Roles of a Cryptochrome in Carbon Fixation and Sucrose Metabolism in the Liverwort Marchantia polymorpha
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Phylogenetic Tree Analysis
2.3. Generation of Transgenic Lines
2.4. RNA Extraction and Transcriptome Sequencing
2.5. Quantification of Gene Expression Levels in Transcriptome Sequencing
2.6. Person Correlation Coefficients Analysis
2.7. Principal Component Analysis
2.8. Differential Expression Analysis
2.9. GO Enrichment Analysis of Different Response Genes (DRGs)
2.10. Processing Subcellar Images
2.11. Quantitative Real-Time PCR (RT-PCR) Analysis
2.12. Measurement of Chlorophyll Content
2.13. Photosynthetic Parameter Analysis
2.14. Quantification of Starch, Sucrose, Glucose
2.15. Quantification and Statistical Analysis
3. Results
3.1. The Sequence Conservation of MpCRY
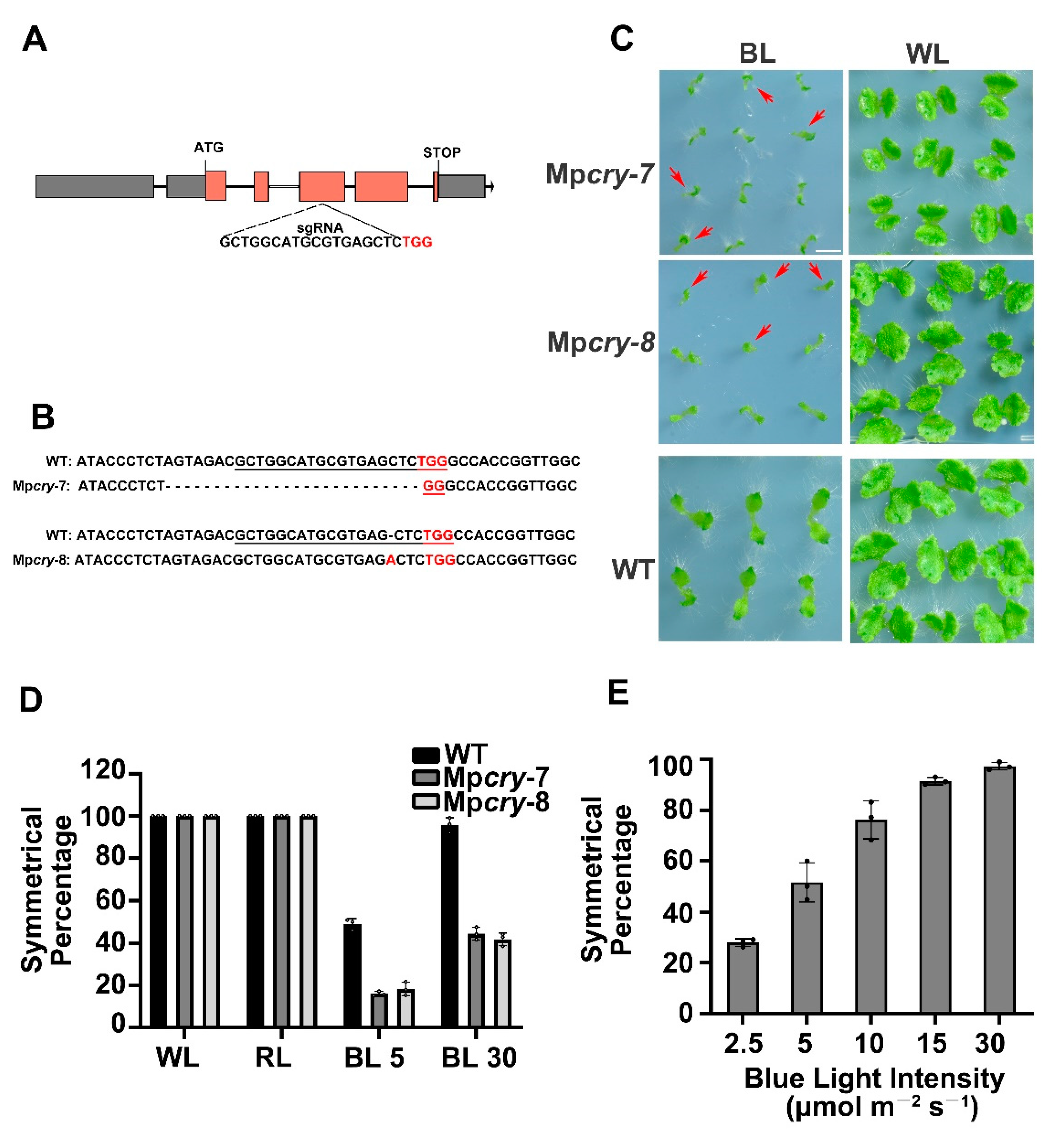
3.2. MpCRY Regulates the Thallus Symmetry of M. polymorpha under Blue Light
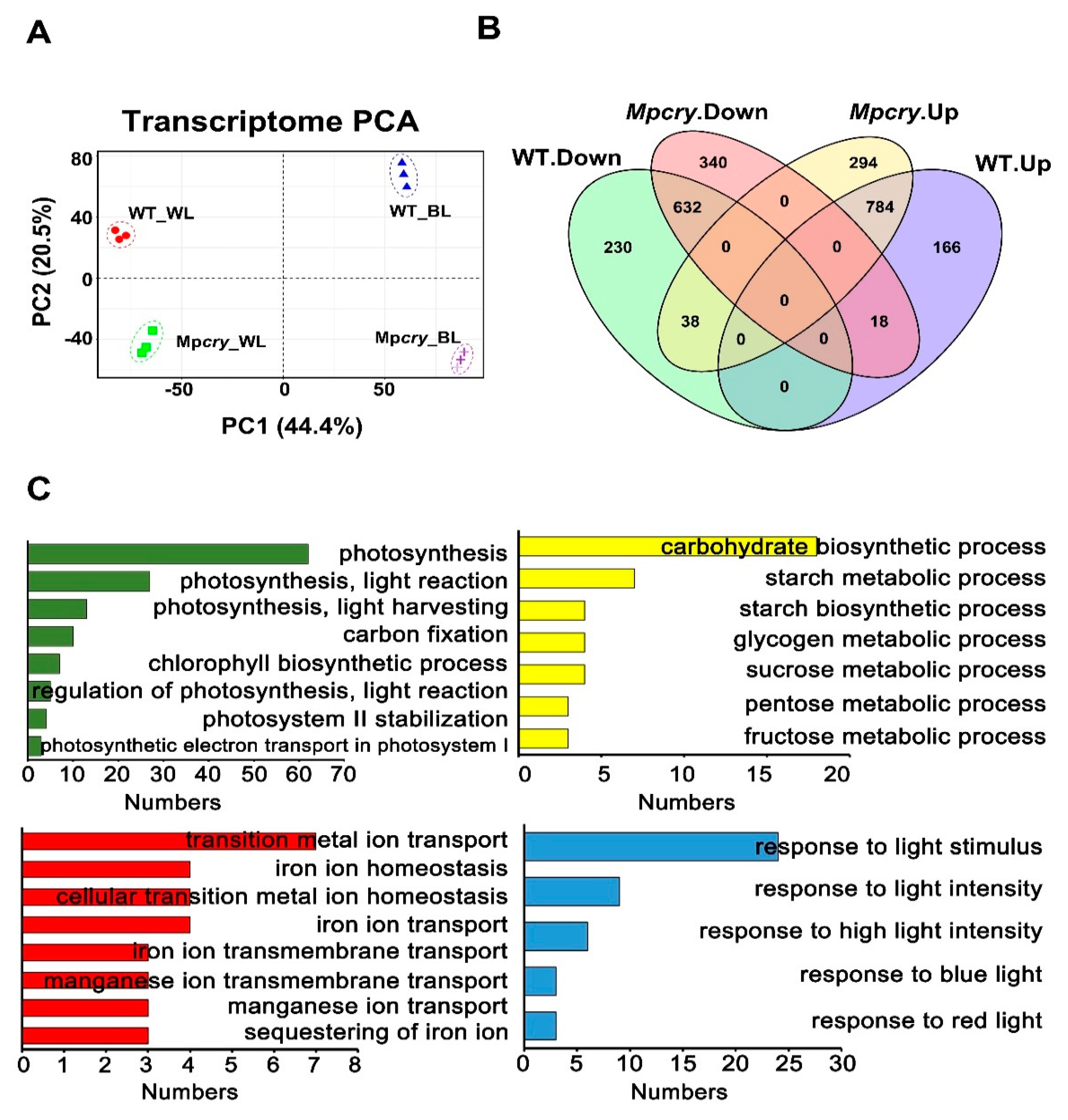
3.3. Transcriptome Analysis of Mpcry Mutants under Blue Light
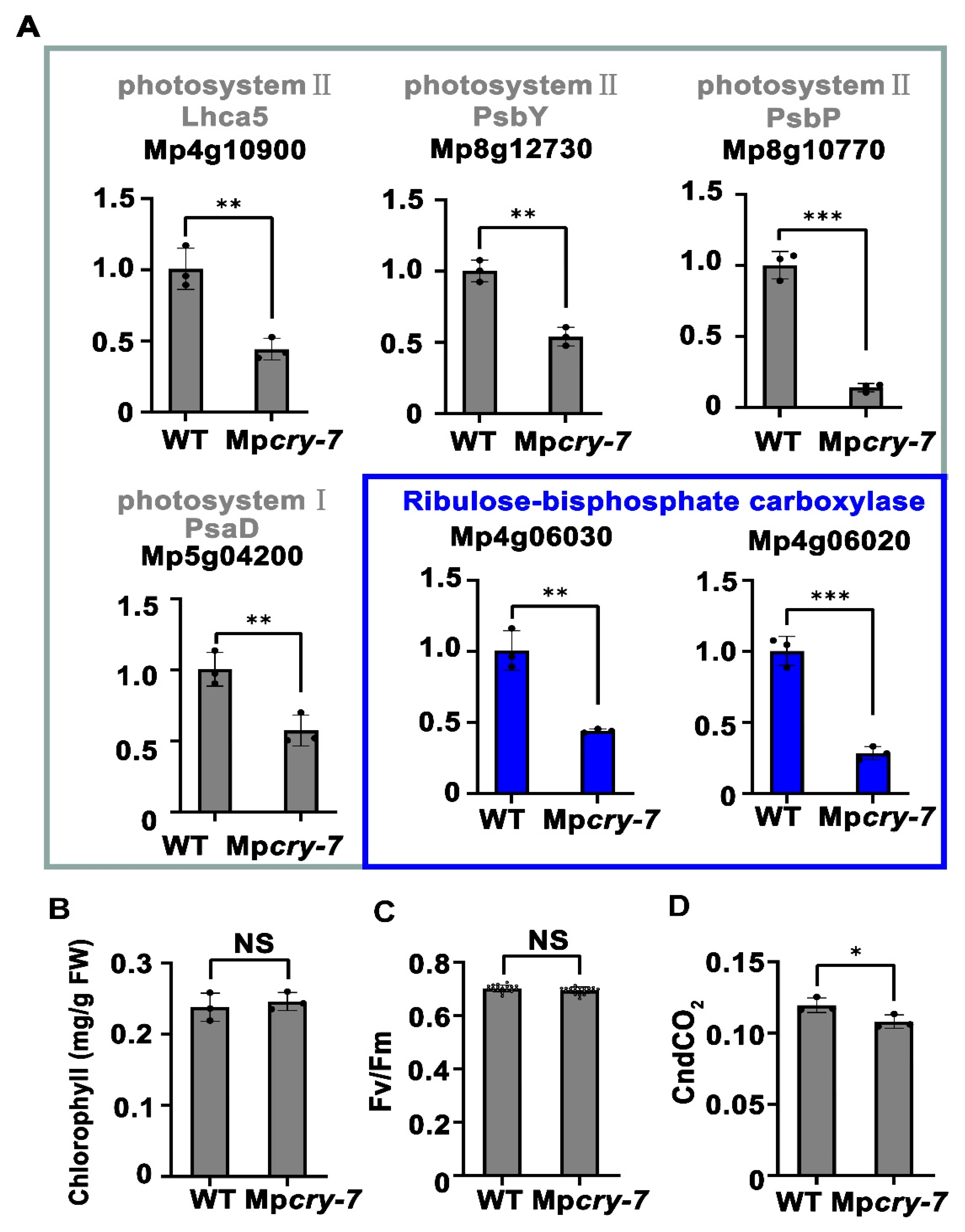
3.4. Mpcry Regulates Transcription of Photosynthesis-Related Genes under Blue Light
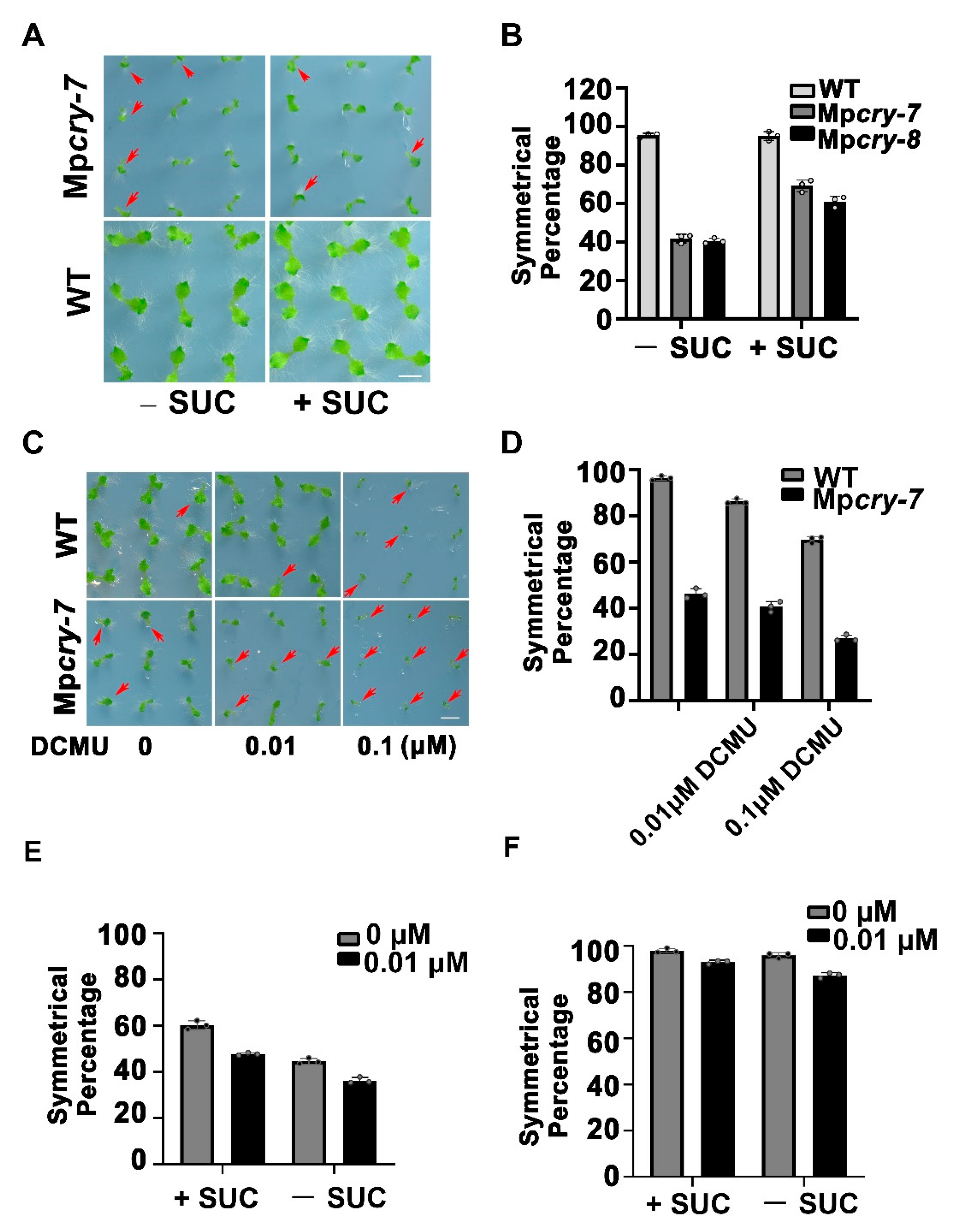
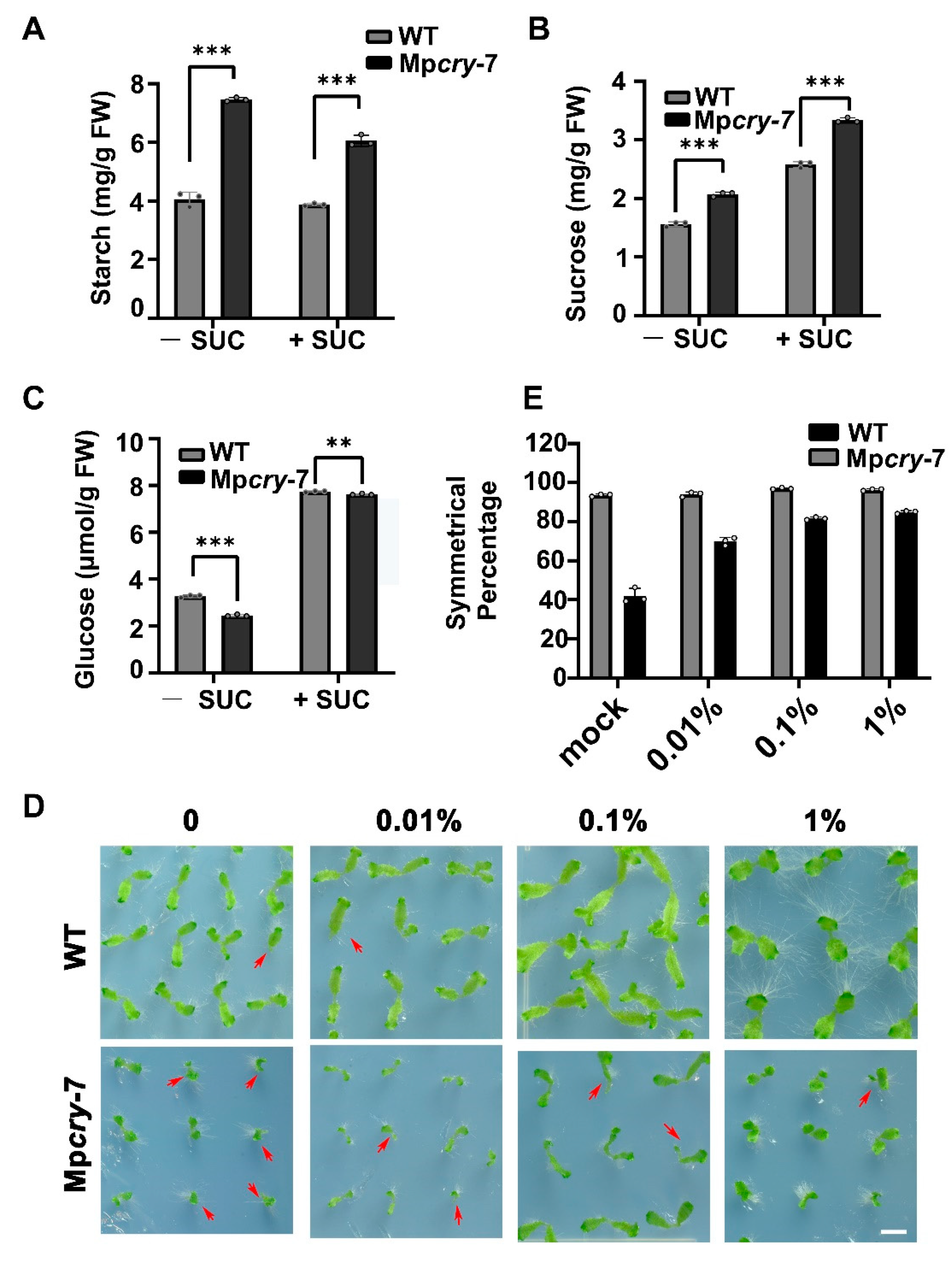
3.5. Sucrose Could Partially Restored the Symmetric Growth of the Mpcry Mutant Thallus
