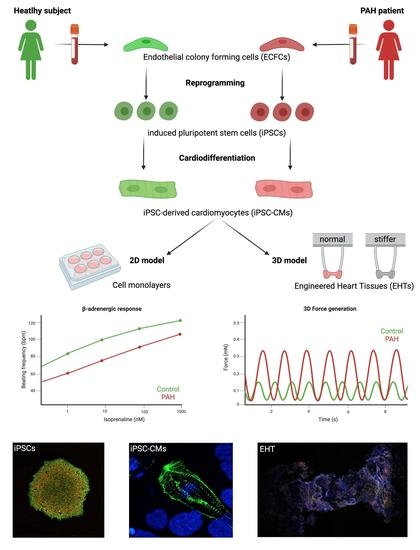
Development of a 3-Dimensional Model to Study Right Heart Dysfunction in Pulmonary Arterial Hypertension: First Observations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Generation of Human iPSCs
2.2. Cardiac Differentiation of iPSCs
2.3. Generation of EHTs
2.4. Development and Characterization of the Stiffer Silicone Posts
2.5. Immunofluorescent Staining
2.6. Quantitative Real-Time PCR Analysis
2.7. Western Blot
2.8. Contractility Measurements
2.9. Statistical Analysis
3. Results
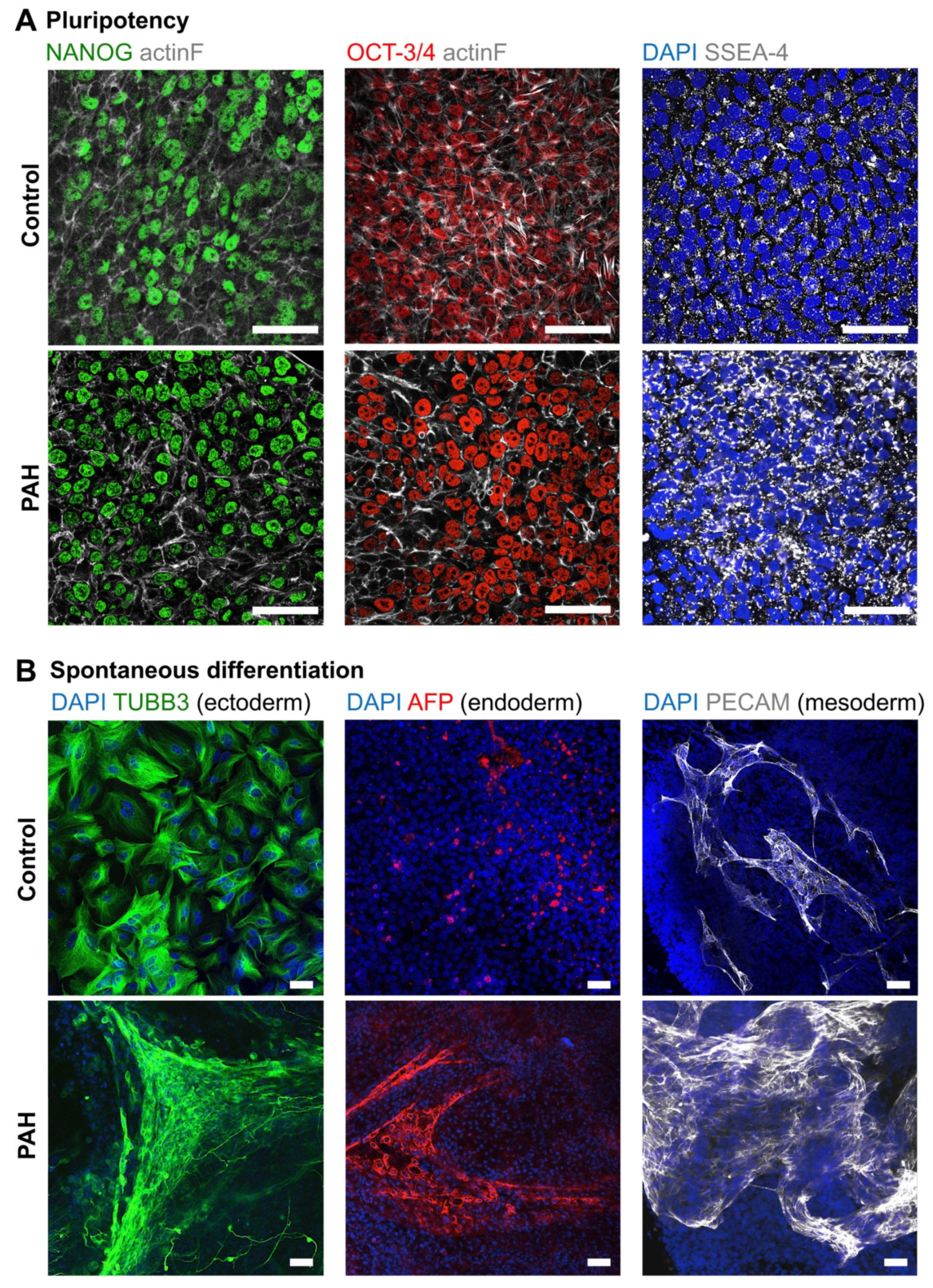
3.1. Successful Production of iPSCs from PAH Patients and Control Subjects
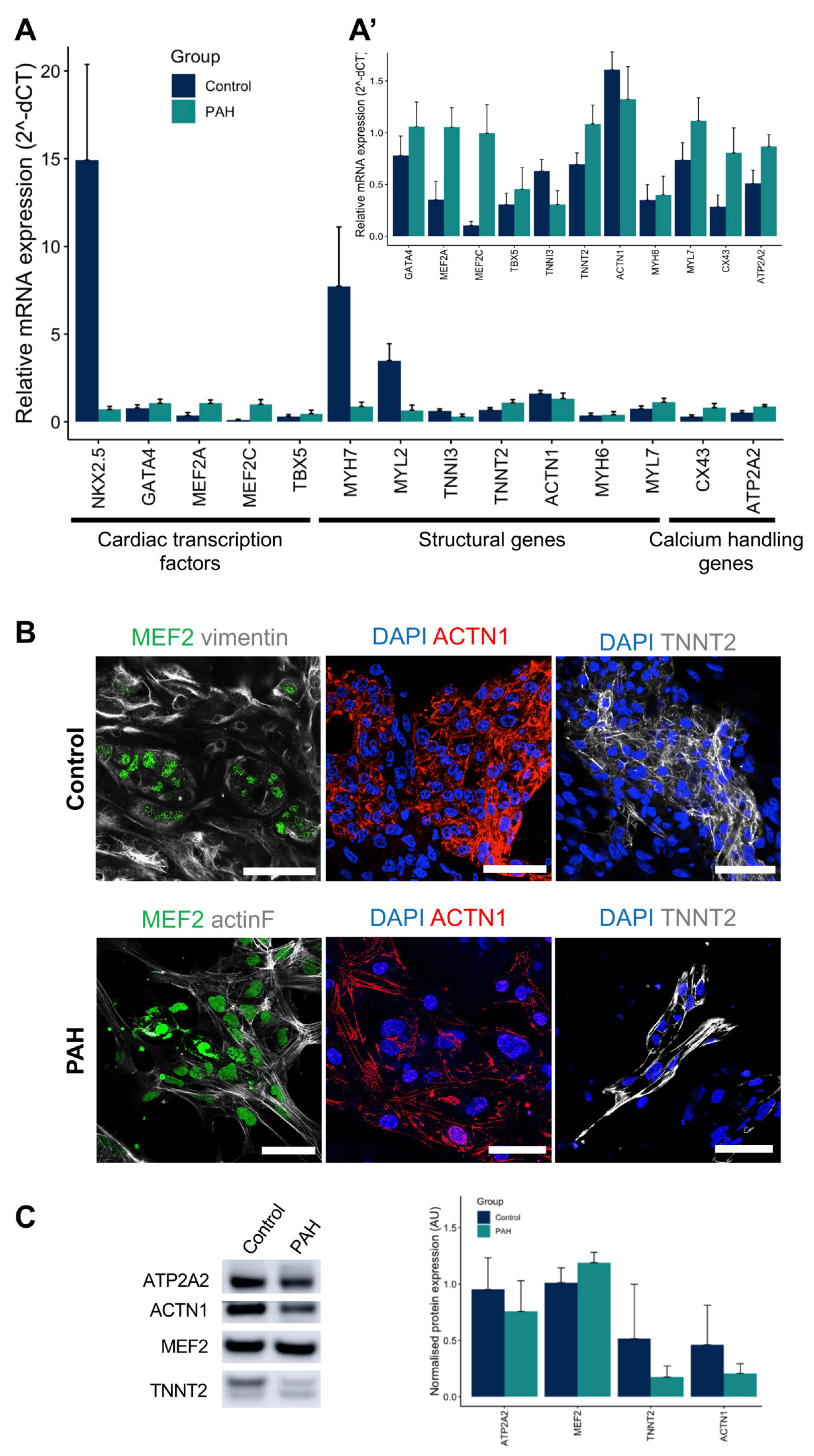
3.2. iPSCs Differentiated into Cardiomyocytes
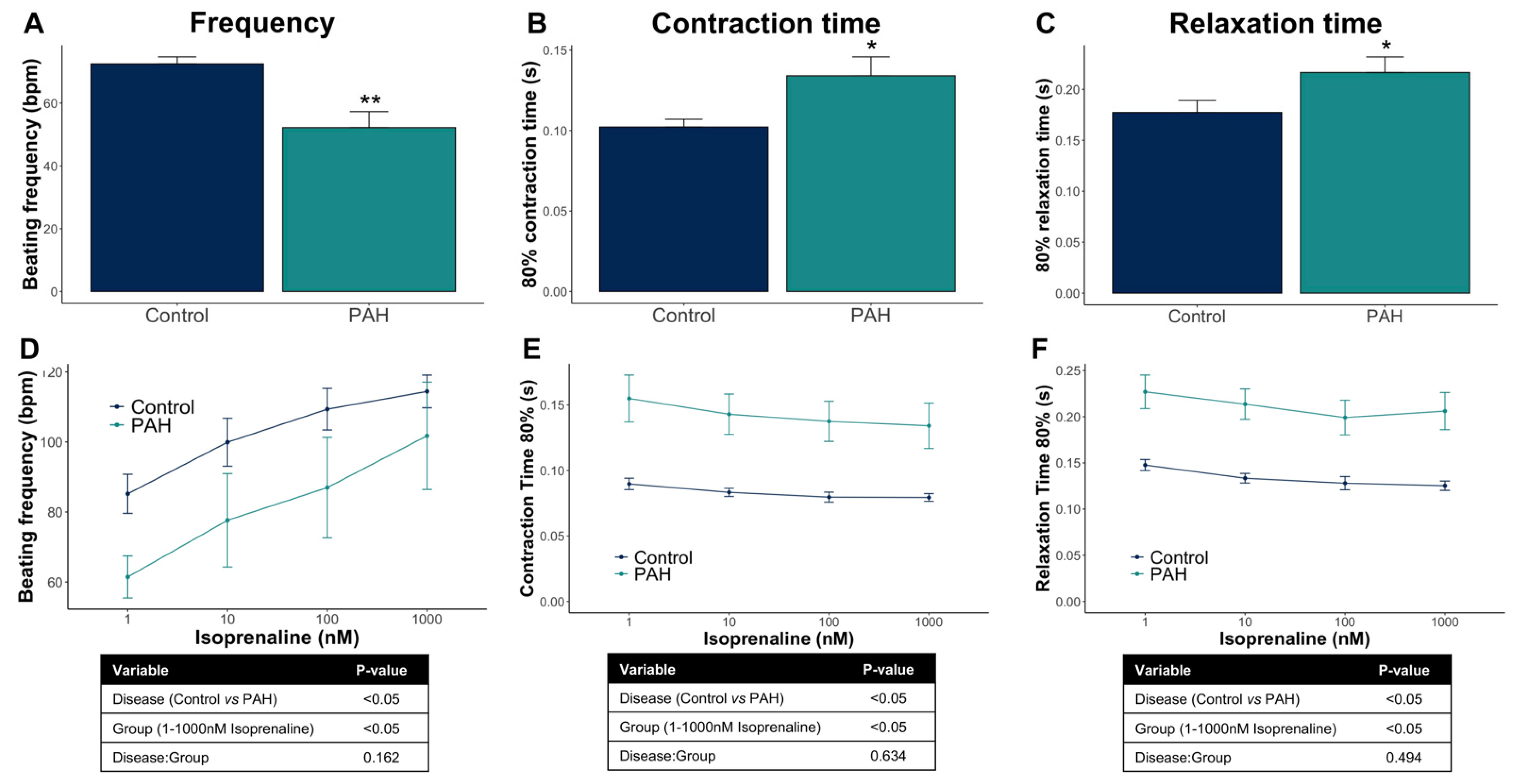
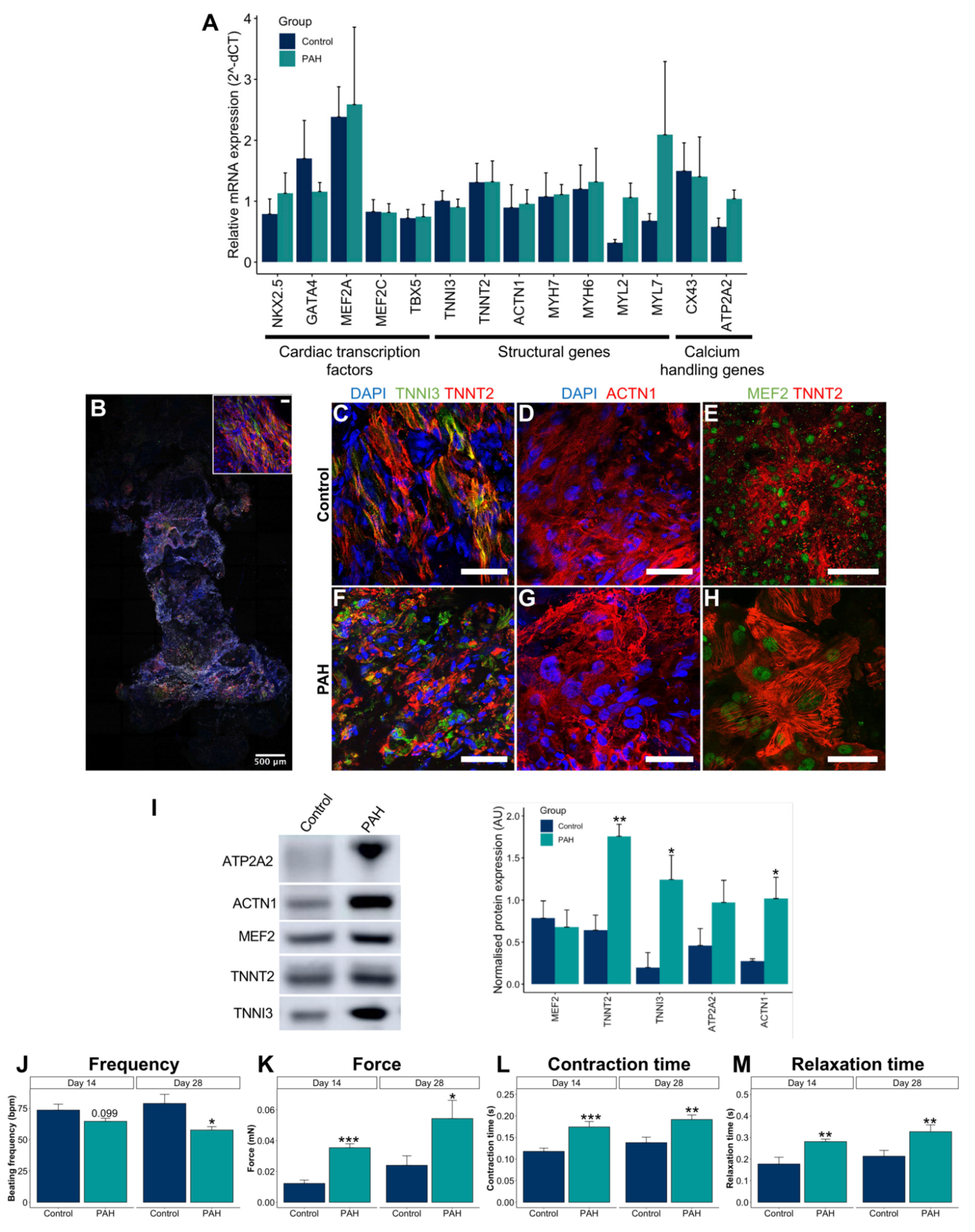
3.3. Control iPSC-CMs and PAH iPSC-CMs Reported Different Functional Properties
3.4. Optimization and Characterization of the 3D EHTs
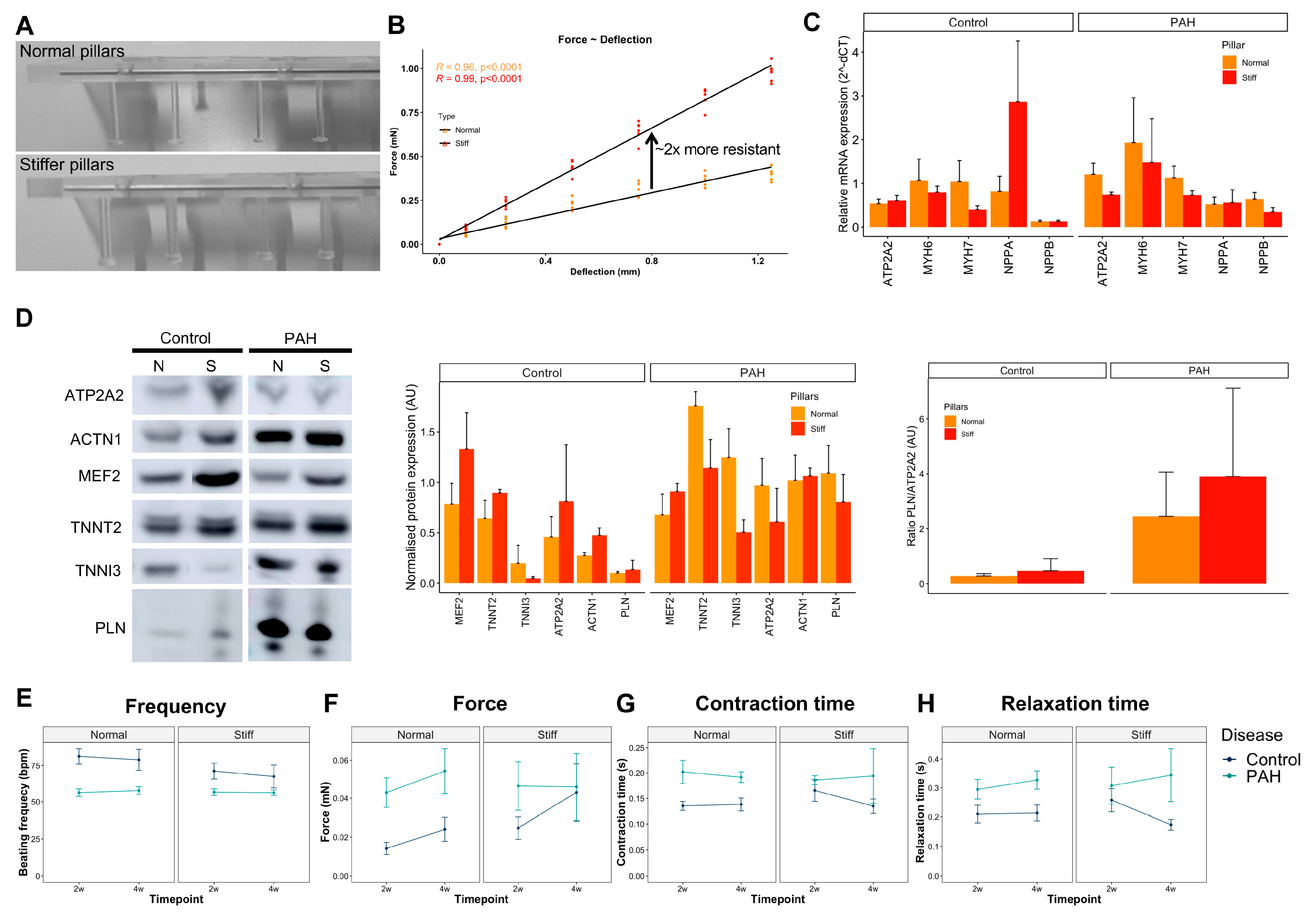
3.5. Increased Afterload on Control and PAH EHTs
4. Discussion
4.1. 2D Cardiac Models and Findings
4.2. 3D Cardiac Models and Findings
4.3. Disease Phenotype
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Vonk Noordegraaf, A.; Westerhof, B.E.; Westerhof, N. The Relationship Between the Right Ventricle and its Load in Pulmonary Hypertension. J. Am. Coll. Cardiol. 2017, 69, 236–243. [Google Scholar] [CrossRef] [PubMed]
