor activity of the rapamycin analog CCI-779 in human primitive neuroectodermal tumor/medulloblastoma models as single agent and in combination chemotherapy. Cancer Res. 2001, 61, 1527–1532. [Google Scholar]
- Guerreiro, A.S.; Fattet, S.; Kulesza, D.W.; Atamer, A.; Elsing, A.N.; Shalaby, T.; Jackson, S.P.; Schoenwaelder, S.M.; Grotzer, M.A.; Delattre, O.; et al. A sensitized RNA interference screen identifies a novel role for the PI3K p110γ isoform in medulloblastoma cell proliferation and chemoresistance. Mol. Cancer Res. 2011, 9, 925–935. [Google Scholar] [CrossRef] [PubMed]
- Ko, T.; Li, S. Genome-wide screening identifies novel genes and biological processes implicated in cisplatin resistance. FASEB J. 2019, 33, 7143–7154. [Google Scholar] [CrossRef] [PubMed]
- Hao, H.-X.; Xie, Y.; Zhang, Y.; Charlat, O.; Oster, E.; Avello, M.; Lei, H.; Mickanin, C.; Liu, D.; Ruffner, H.; et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature 2012, 485, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Petrilli, A.M.; Fernández-Valle, C. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 2016, 35, 537–548. [Google Scholar] [CrossRef] [PubMed]
- MacKay, C.; Carroll, E.; Ibrahim, A.F.M.; Garg, A.; Inman, G.J.; Hay, R.T.; Alpi, A.F. E3 ubiquitin ligase HOIP attenuates apoptotic cell death induced by cisplatin. Cancer Res. 2014, 74, 2246–2257. [Google Scholar] [CrossRef]
- Frisch, S.M.; Francis, H. Disruption of epithelial cell-matrix interactions induces apoptosis. J. Cell Biol. 1994, 124, 619–626. [Google Scholar] [CrossRef]
- Yamanoi, K.; Matsumura, N.; Murphy, S.K.; Baba, T.; Abiko, K.; Hamanishi, J.; Yamaguchi, K.; Koshiyama, M.; Konishi, I.; Mandai, M. Suppression of ABHD2, identified through a functional genomics screen, causes anoikis resistance, chemoresistance and poor prognosis in ovarian cancer. Oncotarget 2016, 7, 47620–47636. [Google Scholar]
- Pommier, Y. Drugging topoisomerases: Lessons and challenges. ACS Chem. Biol. 2013, 8, 82–95. [Google Scholar] [CrossRef]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA topoisomerases and their poisoning by anticancer and antibacterial drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef]
- Bridewell, D.J.; Finlay, G.J.; Baguley, B.C. Differential actions of aclarubicin and doxorubicin: The role of topoisomerase I. Oncol. Res. 1997, 9, 535–542. [Google Scholar] [PubMed]
- Larsen, A.K.; Escargueil, A.E.; Skladanowski, A. Catalytic topoisomerase II inhibitors in cancer therapy. Pharmacol. Ther. 2003, 99, 167–181. [Google Scholar] [CrossRef]
- Fortune, J.M.; Osheroff, N. Merbarone inhibits the catalytic activity of human topoisomerase IIalpha by blocking DNA cleavage. J. Biol. Chem. 1998, 273, 17643–17650. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Yan, T.; Jendrny, C.; Nemecek, A.; Vincetic, M.; Gödtel-Armbrust, U.; Wojnowski, L. Dexrazoxane may prevent doxorubicin-induced DNA damage via depleting both topoisomerase II isoforms. BMC Cancer 2014, 14, 842. [Google Scholar] [CrossRef] [PubMed]
- Roca, J.; Ishida, R.; Berger, J.M.; Andoh, T.; Wang, J.C. Antitumor bisdioxopiperazines inhibit yeast DNA topoisomerase II by trapping the enzyme in the form of a closed protein clamp. Proc. Natl. Acad. Sci. USA 1994, 91, 1781–1785. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.; Yan, T.; Nikolova, T.; Fuhrmann, D.; Nemecek, A.; Gödtel-Armbrust, U.; Kaina, B.; Wojnowski, L. The catalytic topoisomerase II inhibitor dexrazoxane induces DNA breaks, ATF3 and the DNA damage response in cancer cells. Br. J. Pharmacol. 2015, 172, 2246–2257. [Google Scholar] [CrossRef]
- Hsiang, Y.H.; Hertzberg, R.; Hecht, S.; Liu, L.F. Camptothecin induces protein-linked DNA breaks via mammalian DNA topoisomerase I. J. Biol. Chem. 1985, 260, 14873–14878. [Google Scholar]