3.6. Mpcry Is Likely to Regulate the Symmetric Growth of Thallus through Sucrose Metabolism
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Han, X.; Chang, X.; Zhang, Z.; Chen, H.; He, H.; Zhong, B.; Deng, X.W. Origin and evolution of core components responsible for monitoring light environment changes during plant terrestrialization. Mol. Plant 2019, 12, 847–862. [Google Scholar] [CrossRef]
- Cashmore, A.R.; Jarillo, J.A.; Wu, Y.J.; Liu, D. Cryptochromes: Blue light receptors for plants and animals. Science 1999, 284, 760–765. [Google Scholar] [CrossRef] [PubMed]
- Sancar, A. Cryptochrome: The second photoactive pigment in the eye and its role in circadian photoreception. Annu. Rev. Biochem. 2000, 69, 31–67. [Google Scholar] [CrossRef]
- Falciatore, A.; Bowler, C. The evolution and function of blue and red light photoreceptors. Curr. Top. Dev. Biol. 2005, 68, 317–350. [Google Scholar]
- Dodson, C.A.; Hore, P.J.; Wallace, M.I. A radical sense of direction: Signalling and mechanism in cryptochrome magnetoreception. Trends Biochem. Sci. 2013, 38, 435–446. [Google Scholar] [CrossRef] [PubMed]
- Emery, P.; So, W.V.; Kaneko, M.; Hall, J.C.; Rosbash, M. Cry, a drosophila clock and light-regulated cryptochrome, is a major contributor to circadian rhythm resetting and photosensitivity. Cell 1998, 95, 669–679. [Google Scholar] [CrossRef]