- van der Bruggen, C.E.; Happé, C.M.; Dorfmüller, P.; Trip, P.; Spruijt, O.A.; Rol, N.; Hoevenaars, F.P.; Houweling, A.C.; Girerd, B.; Marcus, J.T.; et al. Bone Morphogenetic Protein Receptor Type 2 Mutation in Pulmonary Arterial Hypertension: A View on the Right Ventricle. Circulation 2016, 133, 1747–1760. [Google Scholar] [CrossRef] [Green Version]
- van Wezenbeek, J.; Groeneveldt, J.A.; Llucià-Valldeperas, A.; van der Bruggen, C.E.; Jansen, S.M.; Smits, A.J.; Smal, R.; van Leeuwen, J.W.; dos Remedios, C.; Keogh, A.; et al. Interplay of sex hormones and long-term right ventricular adaptation in a Dutch PAH-cohort. J. Heart Lung Transplant. 2021. [Google Scholar] [CrossRef]
- Usman, A.; Haase, A.; Merkert, S.; Gohring, G.; Hansmann, G.; Gall, H.; Schermuly, R.; Martin, U.; Olmer, R. Generation of pulmonary arterial hypertension patient-specific induced pluripotent stem cell lines from three unrelated patients with a heterozygous missense mutation in exon 12, a heterozygous in-frame deletion in exon 3 and a missense mutation in exon 11 of the BMPR2 gene. Stem Cell Res. 2021, 55, 102488. [Google Scholar] [CrossRef]
- Gu, M.; Shao, N.Y.; Sa, S.; Li, D.; Termglinchan, V.; Ameen, M.; Karakikes, I.; Sosa, G.; Grubert, F.; Lee, J.; et al. Patient-Specific iPSC-Derived Endothelial Cells Uncover Pathways that Protect against Pulmonary Hypertension in BMPR2 Mutation Carriers. Cell Stem Cell 2017, 20, 490–504. