Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi
Abstract
1. Introduction
2. Materials and Methods
2.1. Drosophila Strains and Crosses
2.2. RNA Extraction, Library Preparation, and Sequencing and TE Library Construction
2.3. Small RNA Extraction, Library Preparation, and Sequencing
2.4. TE and Gene Read Mapping and Differential Expression Analysis
2.5. Genome Assembling and Analysis of Evolutionary Rates
2.6. Small RNAs and Ping-Pong Analyses
3. Results
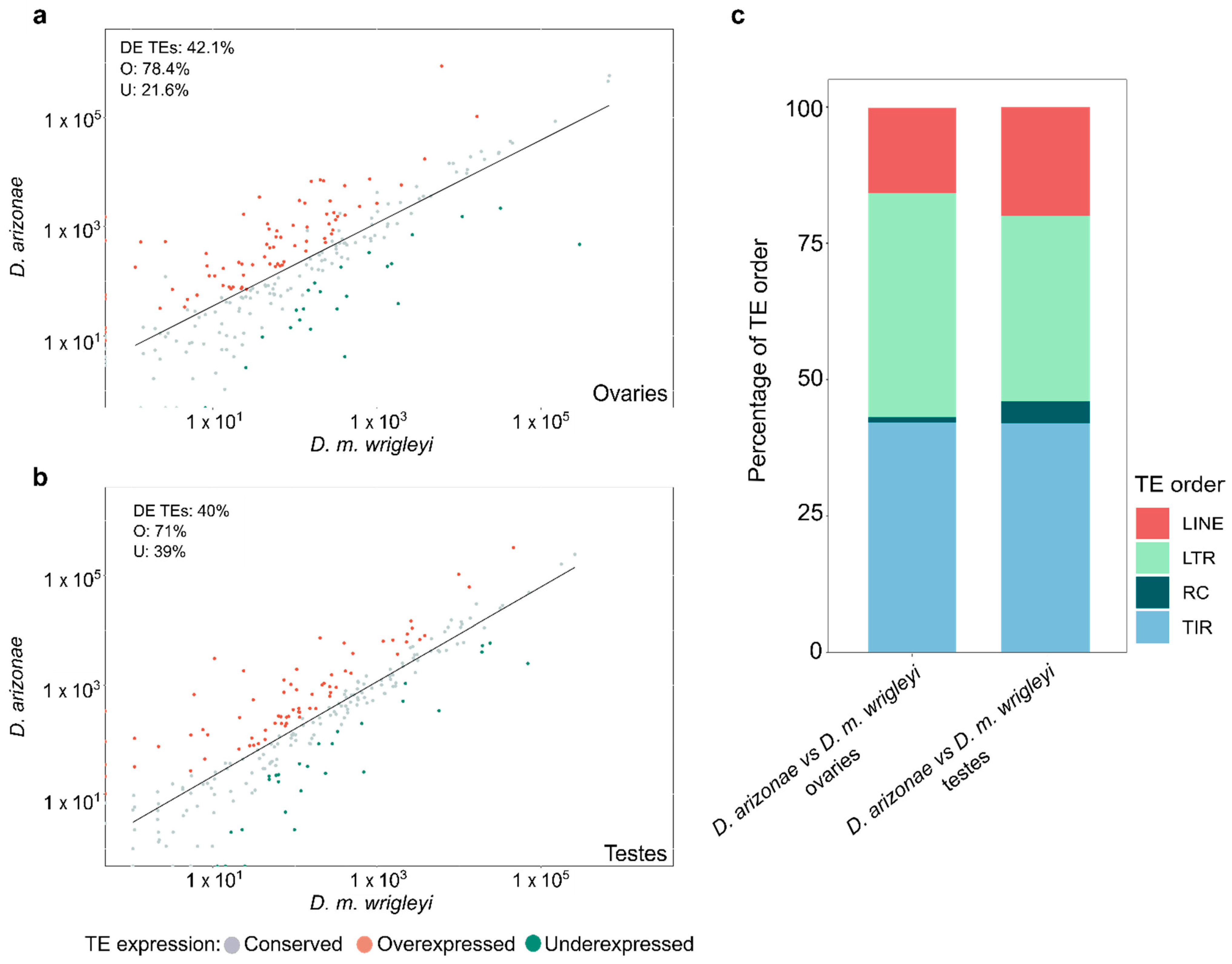
3.1. Differential Expression of TEs in the Comparison D. arizonae vs. D. mojavensis
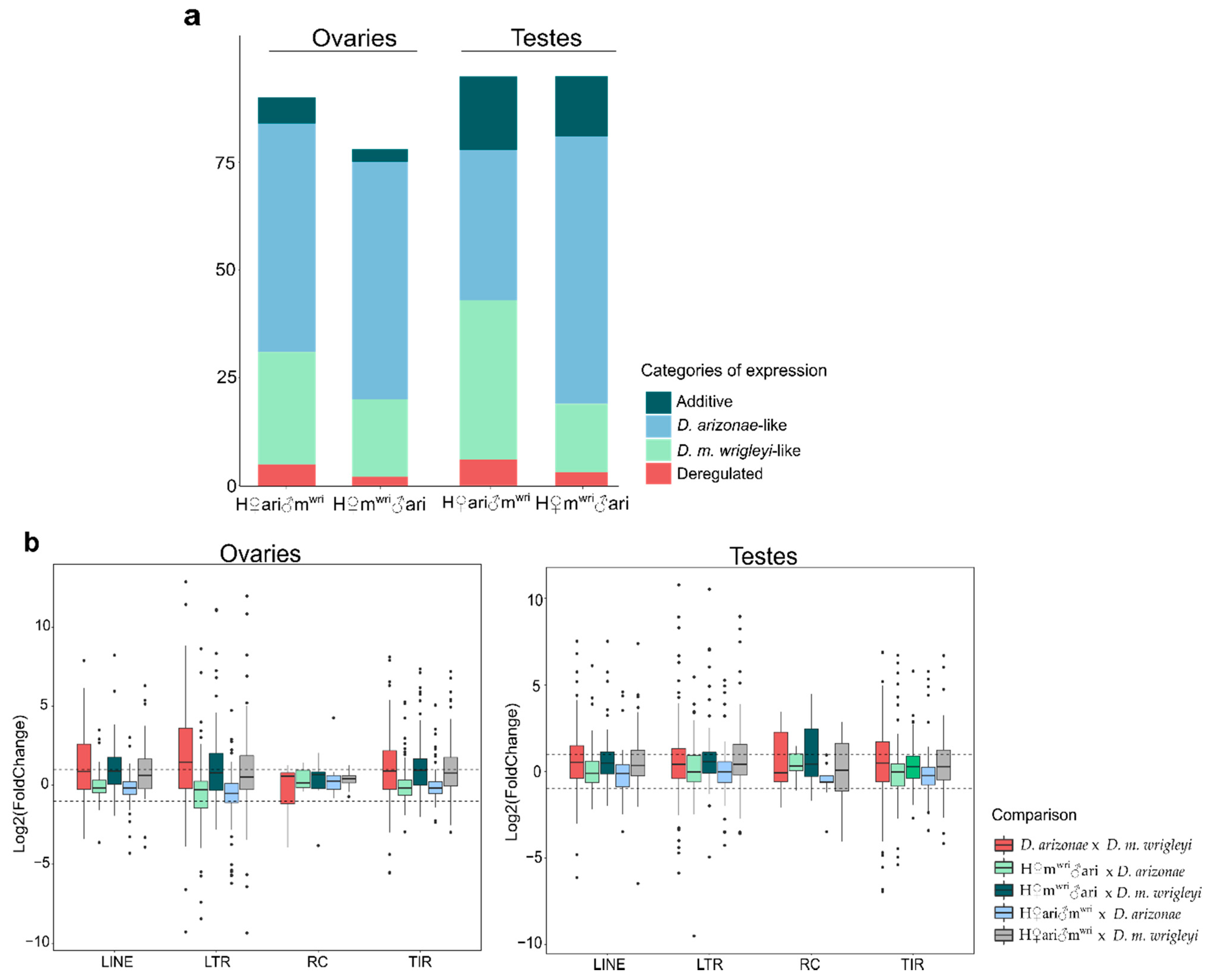
3.2. TE Expression in the Hybrid Germlines Compared with the Parental Lines
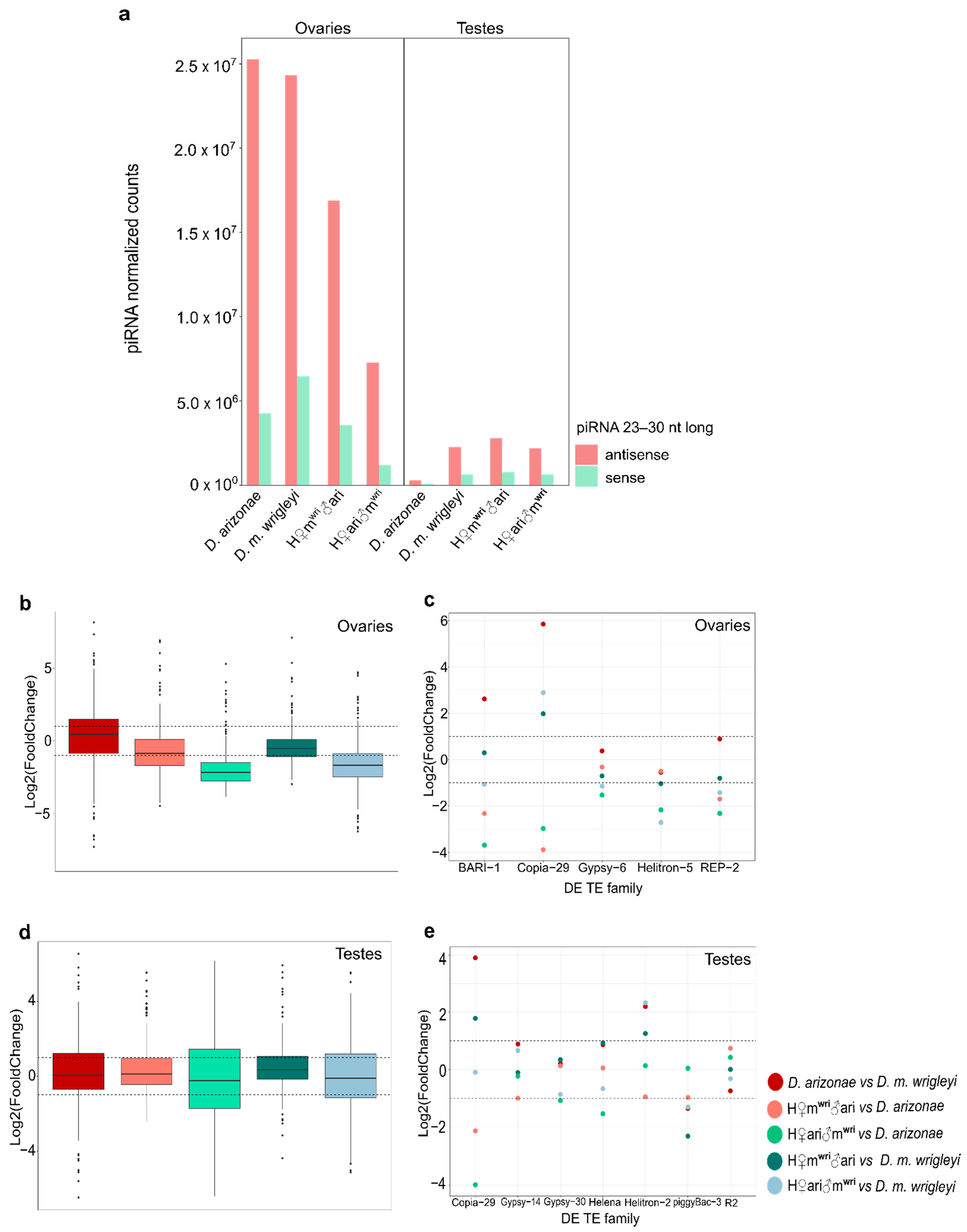
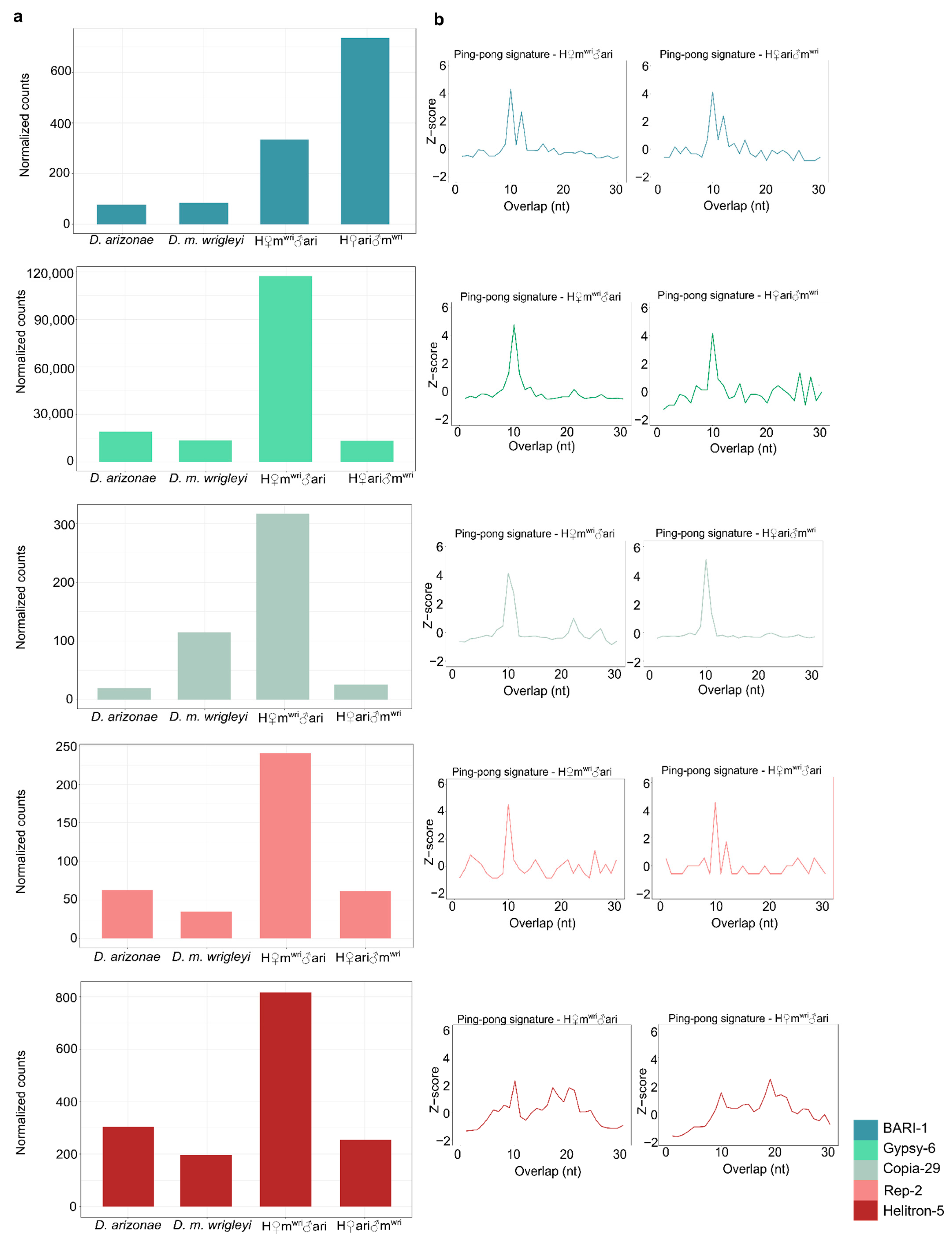
3.3. piRNA Repertoire in Female and Male Hybrid Germlines
3.4. Divergence of Expression and Selective Process Acting on piRNA Pathway Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kidwell, M.G.; Lisch, D.R. Perspective: Transposable elements, parasitic DNA, and genome evolution. Evolution 2001, 55, 1–24. [Google Scholar] [CrossRef]
- Van de Lagemaat, L.N.; Landry, J.R.; Mager, D.L.; Medstrand, P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003, 19, 530–536. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.H.; Choo, K.H. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004, 20, 611–616. [Google Scholar] [CrossRef]
- Capy, P.; Gasperi, G.; Biemont, C.; Bazin, C. Stress and transposable elements: Co-evolution or useful parasites? Heredity 2000, 85, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Fontdevila, A. Hybrid genome evolution by transposition. Cytogenet. Genome 2005, 110, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Garcia Guerreiro, M.P. What makes transposable elements move in the Drosophila genome? Heredity 2012, 108, 461–468. [Google Scholar] [CrossRef]
- Labrador, M.; Farre, M.; Utzet, F.; Fontdevila, A. Interspecific hybridization increases transposition rates of Osvaldo. Mol. Biol. Evol. 1999, 16, 931–937. [Google Scholar] [CrossRef]
- O’Neill, R.J.; O’Neill, M.J.; Graves, J.A. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 1998, 393, 68–72. [Google Scholar] [CrossRef]