- Thresher, R.J.; Vitaterna, M.H.; Miyamoto, Y.; Kazantsev, A.; Hsu, D.S.; Petit, C.; Selby, C.P.; Dawut, L.; Smithies, O.; Takahashi, J.S.; et al. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science 1998, 282, 1490–1494. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, M.; Cashmore, A.R. Hy4 gene of a. Thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 1993, 366, 162–166. [Google Scholar] [CrossRef]
- Liu, H.; Yu, X.; Li, K.; Klejnot, J.; Yang, H.; Lisiero, D.; Lin, C. Photoexcited cry2 interacts with cib1 to regulate transcription and floral initiation in arabidopsis. Science 2008, 322, 1535–1539. [Google Scholar] [CrossRef]
- Lin, C.; Yang, H.; Guo, H.; Mockler, T.; Chen, J.; Cashmore, A.R. Enhancement of blue-light sensitivity of arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc. Natl. Acad. Sci. USA 1998, 95, 2686–2690. [Google Scholar] [CrossRef]
- Pedmale, U.V.; Huang, S.C.; Zander, M.; Cole, B.J.; Hetzel, J.; Ljung, K.; Reis, P.A.B.; Sridevi, P.; Nito, K.; Nery, J.R.; et al. Cryptochromes interact directly with pifs to control plant growth in limiting blue light. Cell 2016, 164, 233–245. [Google Scholar] [CrossRef]
- Somers, D.E.; Devlin, P.F.; Kay, S.A. Phytochromes and cryptochromes in the entrainment of the arabidopsis circadian clock. Science 1998, 282, 1488–1490. [Google Scholar] [CrossRef]