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, Y.; Pu, J. Differentiation and Application of Human Pluripotent Stem Cells Derived Cardiovascular Cells for Treatment of Heart Diseases: Promises and Challenges. Front. Cell Dev. Biol. 2021, 9, 658088. [Google Scholar] [CrossRef] [PubMed]
- Hansen, A.; Eder, A.; Bonstrup, M.; Flato, M.; Mewe, M.; Schaaf, S.; Aksehirlioglu, B.; Schwoerer, A.P.; Uebeler, J.; Eschenhagen, T. Development of a drug screening platform based on engineered heart tissue. Circ. Res. 2010, 107, 35–44. [Google Scholar] [CrossRef] [Green Version]
- Weinberger, F.; Mannhardt, I.; Eschenhagen, T. Engineering Cardiac Muscle Tissue: A Maturating Field of Research. Circ. Res. 2017, 120, 1487–1500. [Google Scholar] [CrossRef]
- Eder, A.; Vollert, I.; Hansen, A.; Eschenhagen, T. Human engineered heart tissue as a model system for drug testing. Adv. Drug Deliv. Rev. 2016, 96, 214–224. [Google Scholar] [CrossRef]
- Smits, J.; Tasev, D.; Andersen, S.; Szulcek, R.; Botros, L.; Ringgaard, S.; Andersen, A.; Vonk-Noordegraaf, A.; Koolwijk, P.; Bogaard, H.J. Blood Outgrowth and Proliferation of Endothelial Colony Forming Cells are Related to Markers of Disease Severity in Patients with Pulmonary Arterial Hypertension. Int. J. Mol. Sci. 2018, 19, 3763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Warlich, E.; Kuehle, J.; Cantz, T.; Brugman, M.H.; Maetzig, T.; Galla, M.; Filipczyk, A.A.; Halle, S.; Klump, H.; Schöler, H.R.; et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol. Ther. J. Am. Soc. Gene