- Kelleher, E.S.; Edelman, N.B.; Barbash, D.A. Drosophila interspecific hybrids phenocopy piRNA-pathway mutants. PLoS Biol. 2012, 10, e1001428. [Google Scholar] [CrossRef]
- Serrato-Capuchina, A.; Matute, D.R. The Role of Transposable Elements in Speciation. Genes 2018, 9, 254. [Google Scholar] [CrossRef]
- Picard, G. Non-mendelian female sterility in Drosophila melanogaster: Hereditary transmission of I factor. Genetics 1976, 83, 107–123. [Google Scholar] [CrossRef]
- Blackman, R.K.; Grimaila, R.; Koehler, M.M.; Gelbart, W.M. Mobilization of hobo elements residing within the decapentaplegic gene complex: Suggestion of a new hybrid dysgenesis system in Drosophila melanogaster. Cell 1987, 49, 497–505. [Google Scholar] [CrossRef]
- Yannopoulos, G.; Stamatis, N.; Monastirioti, M.; Hatzopoulos, P.; Louis, C. Hobo is responsible for the induction of hybrid dysgenesis by strains of Drosophila melanogaster bearing the male recombination factor 23.5MRF. Cell 1987, 49, 487–495. [Google Scholar] [CrossRef]
- Hill, T.; Schlotterer, C.; Betancourt, A.J. Hybrid Dysgenesis in Drosophila simulans Associated with a Rapid Invasion of the P-Element. PLoS Genet. 2016, 12, e1005920. [Google Scholar]
- Kidwell, M.G.; Kidwell, J.F.; Sved, J.A. Hybrid Dysgenesis in Drosophila melanogaster: A Syndrome of Aberrant Traits Including Mutation, Sterility and Male Recombination. Genetics 1977, 86, 813–833. [Google Scholar] [CrossRef]
- Petrov, D.A.; Schutzman, J.L.; Hartl, D.L.; Lozovskaya, E.R. Diverse transposable elements are mobilized in hybrid dysgenesis in Drosophila virilis. Proc. Natl. Acad. Sci. USA 1995, 92, 8050–8054. [Google Scholar] [CrossRef]
- Castillo, D.M.; Moyle, L.C. Transposable elements that cause dysgenesis also contribute to postzygotic isolation in the Drosophila virilis clade. bioRxiv 2019. [Google Scholar] [CrossRef]
- Satyaki, P.R.; Cuykendall, T.N.; Wei, K.H.; Brideau, N.J.; Kwak, H.; Aruna, S.; Ferree, P.M.; Ji, S.; Barbash, D.A. The Hmr and Lhr hybrid incompatibility genes suppress a broad range of heterochromatic repeats. PLoS Genet. 2014, 10, e1004240. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Guerreiro, M.P.G. Interspecific hybridization as a genomic stressor inducing mobilization of transposable elements in Drosophila. Mob. Genet. Elem. 2014, 4, e34394. [Google Scholar] [CrossRef]