- Mao, J.; Zhang, Y.C.; Sang, Y.; Li, Q.H.; Yang, H.Q. From the cover: A role for arabidopsis cryptochromes and cop1 in the regulation of stomatal opening. Proc. Natl. Acad. Sci. USA 2005, 102, 12270–12275. [Google Scholar] [CrossRef] [PubMed]
- Imaizumi, T.; Kadota, A.; Hasebe, M.; Wada, M. Cryptochrome light signals control development to suppress auxin sensitivity in the moss physcomitrella patens. Plant Cell 2002, 14, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Ichikawa, K.; Sugita, M.; Imaizumi, T.; Wada, M.; Aoki, S. Differential expression on a daily basis of plastid sigma factor genes from the moss physcomitrella patens. Regulatory interactions among ppsig5, the circadian clock, and blue light signaling mediated by cryptochromes. Plant Physiol. 2004, 136, 4285–4298. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Riese, M.; Zobell, O.; Saedler, H.; Huijser, P. Sbp-domain transcription factors as possible effectors of cryptochrome-mediated blue light signalling in the moss physcomitrella patens. Planta 2008, 227, 505–515. [Google Scholar] [CrossRef]
- Berger, F.; Bowman, J.L.; Kohchi, T. Marchantia. Curr. Biol. CB 2016, 26, R186–R187. [Google Scholar] [CrossRef][Green Version]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T.; Jenkins, J.; Shu, S.; Ishizaki, K.; Yamaoka, S.; Nishihama, R.; Nakamura, Y.; Berger, F.; et al. Insights into land plant evolution garnered from the marchantia polymorpha genome. Cell 2017, 171, 287–304.e215. [Google Scholar] [CrossRef]
