Unravelling the Impact of the Genetic Variant rs1042058 within the TPL2 Risk Gene Locus on Molecular and Clinical Disease Course Patients with Inflammatory Bowel Disease
Abstract
:1. Introduction
2. Materials and Methods
2.1. Patient Data and Cohort Design
2.2. Genetic Analyses and Clinical Annotations
2.3. Real-Time RT-PCR and Western Blot
2.4. Multiplex Assays of Cytokines in Human Serum
2.5. Statistical Analysis
3. Results
3.1. The Presence of the TPL2 Risk Allele Is Associated with Peripheral Arthritis in CD Patients
3.2. The Presence of the TPL2 Risk Allele Is Associated with Fewer Numbers of Intestinal Surgeries in UC Patients
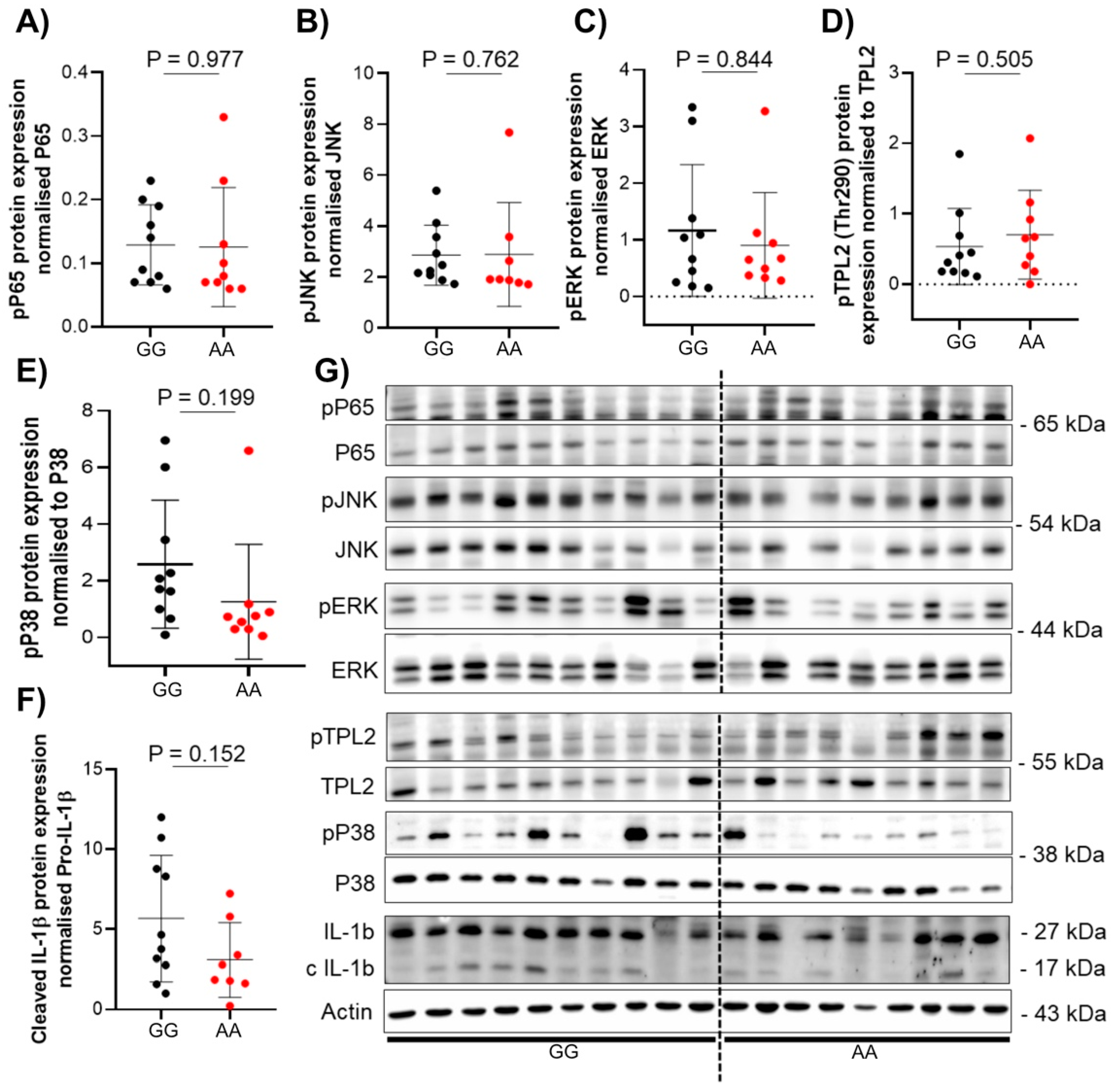
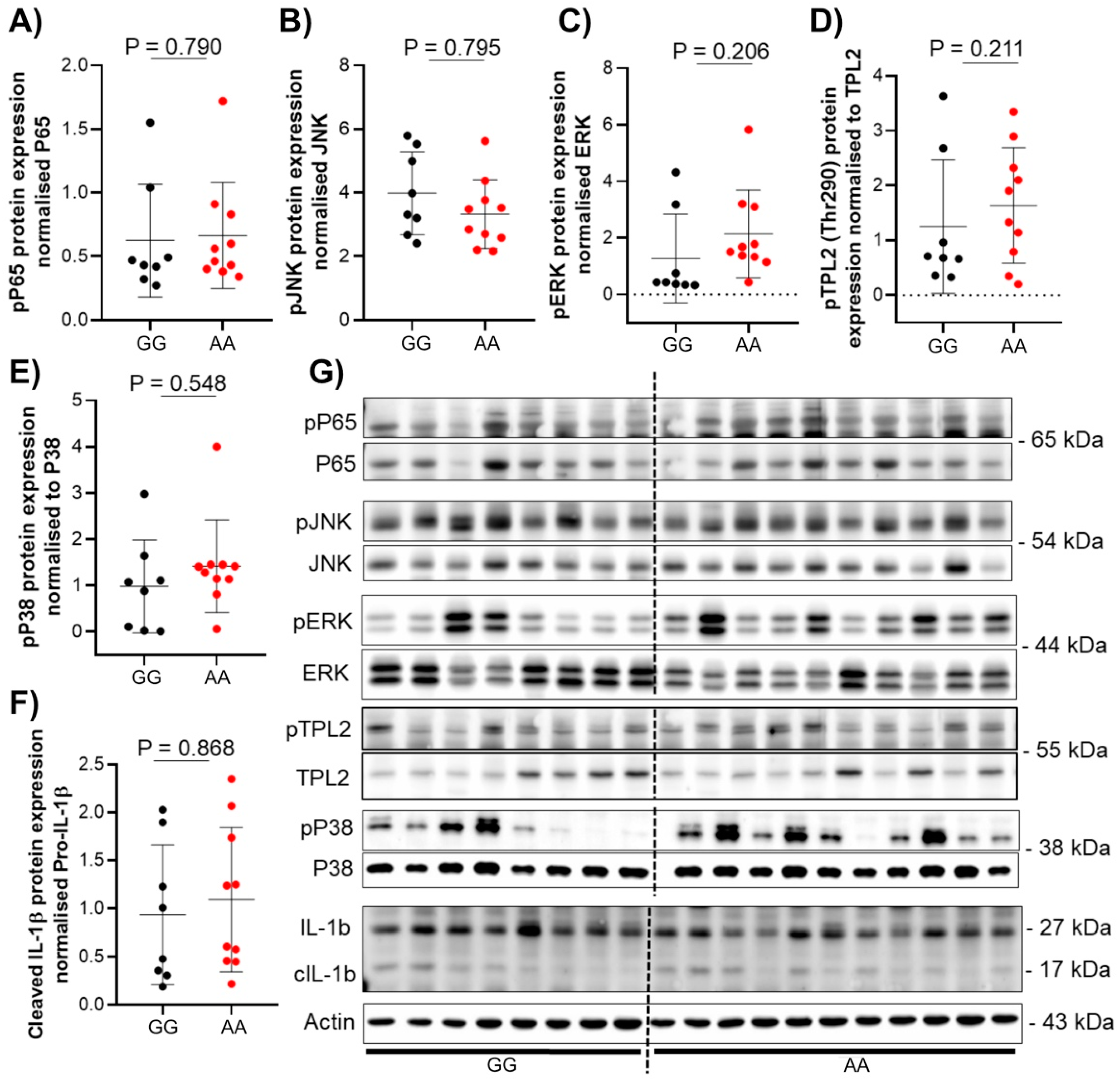
3.3. The Presence of TPL2 Genotypes Had a Low Impact on mRNA Levels of Some Cytokines in IBD Patients
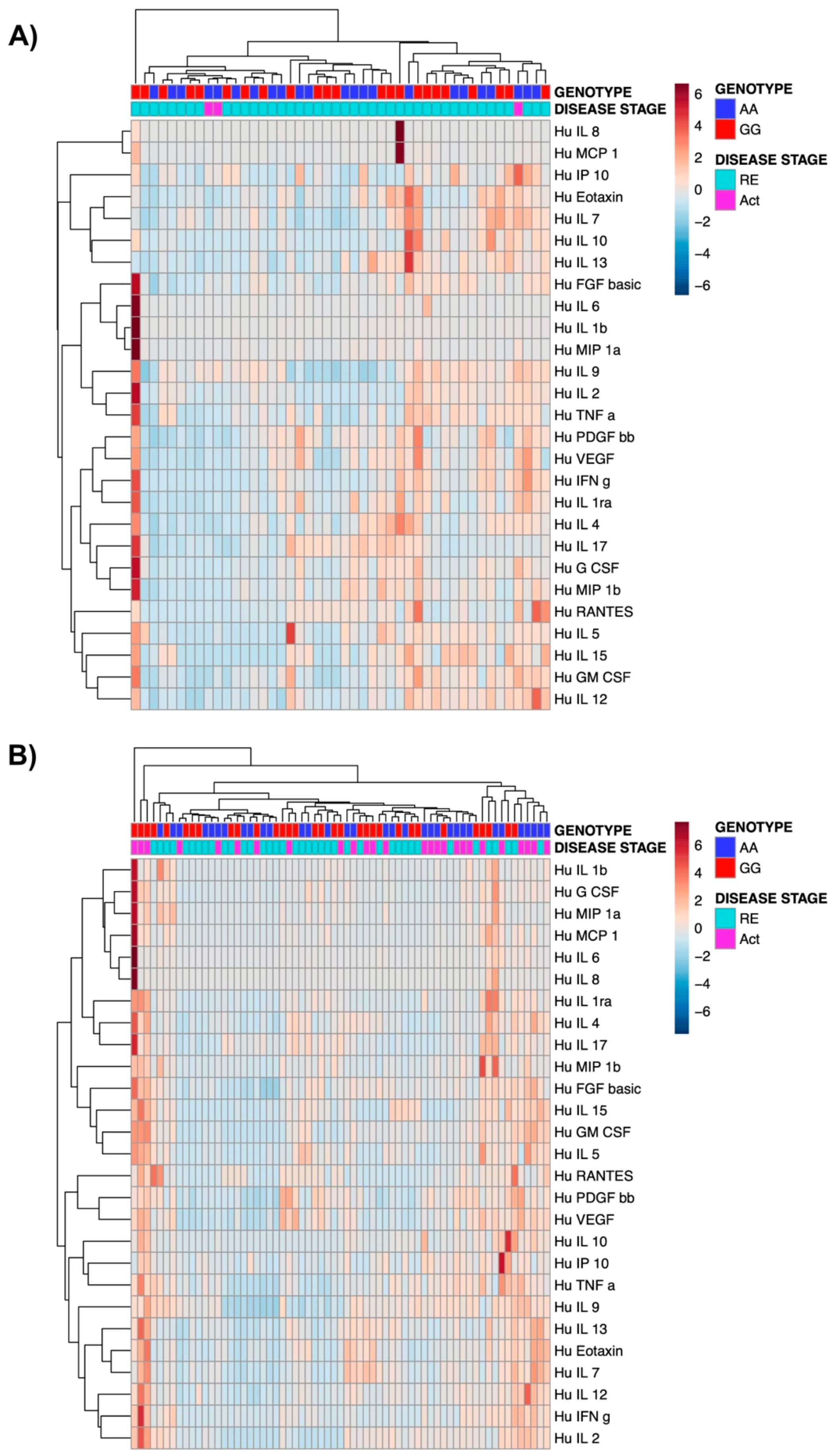
3.4. The Presence of TPL2 Risk Alleles Has No Impact on Serum Cytokine Levels in IBD Patients
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host–microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patriotis, C.; Makris, A.; Bear, S.E.; Tsichlis, P.N. Tumor progression locus 2 (Tpl-2) encodes a protein kinase involved in the progression of rodent T-cell lymphomas and in T-cell activation. Proc. Natl. Acad. Sci. USA 1993, 90, 2251–2255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmeron, A.; Ahmad, T.B.; Carlile, G.W.; Pappin, D.; Narsimhan, R.P.; Ley, S.C. Activation of mek-1 and sek-1 by tpl-2 proto-oncoprotein, a novel map kinase kinase kinase. EMBO J. 1996, 15, 817–826. [Google Scholar] [CrossRef] [PubMed]
