Retromer Complex and PI3K Complex II-Related Genes Mediate the Yeast (Saccharomyces cerevisiae) Sodium Metabisulfite Resistance Response
Abstract
1. Introduction
2. Materials and Methods
2.1. Chemicals and Strains
2.2. Growth Curve Measurement
2.3. Genome-Wide Na2S2O5 Screen and Functional Analysis
2.4. Spot Test
2.5. Fluorescence Observation
2.6. Western Blot
2.7. Methylene Blue Staining
2.8. Use of PI3K Inhibitor
3. Results and Discussion
3.1. Determination of Na2S2O5 Concentration for Yeast Genome-Wide Screening
3.2. Genomic Screens of S. cerevisiae Deletion Mutants Revealed Important Cellular Processes Involved in Na2S2O5 Stress
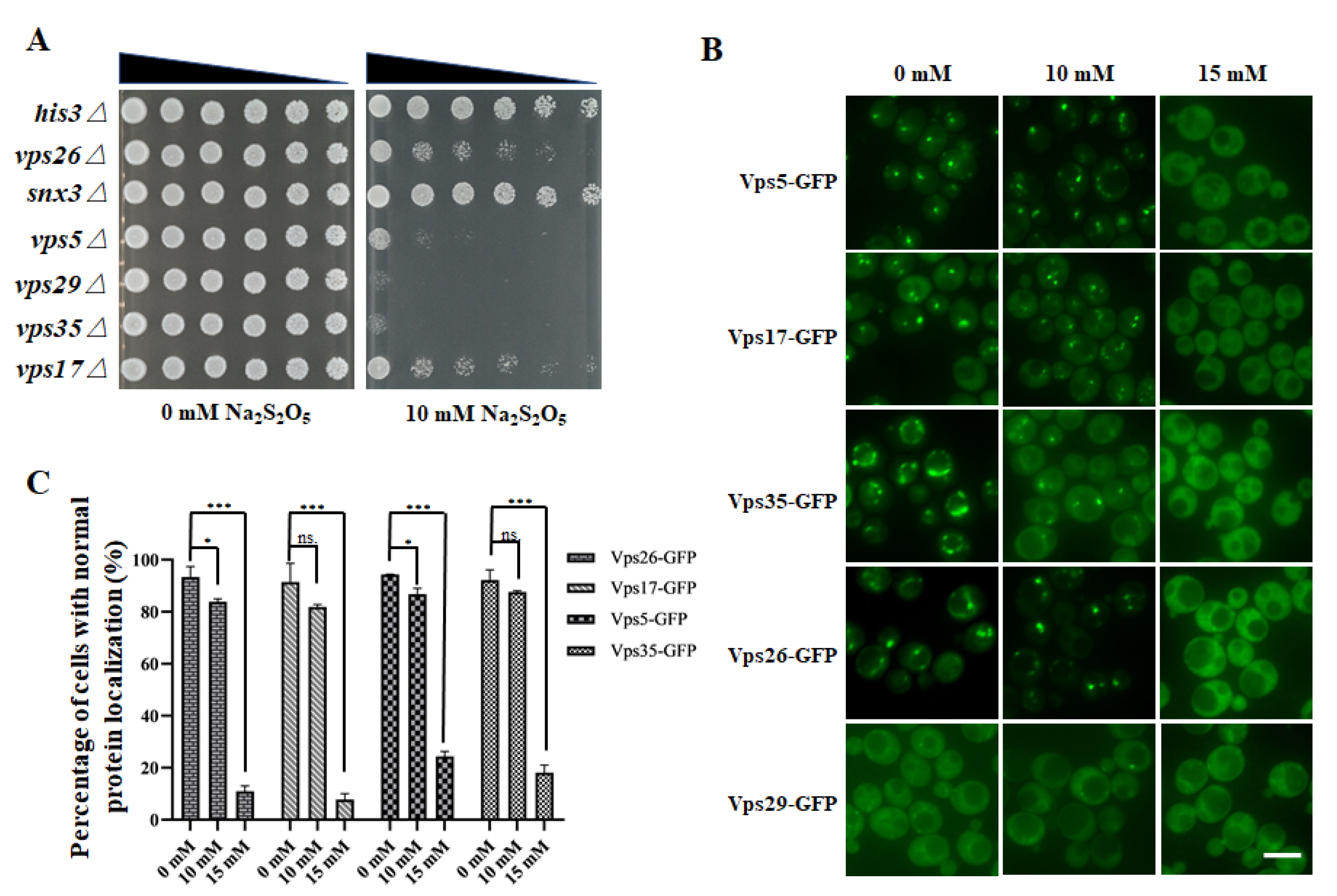
3.3. The Retromer Complex Is Required for Yeast Na2S2O5 Tolerance
3.4. Na2S2O5 Treatment Can Cause Changes in the Localization of the Retromer Complex
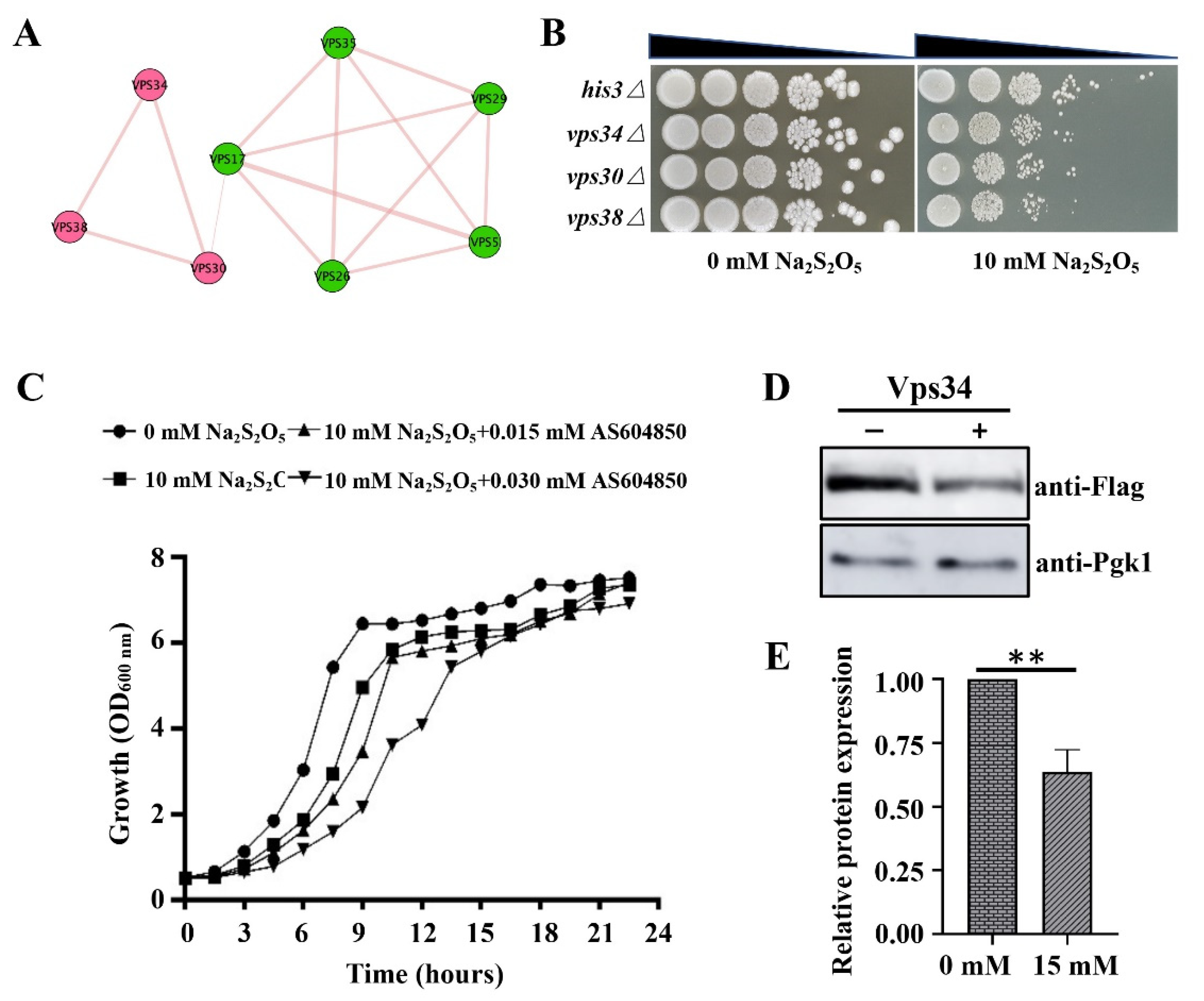
3.5. Phosphatidy-linositol-3-monophosphate Is a Potential Signaling Molecule Mediating the Yeast Na2S2O5 Tolerance Response
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Przybilla, B.; Ring, J. Sulfite hypersensitivity. Hautarzt 1987, 38, 445–448. [Google Scholar] [CrossRef]
- Rencuzogullari, E.; Ila, H.B.; Kayraldiz, A.; Topaktas, M. Chromosome aberrations and sister chromatid exchanges in cultured human lymphocytes treated with sodium metabisulfite, a food preservative. Mutat. Res. 2001, 490, 107–112. [Google Scholar] [CrossRef]
- Wyse, A.T.S.; Grings, M.; Wajner, M.; Leipnitz, G. The role of oxidative stress and bioenergetic dysfunction in sulfite oxidase deficiency: Insights from animal models. Neurotox. Res. 2019, 35, 484–494. [Google Scholar] [CrossRef]
- Gunnison, A.F. Sulphite toxicity: A critical review of in vitro and in vivo data. Food Cosmet. Toxicol. 1981, 19, 667–682. [Google Scholar] [CrossRef]
- Elmas, O.; Aslan, M.; Caglar, S.; Derin, N.; Agar, A.; Aliciguzel, Y.; Yargicoglu, P. The prooxidant effect of sodium metabisulfite in rat liver and kidney. Regul. Toxicol. Pharmacol. 2005, 42, 77–82. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Meng, Z. Effects of sulfur dioxide on apoptosis-related gene expressions in lungs from rats. Regul. Toxicol. Pharmacol. 2005, 43, 272–279. [Google Scholar] [CrossRef]