- Ishizaki, K.; Nishihama, R.; Yamato, K.T.; Kohchi, T. Molecular genetic tools and techniques for marchantia polymorpha research. Plant Cell Physiol. 2016, 57, 262–270. [Google Scholar] [CrossRef]
- Komatsu, A.; Terai, M.; Ishizaki, K.; Suetsugu, N.; Tsuboi, H.; Nishihama, R.; Yamato, K.T.; Wada, M.; Kohchi, T. Phototropin encoded by a single-copy gene mediates chloroplast photorelocation movements in the liverwort marchantia polymorpha. Plant Physiol. 2014, 166, 411–427. [Google Scholar] [CrossRef]
- Suzuki, T.; Takio, S.; Yamamoto, I.; Satoh, T. Characterization of cdna of the liverwort phytochrome gene, and phytochrome involvement in the light-dependent and light-independent protochlorophyllide oxidoreductase gene expression in marchantia paleacea var. Diptera. Plant Cell Physiol. 2001, 42, 576–582. [Google Scholar] [CrossRef]
- Inoue, K.; Nishihama, R.; Kataoka, H.; Hosaka, M.; Manabe, R.; Nomoto, M.; Tada, Y.; Ishizaki, K.; Kohchi, T. Phytochrome signaling is mediated by phytochrome interacting factor in the liverwort marchantia polymorpha. Plant Cell 2016, 28, 1406–1421. [Google Scholar] [CrossRef]
- Soriano, G.; Cloix, C.; Heilmann, M.; Nunez-Olivera, E.; Martinez-Abaigar, J.; Jenkins, G.I. Evolutionary conservation of structure and function of the uvr8 photoreceptor from the liverwort marchantia polymorpha and the moss physcomitrella patens. New Phytol. 2018, 217, 151–162. [Google Scholar] [CrossRef]