- Dumitru, C.D.; Ceci, J.D.; Tsatsanis, C.; Kontoyiannis, D.; Stamatakis, K.; Lin, J.H.; Patriotis, C.; Jenkins, N.A.; Copeland, N.G.; Kollias, G.; et al. Tnf-alpha induction by lps is regulated posttranscriptionally via a tpl2/erk-dependent pathway. Cell 2000, 103, 1071–1083. [Google Scholar] [CrossRef] [Green Version]
- Watford, W.T.; Hissong, B.D.; Durant, L.R.; Yamane, H.; Muul, L.M.; Kanno, Y.; Tato, C.M.; Ramos, H.L.; Berger, A.E.; Mielke, L.; et al. Tpl2 kinase regulates t cell interferon-gamma production and host resistance to toxoplasma gondii. J. Exp. Med. 2008, 205, 2803–2812. [Google Scholar] [CrossRef] [PubMed]
- Senger, K.; Pham, V.C.; Varfolomeev, E.; Hackney, J.A.; Corzo, C.A.; Collier, J.; Lau, V.W.C.; Huang, Z.; Hamidzhadeh, K.; Caplazi, P.; et al. The kinase TPL2 activates ERK and p38 signaling to promote neutrophilic inflammation. Sci. Signal. 2017, 10, eaah4273. [Google Scholar] [CrossRef] [PubMed]
- Kontoyiannis, D.; Boulougouris, G.; Manoloukos, M.; Armaka, M.; Apostolaki, M.; Pizarro, T.; Kotlyarov, A.; Forster, I.; Flavell, R.; Gaestel, M.; et al. Genetic dissection of the cellular pathways and signaling mechanisms in modeled tumor necrosis factor-induced crohn’s-like inflammatory bowel disease. J. Exp. Med. 2002, 196, 1563–1574. [Google Scholar] [CrossRef] [PubMed]