- Romero-Soriano, V.; Modolo, L.; Lopez-Maestre, H.; Mugat, B.; Pessia, E.; Chambeyron, S.; Vieira, C.; Guerreiro, M.P.G. Transposable element misregulation is linked to the divergence between parental piRNA pathways in Drosophila hybrids. GBE 2017, 9, 1450–1470. [Google Scholar] [CrossRef]
- Vela, D.; Fontdevila, A.; Vieira, C.; Garcia Guerreiro, M.P. A genome-wide survey of genetic instability by transposition in Drosophila hybrids. PLoS ONE 2014, 9, e88992. [Google Scholar]
- Klattenhoff, C.; Xi, H.; Li, C.; Lee, S.; Xu, J.; Khurana, J.S.; Zhang, F.; Schultz, N.; Koppetsch, B.S.; Nowosielska, A.; et al. The Drosophila HP1 homolog Rhino is required for transposon silencing and piRNA production by dual-strand clusters. Cell 2009, 138, 1137–1149. [Google Scholar] [CrossRef]
- Luo, S.; Lu, J. Silencing of Transposable Elements by piRNAs in Drosophila: An Evolutionary Perspective. Genomics Proteomics Bioinformatics 2017, 15, 164–176. [Google Scholar] [CrossRef] [PubMed]
- Rozhkov, N.V.; Hammell, M.; Hannon, G.J. Multiple roles for Piwi in silencing Drosophila transposons. Genes Dev. 2013, 27, 400–412. [Google Scholar] [CrossRef] [PubMed]
- Senti, K.A.; Brennecke, J. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet. 2010, 26, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103. [Google Scholar] [CrossRef]
- Dobzhansky, T. Studies on Hybrid Sterility. II. Localization of Sterility Factors in Drosophila pseudoobscura Hybrids. Genetics 1936, 21, 113–135. [Google Scholar] [CrossRef] [PubMed]
- Muller, H.J. Isolating mechanisms evolution and temperature. Biol. Symp. 1942, 6, 71–125. [Google Scholar]
- Sanchez-Flores, A.; Penaloza, F.; Carpinteyro-Ponce, J.; Nazario-Yepiz, N.; Abreu-Goodger, C.; Machado, C.A.; Markow, T.A. Genome evolution in three species of cactophilic Drosophila. G3 2016, 6, 3097–3105. [Google Scholar] [CrossRef]