- Kubota, A.; Kita, S.; Ishizaki, K.; Nishihama, R.; Yamato, K.T.; Kohchi, T. Co-option of a photoperiodic growth-phase transition system during land plant evolution. Nat. Commun. 2014, 5, 3668. [Google Scholar] [CrossRef] [PubMed]
- Sugano, S.S.; Shirakawa, M.; Takagi, J.; Matsuda, Y.; Shimada, T.; Hara-Nishimura, I.; Kohchi, T. Crispr/cas9-mediated targeted mutagenesis in the liverwort marchantia polymorpha l. Plant Cell Physiol. 2014, 55, 475–481. [Google Scholar] [CrossRef]
- Ishizaki, K.; Chiyoda, S.; Yamato, K.T.; Kohchi, T. Agrobacterium-mediated transformation of the haploid liverwort marchantia polymorpha l, an emerging model for plant biology. Plant Cell Physiol. 2008, 49, 1084–1091. [Google Scholar] [CrossRef]
- Gamborg, O.L.; Miller, R.A.; Ojima, K. Nutrient requirements of suspension cultures of soybean root cells. Exp. Cell Res. 1968, 50, 151–158. [Google Scholar] [CrossRef]
- Edgar, R.C. Muscle: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of phyml 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Sugano, S.S.; Nishihama, R. Crispr/cas9-based genome editing of transcription factor genes in marchantia polymorpha. Methods Mol. Biol. 2018, 1830, 109–126. [Google Scholar] [PubMed]