- Hedl, M.; Abraham, C. ATPL2 (MAP3K8) disease-risk polymorphism increases TPL2 expression thereby leading to increased pattern recognition receptor-initiated caspase-1 and caspase-8 activation, signalling and cytokine secretion. Gut 2016, 65, 1799–1811. [Google Scholar] [CrossRef] [Green Version]
- Lawrenz, M.; Visekruna, A.; Kühl, A.; Schmidt, N.; Kaufmann, S.H.; Steinhoff, U. Genetic and pharmacological targeting of tpl-2 kinase ameliorates experimental colitis: A potential target for the treatment of crohn’s disease? Mucosal Immunol. 2012, 5, 129–139. [Google Scholar] [CrossRef]
- Pittet, V.; Juillerat, P.; Mottet, C.; Felley, C.; Ballabeni, P.; Burnand, B.; Michetti, P.; Vader, J.-P.; the Swiss IBD Cohort Study Group. Cohort Profile: The Swiss Inflammatory Bowel Disease Cohort Study (SIBDCS). Int. J. Epidemiol. 2009, 38, 922–931. [Google Scholar] [CrossRef] [Green Version]
- Pittet, V.; Michetti, P.; Mueller, C.; Braegger, C.P.; von Känel, R.; Schoepfer, A.; Macpherson, A.J.; Rogler, G.; Anderegg, C.; Bauerfeind, P.; et al. Cohort Profile Update: The Swiss Inflammatory Bowel Disease Cohort Study (SIBDCS). Int. J. Epidemiol. 2019, 48, 385–386. [Google Scholar] [CrossRef]
- Storm, N.; Darnhofer-Patel, B.; van den Boom, D.; Rodi, C.P. MALDI-TOF mass spectrometry-based SNP genotyping. Methods Mol. Biol. 2003, 212, 241–262. [Google Scholar] [PubMed]
- Spalinger, M.R.; Manzini, R.; Hering, L.; Riggs, J.B.; Gottier, C.; Lang, S.; Atrott, K.; Fettelschoss-Gabriel, A.; Olomski, F.; Kündig, T.M.; et al. PTPN2 Regulates Inflammasome Activation and Controls Onset of Intestinal Inflammation and