- Ross, W.; Rowe, T.; Glisson, B.; Yalowich, J.; Liu, L. Role of topoisomerase II in mediating epipodophyllotoxin-induced DNA cleavage. Cancer Res. 1984, 44, 5857–5860. [Google Scholar]
- Gewirtz, D.A. A critical evaluation of the mechanisms of action proposed for the antitumor effects of the anthracycline antibiotics adriamycin and daunorubicin. Biochem. Pharmacol. 1999, 57, 727–741. [Google Scholar]
- Edwardson, D.W.; Narendrula, R.; Chewchuk, S.; Mispel-Beyer, K.; Mapletoft, J.P.J.; Parissenti, A.M. Role of Drug Metabolism in the Cytotoxicity and Clinical Efficacy of Anthracyclines. Curr. Drug Metab. 2015, 16, 412–426. [Google Scholar] [CrossRef]
- Gammella, E.; Maccarinelli, F.; Buratti, P.; Recalcati, S.; Cairo, G. The role of iron in anthracycline cardiotoxicity. Front. Pharmacol. 2014, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.Y.; Liu, L.F. Mechanisms of resistance to topoisomerase inhibitors. In Anticancer Drug Resistance; Springer: Berlin, Germany, 1994; pp. 263–281. [Google Scholar]
- Ganapathi, R.N.; Ganapathi, M.K. Mechanisms regulating resistance to inhibitors of topoisomerase II. Front. Pharmacol. 2013, 4, 89. [Google Scholar] [CrossRef] [PubMed]
- Rasheed, Z.A.; Rubin, E.H. Mechanisms of resistance to topoisomerase I-targeting drugs. Oncogene 2003, 22, 7296–7304. [Google Scholar] [CrossRef] [PubMed]
- Toyoda, Y.; Hagiya, Y.; Adachi, T.; Hoshijima, K.; Kuo, M.T.; Ishikawa, T. MRP class of human ATP binding cassette (ABC) transporters: Historical background and new research directions. Xenobiotica 2008, 38, 833–862. [Google Scholar] [PubMed]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the role of ABC transporters in multidrug-resistant cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of ATP-dependent transporters. Nat. Rev. Cancer 2002, 2, 48–58. [Google Scholar] [CrossRef]
- Nanayakkara, A.K.; Follit, C.A.; Chen, G.; Williams, N.S.; Vogel, P.D.; Wise, J.G. Targeted inhibitors of P-glycoprotein increase chemotherapeutic-induced mortality of multidrug resistant tumor cells. Sci. Rep. 2018, 8, 967. [Google Scholar] [CrossRef]
- Palmer, A.C.; Chidley, C.; Sorger, P.K. A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity. Elife 2019, 8. [Google Scholar] [CrossRef]
- Burgess, D.J.; Doles, J.; Zender, L.; Xue, W.; Ma, B.; McCombie, W.R.; Hannon, G.J.; Lowe, S.W.; Hemann, M.T. Topoisomerase levels determine chemotherapy response in vitro and in vivo. Proc. Natl. Acad. Sci. USA 2008, 105, 9053–9058. [Google Scholar] [CrossRef]
- Rafiee, R.; Sobh, A.; Elsayed, A.H.; Tagmount, A.; Vulpe, C.D.; Lamba, J.K. Genome-scale CRISPR-Cas9 synthetic lethal screening of AML cell line identified functional modulators of etoposide resistance predictive of clinical outcome in AML patients. Blood 2019, 134, 2685. [Google Scholar] [CrossRef]
- Rubnitz, J.E.; Inaba, H.; Dahl, G.; Ribeiro, R.C.; Bowman, W.P.; Taub, J.; Pounds, S.; Razzouk, B.I.; Lacayo, N.J.; Cao, X.; et al. Minimal residual disease-directed therapy for childhood acute myeloid leukaemia: Results of the AML02 multicentre trial. Lancet Oncol. 2010, 11, 543–552. [Google Scholar]
- Wijdeven, R.H.; Pang, B.; van der Zanden, S.Y.; Qiao, X.; Blomen, V.; Hoogstraat, M.; Lips, E.H.; Janssen, L.; Wessels, L.; Brummelkamp, T.R.; et al. Genome-Wide Identification and Characterization of Novel Factors Conferring Resistance to Topoisomerase II Poisons in Cancer. Cancer Res. 2015, 75, 4176–4187. [Google Scholar] [CrossRef] [PubMed]
- Borys, S.M.; Younger, S.T. Identification of functional regulatory elements in the human genome using pooled CRISPR screens. BMC Genomics 2020, 21, 107. [Google Scholar] [CrossRef] [PubMed]