- Sugano, S.S.; Nishihama, R.; Shirakawa, M.; Takagi, J.; Matsuda, Y.; Ishida, S.; Shimada, T.; Hara-Nishimura, I.; Osakabe, K.; Kohchi, T. Efficient crispr/cas9-based genome editing and its application to conditional genetic analysis in marchantia polymorpha. PLoS ONE 2018, 13, e0205117. [Google Scholar] [CrossRef]
- Ishizaki, K.; Nishihama, R.; Ueda, M.; Inoue, K.; Ishida, S.; Nishimura, Y.; Shikanai, T.; Kohchi, T. Development of gateway binary vector series with four different selection markers for the liverwort marchantia polymorpha. PLoS ONE 2015, 10, e0138876. [Google Scholar]
- Kubota, A.; Ishizaki, K.; Hosaka, M.; Kohchi, T. Efficient agrobacterium-mediated transformation of the liverwort marchantia polymorpha using regenerating thalli. Biosci. Biotechnol. Bioch. 2013, 77, 167–172. [Google Scholar] [CrossRef]
- Yang, X.; Liu, D.; Liu, F.; Wu, J.; Zou, J.; Xiao, X.; Zhao, F.; Zhu, B. Htqc: A fast quality control toolkit for illumina sequencing data. BMC Bioinform. 2013, 14, 33. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. Tophat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by rna-seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. Edger: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Conesa, A.; Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Talon, M.; Robles, M. Blast2go: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 2005, 21, 3674–3676. [Google Scholar] [CrossRef] [PubMed]