- Banho, C.A.; Mérel, V.; Oliveira, T.Y.K.; Carareto, C.M.A.; Vieira, C. Comparative transcriptomics between Drosophila mojavensis and D. arizonae reveals transgressive gene expression and underexpression of spermatogenesis-related genes in hybrid testes. Sci. Rep. 2021, 10, 9844. [Google Scholar] [CrossRef]
- Jennings, J.H.; Etges, W.J. Species hybrids in the laboratory but not in nature: A reanalysis of premating isolation between Drosophila arizonae and D. mojavensis. Evolution 2010, 64, 587–598. [Google Scholar] [CrossRef]
- Reed, L.K.; Markow, T.A. Early events in speciation: Polymorphism for hybrid male sterility in Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 9009–9012. [Google Scholar] [CrossRef]
- Reed, L.K.; LaFlamme, B.A.; Markow, T.A. Genetic architecture of hybrid male sterility in Drosophila: Analysis of intraspecies variation for interspecies isolation. PLoS ONE 2008, 3, e3076. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Maestre, H.; Carnelossi, E.A.; Lacroix, V.; Burlet, N.; Mugat, B.; Chambeyron, S.; Carareto, C.M.; Vieira, C. Identification of misexpressed genetic elements in hybrids between Drosophila-related species. Sci. Rep. 2017, 7, 40618. [Google Scholar] [CrossRef] [PubMed]
- Carnelossi, E.A.; Lerat, E.; Henri, H.; Martinez, S.; Carareto, C.M.; Vieira, C. Specific activation of an I-like element in Drosophila interspecific hybrids. Genome Biol. Evol. 2014, 6, 1806–1817. [Google Scholar] [CrossRef]
- Baffi, M.A.; Ceron, C.R. Molecular analysis of the rDNA ITS-1 intergenic spacer in Drosophila mulleri, D. arizonae, and their hybrids. Biochem. Genet. 2002, 40, 411–421. [Google Scholar] [CrossRef]
- Lerat, E.; Fablet, M.; Modolo, L.; Lopez-Maestre, H.; Vieira, C. TEtools facilitates big data expression analysis of transposable elements and reveals an antagonism between their activity and that of piRNA genes. Nucleic Acids Res. 2017, 45, e17. [Google Scholar] [CrossRef] [PubMed]
- Smit, F.A.; Hubley, R.; Green, P. Repeat-Masker Open-3.0. Available online: http://www. repeatmasker.org (accessed on 15 October 2021).
- Bailly-Bechet, M.; Haudry, A.; Lerat, E. “One code to find them all”: A perl tool to conveniently parse RepeatMasker output files. Mob. DNA 2014, 5, 1–15. [Google Scholar] [CrossRef]