Colon Cancer. Cell Rep. 2018, 22, 1835–1848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteleone, G.; Fina, D.; Caruso, R.; Pallone, F. New mediators of immunity and inflammation in inflammatory bowel disease. Curr. Opin. Gastroenterol. 2006, 22, 361–364. [Google Scholar] [CrossRef] [PubMed]
- Kaser, A.; Zeissig, S.; Blumberg, R.S. Inflammatory bowel disease. Annu. Rev. Immunol. 2010, 28, 573–621. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.Z.; Li, Y.Y. Inflammatory bowel disease: Pathogenesis. World J. Gastroenterol. 2014, 20, 91–99. [Google Scholar] [CrossRef]
- Guan, Q. A Comprehensive Review and Update on the Pathogenesis of Inflammatory Bowel Disease. J. Immunol. Res. 2019, 2019, 7247238. [Google Scholar] [CrossRef] [Green Version]
- Szigethy, E.; McLafferty, L.; Goyal, A. Inflammatory bowel disease. Child Adolesc. Psychiatr. Clin. N. Am. 2010, 19, 301–318, ix. [Google Scholar] [CrossRef] [PubMed]
- Sairenji, T.; Collins, K.L.; Evans, D.V. An Update on Inflammatory Bowel Disease. Prim. Care 2017, 44, 673–692. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.-C.; Stappenbeck, T.S. Genetics and Pathogenesis of Inflammatory Bowel Disease. Annu. Rev. Pathol. Mech. Dis. 2016, 11, 127–148. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Genotype (CD) | AA | AG | GG |
|---|---|---|---|
| Number (%) | 210 (17.5) | 562 (47) | 425 (35.5) |
| CD patients | AA | AG/GG | p-value (Wilcoxon) |
| Age at Diagnosis | |||
| Median, q25–q75, | 23, 17–31, | 23.5, 17–34, | 0.657 |