- Dvash, E.; Har-Tal, M.; Barak, S.; Meir, O.; Rubinstein, M. Leukotriene C4 is the major trigger of stress-induced oxidative DNA damage. Nat. Commun. 2015, 6, 10112. [Google Scholar] [CrossRef] [PubMed]
- Diviani, D.; Osman, H.; Reggi, E. A-Kinase Anchoring Protein-Lbc: A Molecular Scaffold Involved in Cardiac Protection. J Cardiovasc Dev. Dis 2018, 5, 12. [Google Scholar] [CrossRef]
- Ha, J.S.; Byun, J.; Ahn, D.-R. Overcoming doxorubicin resistance of cancer cells by Cas9-mediated gene disruption. Sci. Rep. 2016, 6, 22847. [Google Scholar]
- Wang, K.; Xing, Z.-H.; Jiang, Q.-W.; Yang, Y.; Huang, J.-R.; Yuan, M.-L.; Wei, M.-N.; Li, Y.; Wang, S.-T.; Liu, K.; et al. Targeting uPAR by CRISPR/Cas9 System Attenuates Cancer Malignancy and Multidrug Resistance. Front. Oncol. 2019, 9, 80. [Google Scholar] [CrossRef]
- Xiao, Z.; Wan, J.; Nur, A.A.; Dou, P.; Mankin, H.; Liu, T.; Ouyang, Z. Targeting CD44 by CRISPR-Cas9 in Multi-Drug Resistant Osteosarcoma Cells. Cell. Physiol. Biochem. 2018, 51, 1879–1893. [Google Scholar] [CrossRef]
- Gascoigne, K.E.; Taylor, S.S. How do anti-mitotic drugs kill cancer cells? J. Cell Sci. 2009, 122, 2579–2585. [Google Scholar] [CrossRef]
- Sudo, T.; Nitta, M.; Saya, H.; Ueno, N.T. Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Res. 2004, 64, 2502–2508. [Google Scholar] [CrossRef]
- Peterson, J.R.; Mitchison, T.J. Small molecules, big impact: A history of chemical inhibitors and the cytoskeleton. Chem. Biol. 2002, 9, 1275–1285. [Google Scholar] [CrossRef]
- Shimomura, M.; Yaoi, T.; Itoh, K.; Kato, D.; Terauchi, K.; Shimada, J.; Fushiki, S. Drug resistance to paclitaxel is not only associated with ABCB1 mRNA expression but also with drug accumulation in intracellular compartments in human lung cancer cell lines. Int. J. Oncol. 2012, 40, 995–1004. [Google Scholar] [PubMed]
- Huang, C.; Zhang, X.; Jiang, L.; Zhang, L.; Xiang, M.; Ren, H. FoxM1 Induced Paclitaxel Resistance via Activation of the FoxM1/PHB1/RAF-MEK-ERK Pathway and Enhancement of the ABCA2 Transporter. Mol. Ther. Oncol. 2019, 14, 196–212. [Google Scholar] [CrossRef] [PubMed]
- Borst, P.; Elferink, R.O. Mammalian ABC transporters in health and disease. Annu. Rev. Biochem. 2002, 71, 537–592. [Google Scholar] [CrossRef]
- Orr, G.A.; Verdier-Pinard, P.; McDaid, H.; Horwitz, S.B. Mechanisms of Taxol resistance related to microtubules. Oncogene 2003, 22, 7280–7295. [Google Scholar]
- Minotti, A.M.; Barlow, S.B.; Cabral, F. Resistance to antimitotic drugs in Chinese hamster ovary cells correlates with changes in the level of polymerized tubulin. J. Biol. Chem. 1991, 266, 3987–3994. [Google Scholar]
- Balachandran, R.; Welsh, M.J.; Day, B.W. Altered levels and regulation of stathmin in paclitaxel-resistant ovarian cancer cells. Oncogene 2003, 22, 8924–8930. [Google Scholar] [CrossRef][Green Version]
- Montgomery, R.B.; Guzman, J.; O’Rourke, D.M.; Stahl, W.L. Expression of oncogenic epidermal growth factor receptor family kinases induces paclitaxel resistance and alters beta-tubulin isotype expression. J. Biol. Chem. 2000, 275, 17358–17363. [Google Scholar] [CrossRef]
- Ben-Hamo, R.; Zilberberg, A.; Cohen, H.; Bahar-Shany, K.; Wachtel, C.; Korach, J.; Aviel-Ronen, S.; Barshack, I.; Barash, D.; Levanon, K.; et al. Resistance to paclitaxel is associated with a variant of the gene BCL2 in multiple tumor types. NPJ Precis. Oncol. 2019, 3, 12. [Google Scholar] [CrossRef]
- Srivastava, R.K.; Srivastava, A.R.; Korsmeyer, S.J.; Nesterova, M.; Cho-Chung, Y.S.; Longo, D.L. Involvement of microtubules in the regulation of Bcl2 phosphorylation and apoptosis through cyclic AMP-dependent protein kinase. Mol. Cell. Biol. 1998, 18, 3509–3517. [Google Scholar] [CrossRef]
- Haldar, S.; Basu, A.; Croce, C.M. Bcl2 is the guardian of microtubule integrity. Cancer Res. 1997, 57, 229–233. [Google Scholar]