- Wacholder, A.C.; Cox, C.; Meyer, T.J.; Ruggiero, R.P.; Vemulapalli, V.; Damert, A.; Carbone, L.; Pollock, D.D. Inference of transposable element ancestry. PLoS Genet. 2014, 10, e1004482. [Google Scholar] [CrossRef]
- Grentzinger, T.; Armenise, C.; Pelisson, A.; Brun, C.; Mugat, B.; Chambeyron, S. A user-friendly chromatographic method to purify small regulatory RNAs. Methods 2014, 67, 91–101. [Google Scholar] [CrossRef]
- Modolo, L.; Lerat, E. UrQt: An efficient software for the Unsupervised Quality trimming of NGS data. BMC Bioinform. 2015, 16, 137. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- RC Team. R: A Language and Environment for Statistical Computing; R Fondation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Peng, Y.; Leung, H.C.; Yiu, S.M.; Chin, F.Y. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012, 28, 1420–1428. [Google Scholar] [CrossRef]
- Boetzer, M.; Henkel, C.V.; Jansen, H.J.; Butler, D.; Pirovano, W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 2011, 27, 578–579. [Google Scholar]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Wong, W.S.; Nielsen, R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol. Biol. Evol. 2005, 22, 1107–1118. [Google Scholar] [CrossRef] [PubMed]
- Fablet, M.; Jacquet, A.; Rebollo, R.; Haudry, A.; Rey, C.; Salces-Ortiz, J.; Bajad, P.; Burlet, N.; Jantsch, M.F.; Guerreiro, M.P.G.; et al. Dynamic Interactions Between the Genome and an Endogenous Retrovirus: Tirant in Drosophila simulans Wild-Type Strains. G3 2019, 9, 855–865. [Google Scholar] [CrossRef]
- Kechin, A.; Boyarskikh, U.; Kel, A.; Filipenko, M. CutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput. Biol. 2017, 24, 1138–1143. [Google Scholar] [CrossRef]
- Schmieder, R.; Edwards, R. Quality control and preprocessing of metagenomic datasets. Bioinformatics 2011, 27, 863–864. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10, R25. [Google Scholar] [CrossRef]
- Antoniewski, C. Computing siRNA and piRNA overlap signatures. Methods Mol. Biol. 2014, 1173, 135–146. [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature 2007, 450, 203–218. [Google Scholar] [CrossRef] [PubMed]