| min–max | 5–73 | 1–81 | |
| Genotype: | AA | AG/GG | p-value (chi2) |
| Gender | |||
| Men | 110 (52.4) | 491 (49.7) | 0.488 |
| Women | 100 (47.6) | 496 (50.3) | |
| Peripheral arthritis | |||
| Yes | 21 (14.8) | 232 (23.5) | 0.005 |
| No | 179 (85.2) | 755 (76.5) | |
| Uveitis/Iritis | |||
| Yes | 1 (0.5) | 10 (1) | 0.459 |
| No | 209 (99.5) | 977 (99) | |
| Pyoderma gangrenosum | |||
| Yes | 1 (0.5) | 5 (0.5) | 0.955 |
| No | 209 (99.5) | 982 (99.5) | |
| Erythema nodosum | |||
| Yes | 0 | 8 (0.8) | 0.191 |
| No | 210 (100) | 979 (99.2) | |
| Aphtous oral ulcers | |||
| Yes | 6 (2.9) | 27 (2.7) | 0.922 |
| No | 204 (97.1) | 960 (97.3) | |
| Ankylosing spondylitis | |||
| Yes | 6 (2.9) | 14 (1.4) | 0.140 |
| No | 204 (97.1) | 973 (98.6) | |
| Prim. scler. cholangitis | |||
| Yes | 0 | 5 (0.5) | 0.301 |
| No | 210 (100) | 982 (99.5) | |
| Other | |||
| Yes | 2 (0.9) | 10 (1) | 0.936 |
| No | 208 (99.1) | 977 (99) | |
| Summary of Extraintestinal manifestations | |||
| Yes | 40 (19) | 258 (26.1) | 0.031 |
| No | 170 (81) | 729 (73.9) | |
| Genotype (UC): | AA | AG | GG |
| Number (%) | 173 (18.3) | 449 (47.4) | 326 (34.3) |
| UC patients | AA | AG or GG | p-value (Wilcoxon) |
| Age at Diagnosis | |||
| Median, q25–q75, | 26, 19–37, | 27, 17–36, | 0.404 |
| min–max | 3–75 | 2–79 | |
| Number (%) | AA | AG or GG | p-value (chi2) |
| Gender | |||
| Men | 81 (46.8) | 426 (55) | 0.052 |
| Women | 92 (53.2) | 349 (45) | |
| Surgery | |||
| Yes | 7 (4) | 11 (1.4) | 0.022 |