- Fan, W. Possible mechanisms of paclitaxel-induced apoptosis. Biochem. Pharmacol. 1999, 57, 1215–1221. [Google Scholar] [PubMed]
- Patel, N.M.; Nozaki, S.; Shortle, N.H.; Bhat-Nakshatri, P.; Newton, T.R.; Rice, S.; Gelfanov, V.; Boswell, S.H.; Goulet, R.J., Jr.; Sledge, G.W., Jr.; et al. Paclitaxel sensitivity of breast cancer cells with constitutively active NF-kappaB is enhanced by IkappaBalpha super-repressor and parthenolide. Oncogene 2000, 19, 4159–4169. [Google Scholar] [CrossRef] [PubMed]
- Duan, Z.; Feller, A.J.; Penson, R.T.; Chabner, B.A. Discovery of differentially expressed genes associated with paclitaxel resistance using cDNA array technology: Analysis of interleukin (IL) 6, IL-8, and monocyte …. Clin. Cancer Res. 1999, 5, 3445–3453. [Google Scholar]
- Lamendola, D.E.; Duan, Z.; Yusuf, R.Z.; Seiden, M.V. Molecular description of evolving paclitaxel resistance in the SKOV-3 human ovarian carcinoma cell line. Cancer Res. 2003, 63, 2200–2205. [Google Scholar]
- Aldonza, M.B.D.; Ku, J.; Hong, J.-Y.; Kim, D.; Yu, S.J.; Lee, M.-S.; Prayogo, M.C.; Tan, S.; Kim, D.; Han, J.; et al. Prior acquired resistance to paclitaxel relays diverse EGFR-targeted therapy persistence mechanisms. Sci. Adv. 2020, 6, eaav7416. [Google Scholar] [CrossRef] [PubMed]
- Gerhards, N.M.; Blomen, V.A.; Mutlu, M.; Nieuwenhuis, J.; Howald, D.; Guyader, C.; Jonkers, J.; Brummelkamp, T.R.; Rottenberg, S. Haploid genetic screens identify genetic vulnerabilities to microtubule-targeting agents. Mol. Oncol. 2018, 12, 953–971. [Google Scholar] [CrossRef]
- Whitehurst, A.W.; Bodemann, B.O.; Cardenas, J.; Ferguson, D.; Girard, L.; Peyton, M.; Minna, J.D.; Michnoff, C.; Hao, W.; Roth, M.G.; et al. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature 2007, 446, 815–819. [Google Scholar] [CrossRef]
- Lau, M.-T.; Ghazanfar, S.; Parkin, A.; Chou, A.; Rouaen, J.R.; Littleboy, J.B.; Nessem, D.; Khuong, T.M.; Nevoltris, D.; Schofield, P.; et al. Systematic functional identification of cancer multi-drug resistance genes. Genome Biol. 2020, 21, 27. [Google Scholar]
- Wei, W.; He, Y.; Wu, Y.-M. Identification of genes associated with SiHa cell sensitivity to paclitaxel by CRISPR-Cas9 knockout screening. Int. J. Clin. Exp. Pathol. 2018, 11, 1972–1978. [Google Scholar]
- Lian, B.; Pei, Y.-C.; Jiang, Y.-Z.; Xue, M.-Z.; Li, D.-Q.; Li, X.-G.; Zheng, Y.-Z.; Liu, X.-Y.; Qiao, F.; Sun, W.-L.; et al. Truncated HDAC9 identified by integrated genome-wide screen as the key modulator for paclitaxel resistance in triple-negative breast cancer. Theranostics 2020, 10, 11092–11109. [Google Scholar] [CrossRef]
- Zhao, W.-S.; Yan, W.-P.; Chen, D.-B.; Dai, L.; Yang, Y.-B.; Kang, X.-Z.; Fu, H.; Chen, P.; Deng, K.-J.; Wang, X.-Y.; et al. Genome-scale CRISPR activation screening identifies a role of ELAVL2-CDKN1A axis in paclitaxel resistance in esophageal squamous cell carcinoma. Am. J. Cancer Res. 2019, 9, 1183–1200. [Google Scholar]
- Rushworth, L.K.; Harle, V.; Repiscak, P.; Clark, W.; Shaw, R.; Hall, H.; Bushell, M.; Leung, H.Y.; Patel, R. In vivo CRISPR/Cas9 knockout screen: TCEAL1 silencing enhances docetaxel efficacy in prostate cancer. Life Sci Alliance 2020, 3. [Google Scholar] [CrossRef]
- Gupta, N.; Gupta, P.; Srivastava, S.K. Penfluridol overcomes paclitaxel resistance in metastatic breast cancer. Sci. Rep. 2019, 9, 5066. [Google Scholar] [CrossRef]
- Werkheiser, W.C. Specific Binding of 4-Amino Folic Acid Analogues by Folic Acid Reductase. J. Biol. Chem. 1961, 236, 888–893. [Google Scholar] [CrossRef]