- Kencebay, C.; Derin, N.; Ozsoy, O.; Kipmen-Korgun, D.; Tanriover, G.; Ozturk, N.; Basaranlar, G.; Yargicoglu-Akkiraz, P.; Sozen, B.; Agar, A. Merit of quinacrine in the decrease of ingested sulfite-induced toxic action in rat brain. Food Chem. Toxicol. 2013, 52, 129–136. [Google Scholar] [CrossRef]
- Ercan, S.; Basaranlar, G.; Gungor, N.E.; Kencebay, C.; Sahin, P.; Celik-Ozenci, C.; Derin, N. Ghrelin inhibits sodium metabisulfite induced oxidative stress and apoptosis in rat gastric mucosa. Food Chem. Toxicol. 2013, 56, 154–161. [Google Scholar] [CrossRef]
- Avis, T.J.; Michaud, M.; Tweddell, R.J. Role of lipid composition and lipid peroxidation in the sensitivity of fungal plant pathogens to aluminum chloride and sodium metabisulfite. Appl. Environ. Microbiol. 2007, 73, 2820–2824. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ercan, S.; Kencebay, C.; Basaranlar, G.; Derin, N.; Aslan, M. Induction of xanthine oxidase activity, endoplasmic reticulum stress and caspase activation by sodium metabisulfite in rat liver and their attenuation by Ghrelin. Food Chem. Toxicol. 2015, 76, 27–32. [Google Scholar] [CrossRef]
- Nie, A.; Meng, Z. Sulfur dioxide derivatives modulate Na/Ca exchange currents and cytosolic [Ca2+] i in rat myocytes. Biochem. Biophys. Res. Commun. 2007, 358, 879–884. [Google Scholar] [CrossRef]
- Lai, M.C.; Hung, T.Y.; Lin, K.M.; Sung, P.S.; Wu, S.J.; Yang, C.S.; Wu, Y.J.; Tsai, J.J.; Wu, S.N.; Huang, C.W. Sodium metabisulfite: Effects on ionic currents and excitotoxicity. Neurotox. Res. 2018, 34, 1–15. [Google Scholar] [CrossRef]
- Zhang, Q.; Bai, Y.; Yang, Z.; Tian, J.; Meng, Z. The molecular mechanisms of sodium metabisulfite on the expression of K ATP and L-Ca2+ channels in rat hearts. Regul. Toxicol. Pharmacol. 2015, 72, 440–446. [Google Scholar] [CrossRef]
- Teresa, M.V.; Rekha, K.; Bindu, A. Effect of sodium metabisulphite on germination, growth and yield of Vigna sinensis, Savi. J. Environ. Biol. 2003, 24, 449–452. [Google Scholar] [CrossRef] [PubMed]
- Nadai, C.; Treu, L.; Campanaro, S.; Giacomini, A.; Corich, V. Different mechanisms of resistance modulate sulfite tolerance in wine yeasts. J. Microbiol. Biotechnol. 2016, 100, 797–813. [Google Scholar] [CrossRef]