- Rius, N.; Guillen, Y.; Delprat, A.; Kapusta, A.; Feschotte, C.; Ruiz, A. Exploration of the Drosophila buzzatii transposable element content suggests underestimation of repeats in Drosophila genomes. BMC Genomics 2016, 17, 344. [Google Scholar] [CrossRef]
- Reed, L.K.; Nyboer, M.; Markow, T.A. Evolutionary relationships of Drosophila mojavensis geographic host races and their sister species Drosophila arizonae. Mol. Ecol. 2007, 16, 1007–1022. [Google Scholar] [CrossRef]
- Vieira, C.; Fablet, M.; Lerat, E.; Boulesteix, M.; Rebollo, R.; Burlet, N.; Akkouche, A.; Hubert, B.; Mortada, H.; Biémont, C. A comparative analysis of the amounts and dynamics of transposable elements in natural populations of Drosophila melanogaster and Drosophila simulans. J. Environ. Radioact. 2012, 113, 83–86. [Google Scholar] [CrossRef]
- Saint-Leandre, B.; Capy, P.; Hua-Van, A.; Filée, J. piRNA and Transposon Dynamics in Drosophila: A Female Story. Genome Biol. Evol. 2020, 12, 931–947. [Google Scholar] [CrossRef] [PubMed]
- Song, J.; Liu, J.; Schnakenberg, S.L.; Ha, H.; Xing, J.; Chen, K.C. Variation in piRNA and transposable element content in strains of Drosophila melanogaster. Genome Biol. Evol. 2014, 6, 2786–2798. [Google Scholar] [CrossRef]
- Moshkovich, N.; Lei, E.P. HP1 recruitment in the absence of argonaute proteins in Drosophila. PLoS Genet. 2010, 6, e1000880. [Google Scholar] [CrossRef]
- Olovnikov, I.; Aravin, A.A.; Fejes, T.K. Small RNA in the nucleus: The RNA-chromatin ping-pong. Curr. Opin. Genet. Dev. 2012, 22, 164–171. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sato, K.; Siomi, M.C. The piRNA pathway in Drosophila ovarian germ and somatic cells. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 2020, 96, 32–42. [Google Scholar] [CrossRef]
- Thomae, A.W.; Schade, G.O.; Padeken, J.; Borath, M.; Vetter, I.; Kremmer, E.; Heun, P.; Imhof, A. A pair of centromeric proteins mediates reproductive isolation in Drosophila species. Dev. Cell 2013, 27, 412–424. [Google Scholar] [CrossRef]