- Cerqueira, N.M.F.S.A.; Fernandes, P.A.; Ramos, M.J. Ribonucleotide reductase: A critical enzyme for cancer chemotherapy and antiviral agents. Recent Pat. Anticancer Drug Discov. 2007, 2, 11–29. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Mackey, J.R.; Dumontet, C. Nucleoside analogues: Mechanisms of drug resistance and reversal strategies. Leukemia 2001, 15, 875–890. [Google Scholar] [CrossRef]
- Bepler, G.; Sommers, K.E.; Cantor, A.; Li, X.; Sharma, A.; Williams, C.; Chiappori, A.; Haura, E.; Antonia, S.; Tanvetyanon, T.; et al. Clinical efficacy and predictive molecular markers of neoadjuvant gemcitabine and pemetrexed in resectable non-small cell lung cancer. J. Thorac. Oncol. 2008, 3, 1112–1118. [Google Scholar]
- Moulder, S.; Valkov, N.; Neuger, A.; Choi, J.; Lee, J.H.; Minton, S.; Munster, P.; Gump, J.; Lacevic, M.; Lush, R.; et al. Phase 2 study of gemcitabine and irinotecan in metastatic breast cancer with correlatives to determine topoisomerase I localization as a predictor of response. Cancer 2008, 113, 2646–2654. [Google Scholar]
- Richardson, D.L.; Backes, F.J.; Seamon, L.G.; Zanagnolo, V.; O’Malley, D.M.; Cohn, D.E.; Fowler, J.M.; Copeland, L.J. Combination gemcitabine, platinum, and bevacizumab for the treatment of recurrent ovarian cancer. Gynecol. Oncol. 2008, 111, 461–466. [Google Scholar] [CrossRef]
- Valle, J.; Wasan, H.; Palmer, D.H.; Cunningham, D.; Anthoney, A.; Maraveyas, A.; Madhusudan, S.; Iveson, T.; Hughes, S.; Pereira, S.P.; et al. Cisplatin plus gemcitabine versus gemcitabine for biliary tract cancer. N. Engl. J. Med. 2010, 362, 1273–1281. [Google Scholar] [CrossRef]
- Burris, H.A., 3rd; Moore, M.J.; Andersen, J.; Green, M.R.; Rothenberg, M.L.; Modiano, M.R.; Cripps, M.C.; Portenoy, R.K.; Storniolo, A.M.; Tarassoff, P.; et al. Improvements in survival and clinical benefit with gemcitabine as first-line therapy for patients with advanced pancreas cancer: A randomized trial. J. Clin. Oncol. 1997, 15, 2403–2413. [Google Scholar]
- Mini, E.; Nobili, S.; Caciagli, B.; Landini, I.; Mazzei, T. Cellular pharmacology of gemcitabine. Ann. Oncol. 2006, 17 (Suppl. 5), v7–v12. [Google Scholar] [CrossRef] [PubMed]
- Sarvepalli, D.; Rashid, M.U.; Rahman, A.U.; Ullah, W.; Hussain, I.; Hasan, B.; Jehanzeb, S.; Khan, A.K.; Jain, A.G.; Khetpal, N.; et al. Gemcitabine: A Review of Chemoresistance in Pancreatic Cancer. Crit. Rev. Oncog. 2019, 24, 199–212. [Google Scholar] [CrossRef]
- Nakano, Y.; Tanno, S.; Koizumi, K.; Nishikawa, T.; Nakamura, K.; Minoguchi, M.; Izawa, T.; Mizukami, Y.; Okumura, T.; Kohgo, Y. Gemcitabine chemoresistance and molecular markers associated with gemcitabine transport and metabolism in human pancreatic cancer cells. Br. J. Cancer 2007, 96, 457–463. [Google Scholar] [CrossRef]
- Costantino, C.L.; Witkiewicz, A.K.; Kuwano, Y.; Cozzitorto, J.A.; Kennedy, E.P.; Dasgupta, A.; Keen, J.C.; Yeo, C.J.; Gorospe, M.; Brody, J.R. The role of HuR in gemcitabine efficacy in pancreatic cancer: HuR Up-regulates the expression of the gemcitabine metabolizing enzyme deoxycytidine kinase. Cancer Res. 2009, 69, 4567–4572. [Google Scholar] [CrossRef]
- Arlt, A.; Gehrz, A.; Müerköster, S.; Vorndamm, J.; Kruse, M.-L.; Fölsch, U.R.; Schäfer, H. Role of NF-kappaB and Akt/PI3K in the resistance of pancreatic carcinoma cell lines against gemcitabine-induced cell death. Oncogene 2003, 22, 3243–3251. [Google Scholar] [CrossRef]
- Tréhoux, S.; Duchêne, B.; Jonckheere, N.; Van Seuningen, I. The MUC1 oncomucin regulates pancreatic cancer cell biological properties and chemoresistance. Implication of p42-44 MAPK, Akt, Bcl-2 and MMP13 pathways. Biochem. Biophys. Res. Commun. 2015, 456, 757–762. [Google Scholar] [CrossRef]
- Yang, X.L.; Lin, F.J.; Guo, Y.J.; Shao, Z.M.; Ou, Z.L. Gemcitabine resistance in breast cancer cells regulated by PI3K/AKT-mediated cellular proliferation exerts negative feedback via the MEK/MAPK and mTOR pathways. Onco. Targets. Ther. 2014, 7, 1033–1042. [Google Scholar]