| No | 166 (96) | 764 (98.6) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morsy, Y.; Brillant, N.; Franc, Y.; Scharl, M.; Wawrzyniak, M.; on behalf of the Swiss IBD Cohort Study Group. Unravelling the Impact of the Genetic Variant rs1042058 within the TPL2 Risk Gene Locus on Molecular and Clinical Disease Course Patients with Inflammatory Bowel Disease. Cells 2021, 10, 3589. https://doi.org/10.3390/cells10123589
Morsy Y, Brillant N, Franc Y, Scharl M, Wawrzyniak M, on behalf of the Swiss IBD Cohort Study Group. Unravelling the Impact of the Genetic Variant rs1042058 within the TPL2 Risk Gene Locus on Molecular and Clinical Disease Course Patients with Inflammatory Bowel Disease. Cells. 2021; 10(12):3589. https://doi.org/10.3390/cells10123589
Chicago/Turabian StyleMorsy, Yasser, Nathalie Brillant, Yannick Franc, Michael Scharl, Marcin Wawrzyniak, and on behalf of the Swiss IBD Cohort Study Group. 2021. "Unravelling the Impact of the Genetic Variant rs1042058 within the TPL2 Risk Gene Locus on Molecular and Clinical Disease Course Patients with Inflammatory Bowel Disease" Cells 10, no. 12: 3589. https://doi.org/10.3390/cells10123589
APA StyleMorsy, Y., Brillant, N., Franc, Y., Scharl, M., Wawrzyniak, M., & on behalf of the Swiss IBD Cohort Study Group. (2021). Unravelling the Impact of the Genetic Variant rs1042058 within the TPL2 Risk Gene Locus on Molecular and Clinical Disease Course Patients with Inflammatory Bowel Disease. Cells, 10(12), 3589. https://doi.org/10.3390/cells10123589