- Divol, B.; du Toit, M.; Duckitt, E. Surviving in the presence of sulphur dioxide: Strategies developed by wine yeasts. Appl. Microbiol. Biotechnol. 2012, 95, 601–613. [Google Scholar] [CrossRef]
- Donalies, U.E.; Stahl, U. Increasing sulphite formation in Saccharomyces cerevisiae by overexpression of MET14 and SSU1. Yeast 2002, 19, 475–484. [Google Scholar] [CrossRef]
- Teixeira, M.C.; Monteiro, P.T.; Palma, M.; Costa, C.; Godinho, C.P.; Pais, P.; Cavalheiro, M.; Antunes, M.; Lemos, A.; Pedreira, T.; et al. YEASTRACT: An upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 2018, 46, D348–D353. [Google Scholar] [CrossRef]
- Valero, E.; Tronchoni, J.; Morales, P.; Gonzalez, R. Autophagy is required for sulfur dioxide tolerance in Saccharomyces cerevisiae. Microb. Biotechnol. 2020, 13, 599–604. [Google Scholar] [CrossRef]
- Barberis, A.; Gunde, T.; Berset, C.; Audetat, S.; Luthi, U. Yeast as a screening tool. Drug Discov. Today Technol. 2005, 2, 187–192. [Google Scholar] [CrossRef]
- Zimmermann, A.; Hofer, S.; Pendl, T.; Kainz, K.; Madeo, F.; Carmona-Gutierrez, D. Yeast as a tool to identify anti-aging compounds. FEMS Yeast Res. 2018, 18, foy020. [Google Scholar] [CrossRef] [PubMed]
- Hillenmeyer, M.E.; Fung, E.; Wildenhain, J.; Pierce, S.E.; Hoon, S.; Lee, W.; Proctor, M.; St Onge, R.P.; Tyers, M.; Koller, D.; et al. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Science 2008, 320, 362–365. [Google Scholar] [CrossRef]
- Huh, W.K.; Falvo, J.V.; Gerke, L.C.; Carroll, A.S.; Howson, R.W.; Weissman, J.S.; O’Shea, E.K. Global analysis of protein localization in budding yeast. Nature 2003, 425, 686–691. [Google Scholar] [CrossRef]
- Jin, X.; Zhang, J.; An, T.; Zhao, H.; Fu, W.; Li, D.; Liu, S.; Cao, X.; Liu, B. A genome-wide screen in Saccharomyces cerevisiae reveals a critical role for oxidative phosphorylation in cellular tolerance to lithium hexafluorophosphate. Cells 2021, 10, 888. [Google Scholar] [CrossRef]
- Wagih, O.; Usaj, M.; Baryshnikova, A.; VanderSluis, B.; Kuzmin, E.; Costanzo, M.; Myers, C.L.; Andrews, B.J.; Boone, C.M.; Parts, L. SGAtools: One-stop analysis and visualization of array-based genetic interaction screens. Nucleic Acids Res. 2013, 41, W591–W596. [Google Scholar] [CrossRef]