- Wang, R.; Cheng, L.; Xia, J.; Wang, Z.; Wu, Q.; Wang, Z. Gemcitabine resistance is associated with epithelial-mesenchymal transition and induction of HIF-1α in pancreatic cancer cells. Curr. Cancer Drug Targets 2014, 14, 407–417. [Google Scholar] [CrossRef]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.-C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef]
- Fang, Y.; Zhou, W.; Rong, Y.; Kuang, T.; Xu, X.; Wu, W.; Wang, D.; Lou, W. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp. Cell Res. 2019, 383, 111543. [Google Scholar] [CrossRef]
- Yang, Z.; Zhao, N.; Cui, J.; Wu, H.; Xiong, J.; Peng, T. Exosomes derived from cancer stem cells of gemcitabine-resistant pancreatic cancer cells enhance drug resistance by delivering miR-210. Cell. Oncol. 2020, 43, 123–136. [Google Scholar] [CrossRef]
- Giroux, V.; Iovanna, J.; Dagorn, J.-C. Probing the human kinome for kinases involved in pancreatic cancer cell survival and gemcitabine resistance. FASEB J. 2006, 20, 1982–1991. [Google Scholar] [CrossRef]
- Ng, S.S.W.; Tsao, M.S.; Chow, S.; Hedley, D.W. Inhibition of phosphatidylinositide 3-kinase enhances gemcitabine-induced apoptosis in human pancreatic cancer cells. Cancer Res. 2000, 60, 5451–5455. [Google Scholar]
- Bondar, V.M.; Sweeney-Gotsch, B.; Andreeff, M.; Mills, G.B.; McConkey, D.J. Inhibition of the phosphatidylinositol 3’-kinase-AKT pathway induces apoptosis in pancreatic carcinoma cells in vitro and in vivo. Mol. Cancer Ther. 2002, 1, 989–997. [Google Scholar]
- Azorsa, D.O.; Gonzales, I.M.; Basu, G.D.; Choudhary, A.; Arora, S.; Bisanz, K.M.; Kiefer, J.A.; Henderson, M.C.; Trent, J.M.; Von Hoff, D.D.; et al. Synthetic lethal RNAi screening identifies sensitizing targets for gemcitabine therapy in pancreatic cancer. J. Transl. Med. 2009, 7, 43. [Google Scholar] [CrossRef]
- Fredebohm, J.; Wolf, J.; Hoheisel, J.D.; Boettcher, M. Depletion of RAD17 sensitizes pancreatic cancer cells to gemcitabine. J. Cell Sci. 2013, 126, 3380–3389. [Google Scholar] [CrossRef] [PubMed]
- Masoudi, M.; Seki, M.; Yazdanparast, R.; Yachie, N.; Aburatani, H. A genome-scale CRISPR/Cas9 knockout screening reveals SH3D21 as a sensitizer for gemcitabine. Sci. Rep. 2019, 9, 19188. [Google Scholar] [CrossRef]
- Wei, X.; Yang, J.; Adair, S.J.; Ozturk, H.; Kuscu, C.; Lee, K.Y.; Kane, W.J.; O’Hara, P.E.; Liu, D.; Demirlenk, Y.M.; et al. Targeted CRISPR screening identifies PRMT5 as synthetic lethality combinatorial target with gemcitabine in pancreatic cancer cells. Proc. Natl. Acad. Sci. USA 2020, 117, 28068–28079. [Google Scholar] [CrossRef]
- Xu, Y.; Karlsson, A.; Johansson, M. Identification of genes associated to 2’,2’-difluorodeoxycytidine resistance in HeLa cells with a lentiviral short-hairpin RNA library. Biochem. Pharmacol. 2011, 82, 210–215. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Zhan, M.; Jiang, C.; He, M.; Yang, L.; Shen, H.; Huang, S.; Huang, X.; Lin, R.; Shi, Y.; et al. Genome-wide CRISPR screen identifies ELP5 as a determinant of gemcitabine sensitivity in gallbladder cancer. Nat. Commun. 2019, 10, 5492. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.S. Cross-resistance of vinblastine- and taxol-resistant mutants of Chinese hamster ovary cells to other anticancer drugs. Cancer Treat. Rep. 1985, 69, 515–521. [Google Scholar] [PubMed]
- Jensen, P.B.; Holm, B.; Sorensen, M.; Christensen, I.J.; Sehested, M. In vitro cross-resistance and collateral sensitivity in seven resistant small-cell lung cancer cell lines: Preclinical identification of suitable drug partners to taxotere, taxol, topotecan and gemcitabin. Br. J. Cancer 1997, 75, 869–877. [Google Scholar] [CrossRef] [PubMed]