Ther. 2011, 19, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Ronaldson-Bouchard, K.; Ma, S.P.; Yeager, K.; Chen, T.; Song, L.; Sirabella, D.; Morikawa, K.; Teles, D.; Yazawa, M.; Vunjak-Novakovic, G. Advanced maturation of human cardiac tissue grown from pluripotent stem cells. Nature 2018, 556, 239–243. [Google Scholar] [CrossRef]
- Giacomelli, E.; Bellin, M.; Sala, L.; van Meer, B.J.; Tertoolen, L.G.; Orlova, V.V.; Mummery, C.L. Three-dimensional cardiac microtissues composed of cardiomyocytes and endothelial cells co-differentiated from human pluripotent stem cells. Development 2017, 144, 1008–1017. [Google Scholar] [CrossRef] [Green Version]
- Breckwoldt, K.; Letuffe-Breniere, D.; Mannhardt, I.; Schulze, T.; Ulmer, B.; Werner, T.; Benzin, A.; Klampe, B.; Reinsch, M.C.; Laufer, S.; et al. Differentiation of cardiomyocytes and generation of human engineered heart tissue. Nat. Protoc. 2017, 12, 1177–1197. [Google Scholar] [CrossRef] [PubMed]
- Hirt, M.N.; Sorensen, N.A.; Bartholdt, L.M.; Boeddinghaus, J.; Schaaf, S.; Eder, A.; Vollert, I.; Stohr, A.; Schulze, T.; Witten, A.; et al. Increased afterload induces pathological cardiac hypertrophy: A new in vitro model. Basic Res. Cardiol. 2012, 107, 307. [Google Scholar] [CrossRef] [Green Version]