- Tong, A.H.; Evangelista, M.; Parsons, A.B.; Xu, H.; Bader, G.D.; Page, N.; Robinson, M.; Raghibizadeh, S.; Hogue, C.W.; Bussey, H.; et al. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 2001, 294, 2364–2368. [Google Scholar] [CrossRef]
- Chen, K.E.; Healy, M.D.; Collins, B.M. Towards a molecular understanding of endosomal trafficking by retromer and retriever. Traffic 2019, 20, 465–478. [Google Scholar] [CrossRef] [PubMed]
- Abubakar, Y.S.; Zheng, W.; Olsson, S.; Zhou, J. Updated insight into the physiological and pathological roles of the retromer complex. Int. J. Mol. Sci. 2017, 18, 1601. [Google Scholar] [CrossRef]
- Strochlic, T.I.; Setty, T.G.; Sitaram, A.; Burd, C.G. Grd19/Snx3p functions as a cargo-specific adapter for retromer-dependent endocytic recycling. J. Cell Biol. 2007, 177, 115–125. [Google Scholar] [CrossRef]
- Hettema, E.H.; Lewis, M.J.; Black, M.W.; Pelham, H.R. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003, 22, 548–557. [Google Scholar] [CrossRef] [PubMed]
- Nothwehr, S.F.; Ha, S.A.; Bruinsma, P. Sorting of yeast membrane proteins into an endosome-to-Golgi pathway involves direct interaction of their cytosolic domains with Vps35p. J. Cell Biol. 2000, 151, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Padilla, S.M.; Sarkar, S.; Emr, S.D. Retromer function in endosome-to-Golgi retrograde transport is regulated by the yeast Vps34 PtdIns 3-kinase. J. Cell Biol. 2002, 115, 3889–3900. [Google Scholar] [CrossRef]
- Purushothaman, L.K.; Arlt, H.; Kuhlee, A.; Raunser, S.; Ungermann, C. Retromer-driven membrane tubulation separates endosomal recycling from Rab7/Ypt7-dependent fusion. Mol. Biol. Cell 2017, 28, 783–791. [Google Scholar] [CrossRef]
- Gillooly, D.J.; Morrow, I.C.; Lindsay, M.; Gould, R.; Bryant, N.J.; Gaullier, J.M.; Parton, R.G.; Stenmark, H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000, 19, 4577–4588. [Google Scholar] [CrossRef]
- Burman, C.; Ktistakis, N.T. Regulation of autophagy by phosphatidylinositol 3-phosphate. FEBS Lett. 2010, 584, 1302–1312. [Google Scholar] [CrossRef]
- Burman, C.; Ktistakis, N.T. Autophagosome formation in mammalian cells. Semin. Immunopathol. 2010, 32, 397–413. [Google Scholar] [CrossRef]
- Seet, L.F.; Hong, W. The Phox (PX) domain proteins and membrane traffic. Biochim. Biophys. Acta. 2006, 1761, 878–896. [Google Scholar] [CrossRef]