- Zaman, G.J.; Lankelma, J.; van Tellingen, O.; Beijnen, J.; Dekker, H.; Paulusma, C.; Oude Elferink, R.P.; Baas, F.; Borst, P. Role of glutathione in the export of compounds from cells by the multidrug-resistance-associated protein. Proc. Natl. Acad. Sci. USA 1995, 92, 7690–7694. [Google Scholar] [CrossRef] [PubMed]
- Paulusma, C.C.; van Geer, M.A.; Evers, R.; Heijn, M.; Ottenhoff, R.; Borst, P.; Oude Elferink, R.P. Canalicular multispecific organic anion transporter/multidrug resistance protein 2 mediates low-affinity transport of reduced glutathione. Biochem. J 1999, 338 Pt 2, 393–401. [Google Scholar] [CrossRef]
- Wortelboer, H.M.; Usta, M.; van der Velde, A.E.; Boersma, M.G.; Spenkelink, B.; van Zanden, J.J.; Rietjens, I.M.C.M.; van Bladeren, P.J.; Cnubben, N.H.P. Interplay between MRP inhibition and metabolism of MRP inhibitors: The case of curcumin. Chem. Res. Toxicol. 2003, 16, 1642–1651. [Google Scholar] [CrossRef]
- Leslie, E.M.; Deeley, R.G.; Cole, S.P.C. Bioflavonoid stimulation of glutathione transport by the 190-kDa multidrug resistance protein 1 (MRP1). Drug Metab. Dispos. 2003, 31, 11–15. [Google Scholar] [CrossRef]
- Perrotton, T.; Trompier, D.; Chang, X.-B.; Di Pietro, A.; Baubichon-Cortay, H. (R)- and (S)-verapamil differentially modulate the multidrug-resistant protein MRP1. J. Biol. Chem. 2007, 282, 31542–31548. [Google Scholar] [CrossRef]
- Trompier, D.; Chang, X.-B.; Barattin, R.; du Moulinet D’Hardemare, A.; Di Pietro, A.; Baubichon-Cortay, H. Verapamil and its derivative trigger apoptosis through glutathione extrusion by multidrug resistance protein MRP1. Cancer Res. 2004, 64, 4950–4956. [Google Scholar] [CrossRef]
- Lorendeau, D.; Dury, L.; Genoux-Bastide, E.; Lecerf-Schmidt, F.; Simões-Pires, C.; Carrupt, P.-A.; Terreux, R.; Magnard, S.; Di Pietro, A.; Boumendjel, A.; et al. Collateral sensitivity of resistant MRP1-overexpressing cells to flavonoids and derivatives through GSH efflux. Biochem. Pharmacol. 2014, 90, 235–245. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Villani, R.M.; Wang, H.; Simpson, M.J.; Roberts, M.S.; Tang, M.; Liang, X. The role of cellular reactive oxygen species in cancer chemotherapy. J. Exp. Clin. Cancer Res. 2018, 37, 266. [Google Scholar] [CrossRef] [PubMed]
- Weijl, N.I.; Hopman, G.D.; Wipkink-Bakker, A.; Lentjes, E.G.; Berger, H.M.; Cleton, F.J.; Osanto, S. Cisplatin combination chemotherapy induces a fall in plasma antioxidants of cancer patients. Ann. Oncol. 1998, 9, 1331–1337. [Google Scholar] [CrossRef]
- Cui, Q.; Wang, J.-Q.; Assaraf, Y.G.; Ren, L.; Gupta, P.; Wei, L.; Ashby, C.R., Jr.; Yang, D.-H.; Chen, Z.-S. Modulating ROS to overcome multidrug resistance in cancer. Drug Resist. Updat. 2018, 41, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Huijberts, S.; Wang, L.; de Oliveira, R.L.; Rosing, H.; Nuijen, B.; Beijnen, J.; Bernards, R.; Schellens, J.; Wilgenhof, S. Vorinostat in patients with resistant BRAFV600E mutated advanced melanoma: A proof of concept study. Future Oncol. 2020, 16, 619–629. [Google Scholar] [CrossRef]
- Parsels, L.A.; Qian, Y.; Tanska, D.M.; Gross, M.; Zhao, L.; Hassan, M.C.; Arumugarajah, S.; Parsels, J.D.; Hylander-Gans, L.; Simeone, D.M.; et al. Assessment of chk1 phosphorylation as a pharmacodynamic biomarker of chk1 inhibition. Clin. Cancer Res. 2011, 17, 3706–3715. [Google Scholar] [CrossRef]
- Venkatesha, V.A.; Parsels, L.A.; Parsels, J.D.; Zhao, L.; Zabludoff, S.D.; Simeone, D.M.; Maybaum, J.; Lawrence, T.S.; Morgan, M.A. Sensitization of pancreatic cancer stem cells to gemcitabine by Chk1 inhibition. Neoplasia 2012, 14, 519–525. [Google Scholar] [CrossRef]