- Vandenburgh, H.; Shansky, J.; Benesch-Lee, F.; Barbata, V.; Reid, J.; Thorrez, L.; Valentini, R.; Crawford, G. Drug-screening platform based on the contractility of tissue-engineered muscle. Muscle Nerve 2008, 37, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Voigt, N.; Pearman, C.M.; Dobrev, D.; Dibb, K.M. Methods for isolating atrial cells from large mammals and humans. J. Mol. Cell Cardiol. 2015, 86, 187–198. [Google Scholar] [CrossRef]
- Eschenhagen, T.; Carrier, L. Cardiomyopathy phenotypes in human-induced pluripotent stem cell-derived cardiomyocytes-a systematic review. Pflug. Arch. Eur. J. Physiol. 2019, 471, 755–768. [Google Scholar] [CrossRef] [Green Version]
- Moretti, A.; Laugwitz, K.L.; Dorn, T.; Sinnecker, D.; Mummery, C. Pluripotent stem cell models of human heart disease. Cold Spring Harb. Perspect. Med. 2013, 3, a014027. [Google Scholar] [CrossRef]
- Goumans, M.J.; Zwijsen, A.; Ten Dijke, P.; Bailly, S. Bone Morphogenetic Proteins in Vascular Homeostasis and Disease. Cold Spring Harb. Perspect. Biol. 2018, 10, a031989. [Google Scholar] [CrossRef] [PubMed]
- Owen, T.J.; Harding, S.E. Multi-cellularity in cardiac tissue engineering, how close are we to native heart tissue? J. Muscle Res. Cell Motil. 2019, 40, 151–157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sacchetto, C.; Vitiello, L.; de Windt, L.J.; Rampazzo, A.; Calore, M. Modeling Cardiovascular Diseases with hiPSC-Derived Cardiomyocytes in 2D and 3D Cultures. Int. J. Mol. Sci. 2020, 21, 3404. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, C.W.; Okawa, S.; Chuva de Sousa Lopes, S.M.; van Iperen, L.; Passier, R.; Braam, S.R.; Tertoolen, L.G.; del Sol, A.; Davis, R.P.; Mummery, C.L. Transcriptome of human foetal heart compared with cardiomyocytes from pluripotent stem cells. Development 2015, 142, 3231–3238. [Google Scholar] [CrossRef] [Green Version]
- Karbassi, E.; Fenix, A.; Marchiano, S.; Muraoka, N.; Nakamura, K.; Yang, X.; Murry, C.E. Cardiomyocyte maturation: Advances in knowledge and implications for regenerative medicine. Nat. Rev. Cardiol. 2020, 17, 341–359. [Google Scholar] [CrossRef]
- Rain, S.; Handoko, M.L.; Trip, P.; Gan, C.T.; Westerhof, N.; Stienen, G.J.; Paulus, W.J.; Ottenheijm, C.A.; Marcus, J.T.; Dorfmüller, P.; et al. Right ventricular diastolic impairment in patients with pulmonary arterial hypertension. Circulation 2013, 128, 2016–2025. [Google Scholar] [CrossRef] [Green Version]
- Rain, S.; Bos Dda, S.; Handoko, M.L.; Westerhof, N.; Stienen, G.; Ottenheijm, C.; Goebel, M.; Dorfmüller, P.; Guignabert, C.; Humbert, M.; et al. Protein changes contributing to right ventricular cardiomyocyte diastolic dysfunction in pulmonary arterial hypertension. J. Am. Heart Assoc. 2014, 3, e000716. [Google Scholar] [CrossRef] [Green Version]
- Marcus, J.T.; Gan, C.T.; Zwanenburg, J.J.; Boonstra, A.; Allaart, C.P.; Götte, M.J.; Vonk-Noordegraaf, A. Interventricular mechanical asynchrony in pulmonary arterial hypertension: Left-to-right delay in peak shortening is related to right ventricular overload and left ventricular underfilling. J. Am. Coll. Cardiol. 2008, 51, 750–757. [Google Scholar] [CrossRef] [Green Version]