- Nagao, A.; Mituyama, T.; Huang, H.; Chen, D.; Siomi, M.C.; Siomi, H. Biogenesis pathways of piRNAs loaded onto AGO3 in the Drosophila testis. RNA 2010, 16, 2503–2515. [Google Scholar] [CrossRef] [PubMed]
- Kotov, A.A.; Adashev, V.E.; Godneeva, B.K.; Ninova, M.; Shatskikh, A.S.; Bazylev, S.S.; Aravin, A.A.; Olenina, L.V. piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res. 2019, 47, 4255–4271. [Google Scholar] [CrossRef]
- Quenerch’du, E.; Anand, A.; Kai, T. The piRNA pathway is developmentally regulated during spermatogenesis in Drosophila. RNA 2016, 22, 1044–1054. [Google Scholar] [CrossRef]
- Vagin, V.V.; Klenov, M.S.; Kalmykova, A.I.; Stolyarenko, A.D.; Kotelnikov, R.N.; Gvozdev, V.A. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol. 2004, 1, 54–58. [Google Scholar] [CrossRef]
- Savitsky, M.; Kwon, D.; Georgiev, P.; Kalmykova, A.; Gvozdev, V. Telomere elongation is under the control of the RNAi-based mechanism in the Drosophila germline. Genes Dev. 2006, 20, 345–354. [Google Scholar] [CrossRef]
- Aravin, A.A.; Klenov, M.S.; Vagin, V.V.; Bantignies, F.; Cavalli, G.; Gvozdev, V.A. Dissection of a natural RNA silencing process in the Drosophila melanogaster germ line. Mol. Cell. Biol. 2004, 24, 6742–6750. [Google Scholar] [CrossRef] [PubMed]
- Pane, A.; Jiang, P.; Zhao, D.Y.; Singh, M.; Schupbach, T. The Cutoff protein regulates piRNA cluster expression and piRNA production in the Drosophila germline. EMBO J. 2011, 30, 4601–4615. [Google Scholar] [CrossRef]
- Goriaux, C.; Desset, S.; Renaud, Y.; Vaury, C.; Brasset, E. Transcriptional properties and splicing of the flamenco piRNA cluster. EMBO Rep. 2014, 15, 411–418. [Google Scholar] [CrossRef]
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108. [Google Scholar] [CrossRef]
- Phalke, S.; Nickel, O.; Walluscheck, D.; Hortig, F.; Onorati, M.C.; Reuter, G. Retrotransposon silencing and telomere integrity in somatic cells of Drosophila depends on the cytosine-5 methyltransferase DNMT2. Nat. Genet. 2009, 41, 696–702. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Gu, J.; Jin, Y.; Luo, Y.; Preall, J.B.; Ma, J.; Czech, B.; Hannon, G.J. Panoramix enforces piRNA-dependent cotranscriptional silencing. Science 2015, 350, 339–342. [Google Scholar] [CrossRef]