- Mano, Y.; Kikuchi, Y.; Yamamoto, K.; Kita, T.; Hirata, J.; Tode, T.; Ishii, K.; Nagata, I. Bcl-2 as a predictor of chemosensitivity and prognosis in primary epithelial ovarian cancer. Eur. J. Cancer 1999, 35, 1214–1219. [Google Scholar] [CrossRef]
- Sartorius, U.A.; Krammer, P.H. Upregulation of Bcl-2 is involved in the mediation of chemotherapy resistance in human small cell lung cancer cell lines. Int. J. Cancer 2002, 97, 584–592. [Google Scholar] [CrossRef]
- Thomas, S.; Quinn, B.A.; Das, S.K.; Dash, R.; Emdad, L.; Dasgupta, S.; Wang, X.-Y.; Dent, P.; Reed, J.C.; Pellecchia, M.; et al. Targeting the Bcl-2 family for cancer therapy. Expert Opin. Ther. Targets 2013, 17, 61–75. [Google Scholar] [CrossRef]
- García-Aranda, M.; Pérez-Ruiz, E.; Redondo, M. Bcl-2 Inhibition to Overcome Resistance to Chemo- and Immunotherapy. Int. J. Mol. Sci. 2018, 19, 3950. [Google Scholar] [CrossRef]
- Pearce, M.C.; Gamble, J.T.; Kopparapu, P.R.; O’Donnell, E.F.; Mueller, M.J.; Jang, H.S.; Greenwood, J.A.; Satterthwait, A.C.; Tanguay, R.L.; Zhang, X.-K.; et al. Induction of apoptosis and suppression of tumor growth by Nur77-derived Bcl-2 converting peptide in chemoresistant lung cancer cells. Oncotarget 2018, 9, 26072–26085. [Google Scholar] [CrossRef]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef]
- Diehl, V.; Wegner, M.; Grumati, P.; Husnjak, K.; Schaubeck, S.; Gubas, A.; Shah, V.J.; Langschied, F.; Kalousi, A.; Ebersberger, I.; et al. Combinatorial CRISPR screening reveals functional buffering in autophagy. bioRxiv 2020. [Google Scholar] [CrossRef]
- Dede, M.; McLaughlin, M.; Kim, E.; Hart, T. Multiplex enCas12a screens detect functional buffering among paralogs otherwise masked in monogenic Cas9 knockout screens. Genome Biol. 2020, 21, 262. [Google Scholar] [CrossRef]

| RNAi | CRISPR | |
|---|---|---|
| Strengths | Selective targeting of different transcripts of the same gene [67] Independent of cell ploidy or chromatin conformation [68] Knockdown effect is more similar to drug effect [68] Can target any mRNA sequence [68] | High knockdown efficiency [69] Identification of essential genes with low expression [55,70] High on-target rates [48,50] Targeting of non-transcribed elements [60,63] Low immune response [71,72] Stable gene disruption [51] Highly modular system (dCas9 with effector domains) [34] Pooled combinatorial designs available [73] |
| Weaknesses | Considerable off-target effects [48,49] Incomplete silencing of the target gene (knockdown) [52] Stimulate immune response [53,54] Transient gene silencing [74] | Residual protein expression (incomplete KO/truncated transcripts) [75,76] Target space depends on PAM [57] Stochastic mutational outcome (dependent on DNA repair) [77] Depends on chromatin structure and ploidy of the cell [47] Requires expression of a Cas-enzyme [20,29] Induces DNA damage (DSB) [58,59] Performance is influenced by a cells’ TP53 status [78,79,80] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cetin, R.; Quandt, E.; Kaulich, M. Functional Genomics Approaches to Elucidate Vulnerabilities of Intrinsic and Acquired Chemotherapy Resistance. Cells 2021, 10, 260. https://doi.org/10.3390/cells10020260
Cetin R, Quandt E, Kaulich M. Functional Genomics Approaches to Elucidate Vulnerabilities of Intrinsic and Acquired Chemotherapy Resistance. Cells. 2021; 10(2):260. https://doi.org/10.3390/cells10020260
Chicago/Turabian StyleCetin, Ronay, Eva Quandt, and Manuel Kaulich. 2021. "Functional Genomics Approaches to Elucidate Vulnerabilities of Intrinsic and Acquired Chemotherapy Resistance" Cells 10, no. 2: 260. https://doi.org/10.3390/cells10020260
APA StyleCetin, R., Quandt, E., & Kaulich, M. (2021). Functional Genomics Approaches to Elucidate Vulnerabilities of Intrinsic and Acquired Chemotherapy Resistance. Cells, 10(2), 260. https://doi.org/10.3390/cells10020260