- Mauritz, G.J.; Marcus, J.T.; Westerhof, N.; Postmus, P.E.; Vonk-Noordegraaf, A. Prolonged right ventricular post-systolic isovolumic period in pulmonary arterial hypertension is not a reflection of diastolic dysfunction. Heart 2011, 97, 473–478. [Google Scholar] [CrossRef]
- Godini, R.; Lafta, H.Y.; Fallahi, H. Epigenetic modifications in the embryonic and induced pluripotent stem cells. Gene Expr Patterns 2018, 29, 1–9. [Google Scholar] [CrossRef]
- Takeda, N.; Manabe, I.; Uchino, Y.; Eguchi, K.; Matsumoto, S.; Nishimura, S.; Shindo, T.; Sano, M.; Otsu, K.; Snider, P.; et al. Cardiac fibroblasts are essential for the adaptive response of the murine heart to pressure overload. J. Clin. Investig. 2010, 120, 254–265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujiu, K.; Shibata, M.; Nakayama, Y.; Ogata, F.; Matsumoto, S.; Noshita, K.; Iwami, S.; Nakae, S.; Komuro, I.; Nagai, R.; et al. A heart-brain-kidney network controls adaptation to cardiac stress through tissue macrophage activation. Nat. Med. 2017, 23, 611–622. [Google Scholar] [CrossRef] [PubMed]
- de Man, F.S.; Vonk-Noordegraaf, A. The magic of communication: The need to study organ and cell communication in pulmonary arterial hypertension induced right heart failure. Am. J. Physiol. Lung Cell Mol. Physiol. 2021, 321, L634–L636. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Llucià-Valldeperas, A.; Smal, R.; Bekedam, F.T.; Cé, M.; Pan, X.; Manz, X.D.; Wijnker, P.J.M.; Vonk-Noordegraaf, A.; Bogaard, H.J.; Goumans, M.-J.; et al. Development of a 3-Dimensional Model to Study Right Heart Dysfunction in Pulmonary Arterial Hypertension: First Observations. Cells 2021, 10, 3595. https://doi.org/10.3390/cells10123595
Llucià-Valldeperas A, Smal R, Bekedam FT, Cé M, Pan X, Manz XD, Wijnker PJM, Vonk-Noordegraaf A, Bogaard HJ, Goumans M-J, et al. Development of a 3-Dimensional Model to Study Right Heart Dysfunction in Pulmonary Arterial Hypertension: First Observations. Cells. 2021; 10(12):3595. https://doi.org/10.3390/cells10123595
Chicago/Turabian StyleLlucià-Valldeperas, Aida, Rowan Smal, Fjodor T. Bekedam, Margaux Cé, Xiaoke Pan, Xue D. Manz, Paul J. M. Wijnker, Anton Vonk-Noordegraaf, Harm J. Bogaard, Marie-Jose Goumans, and et al. 2021. "Development of a 3-Dimensional Model to Study Right Heart Dysfunction in Pulmonary Arterial Hypertension: First Observations" Cells 10, no. 12: 3595. https://doi.org/10.3390/cells10123595
APA StyleLlucià-Valldeperas, A., Smal, R., Bekedam, F. T., Cé, M., Pan, X., Manz, X. D., Wijnker, P. J. M., Vonk-Noordegraaf, A., Bogaard, H. J., Goumans, M.-J., & Man, F. S. d. (2021). Development of a 3-Dimensional Model to Study Right Heart Dysfunction in Pulmonary Arterial Hypertension: First Observations. Cells, 10(12), 3595. https://doi.org/10.3390/cells10123595