- Teasdale, R.D.; Collins, B.M. Insights into the PX (phox-homology) domain and SNX (sorting nexin) protein families: Structures, functions and roles in disease. Biochem. J. 2012, 441, 39–59. [Google Scholar] [CrossRef]
- Schu, P.V.; Takegawa, K.; Fry, M.J.; Stack, J.H.; Waterfield, M.D.; Emr, S.D. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science 1993, 260, 88–91. [Google Scholar] [CrossRef]
- Kihara, A.; Noda, T.; Ishihara, N.; Ohsumi, Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J. Cell Biol. 2001, 152, 519–530. [Google Scholar] [CrossRef]
- Fassy, F.; Dureuil, C.; Lamberton, A.; Mathieu, M.; Michot, N.; Ronan, B.; Pasquier, B. In vitro characterization of vps34 lipid kinase inhibition by small molecules. Methods Enzymol. 2017, 587, 447–464. [Google Scholar] [CrossRef] [PubMed]
- Feng, Z.; Xu, K.; Kovalev, N.; Nagy, P.D. Recruitment of Vps34 PI3K and enrichment of PI3P phosphoinositide in the viral replication compartment is crucial for replication of a positive-strand RNA virus. PLoS Pathog. 2019, 15, e1007530. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, X.; Zhao, H.; Zhou, M.; Zhang, J.; An, T.; Fu, W.; Li, D.; Cao, X.; Liu, B. Retromer Complex and PI3K Complex II-Related Genes Mediate the Yeast (Saccharomyces cerevisiae) Sodium Metabisulfite Resistance Response. Cells 2021, 10, 3512. https://doi.org/10.3390/cells10123512
Jin X, Zhao H, Zhou M, Zhang J, An T, Fu W, Li D, Cao X, Liu B. Retromer Complex and PI3K Complex II-Related Genes Mediate the Yeast (Saccharomyces cerevisiae) Sodium Metabisulfite Resistance Response. Cells. 2021; 10(12):3512. https://doi.org/10.3390/cells10123512
Chicago/Turabian StyleJin, Xuejiao, Huihui Zhao, Min Zhou, Jie Zhang, Tingting An, Wenhao Fu, Danqi Li, Xiuling Cao, and Beidong Liu. 2021. "Retromer Complex and PI3K Complex II-Related Genes Mediate the Yeast (Saccharomyces cerevisiae) Sodium Metabisulfite Resistance Response" Cells 10, no. 12: 3512. https://doi.org/10.3390/cells10123512
APA StyleJin, X., Zhao, H., Zhou, M., Zhang, J., An, T., Fu, W., Li, D., Cao, X., & Liu, B. (2021). Retromer Complex and PI3K Complex II-Related Genes Mediate the Yeast (Saccharomyces cerevisiae) Sodium Metabisulfite Resistance Response. Cells, 10(12), 3512. https://doi.org/10.3390/cells10123512