- Zhao, K.; Cheng, S.; Miao, N.; Xu, P.; Lu, X.; Zhang, Y.; Wang, M.; Ouyang, X.; Yuan, X.; Liu, W.; et al. A Pandas complex adapted for piRNA-guided transcriptional silencing and heterochromatin formation. Nat. Cell Biol. 2019, 21, 1261–1272. [Google Scholar] [CrossRef] [PubMed]
- Han, B.W.; Wang, W.; Li, C.; Weng, Z.; Zamore, P.D. Noncoding RNA. piRNA-guided transposon cleavage initiates Zucchini-dependent, phased piRNA production. Science 2015, 348, 817–821. [Google Scholar] [CrossRef] [PubMed]
- Cappucci, U.; Noro, F.; Casale, A.M.; Fanti, L.; Berloco, M.; Alagia, A.A.; Grassi, L.; Le Pera, L.; Piacentini, L.; Pimpinelli, S. The Hsp70 chaperone is a major player in stress-induced transposable element activation. Proc. Natl. Acad. Sci. USA 2019, 116, 17943–17950. [Google Scholar] [CrossRef]
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299. [Google Scholar] [CrossRef]
- Karam, J.A.; Parikh, R.Y.; Nayak, D.; Rosenkranz, D.; Gangaraju, V.K. Co-chaperone Hsp70/Hsp90-organizing protein (Hop) is required for transposon silencing and Piwi-interacting RNA (piRNA) biogenesis. J. Biol. Chem. 2017, 292, 6039–6046. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Banho, C.A.; Oliveira, D.S.; Haudry, A.; Fablet, M.; Vieira, C.; Carareto, C.M.A. Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi. Cells 2021, 10, 3574. https://doi.org/10.3390/cells10123574
Banho CA, Oliveira DS, Haudry A, Fablet M, Vieira C, Carareto CMA. Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi. Cells. 2021; 10(12):3574. https://doi.org/10.3390/cells10123574
Chicago/Turabian StyleBanho, Cecília Artico, Daniel Siqueira Oliveira, Annabelle Haudry, Marie Fablet, Cristina Vieira, and Claudia Marcia Aparecida Carareto. 2021. "Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi" Cells 10, no. 12: 3574. https://doi.org/10.3390/cells10123574
APA StyleBanho, C. A., Oliveira, D. S., Haudry, A., Fablet, M., Vieira, C., & Carareto, C. M. A. (2021). Transposable Element Expression and Regulation Profile in Gonads of Interspecific Hybrids of Drosophila arizonae and Drosophila mojavensis wrigleyi. Cells, 10(12), 3574. https://doi.org/10.3390/cells10123574