- Sandhu, L.C.; Warters, R.L.; Dethlefsen, L.A. Fluorescence studies of hoechst 33342 with supercoiled and relaxed plasmid pbr322 DNA. Cytometry 1985, 6, 191–194. [Google Scholar] [CrossRef] [PubMed]
- Tawakoli, P.N.; Al-Ahmad, A.; Hoth-Hannig, W.; Hannig, M.; Hannig, C. Comparison of different live/dead stainings for detection and quantification of adherent microorganisms in the initial oral biofilm. Clin. Oral Investig. 2013, 17, 841–850. [Google Scholar] [CrossRef]
- Saint-Marcoux, D.; Proust, H.; Dolan, L.; Langdale, J.A. Identification of reference genes for real-time quantitative pcr experiments in the liverwort marchantia polymorpha. PLoS ONE 2015, 10, e0118678. [Google Scholar]
- Wang, Q.; Lin, C. Mechanisms of cryptochrome-mediated photoresponses in plants. Annu. Rev. Plant Biol. 2020, 71, 103–129. [Google Scholar] [CrossRef]
- Selby, C.P.; Sancar, A. A cryptochrome/photolyase class of enzymes with single-stranded DNA-specific photolyase activity. Proc. Natl. Acad. Sci. USA 2006, 103, 17696–17700. [Google Scholar] [CrossRef]
- Lin, C.; Shalitin, D. Cryptochrome structure and signal transduction. Annu. Rev. Plant Biol. 2003, 54, 469–496. [Google Scholar] [CrossRef]
- Nakazato, T.; Kadota, A.; Wada, M. Photoinduction of spore germination in Marchantia polymorpha L. is mediated by photosynthesis. Plant Cell Physiol. 1999, 40, 1014–1020. [Google Scholar] [CrossRef][Green Version]
- Inoue, K.; Nishihama, R.; Kohchi, T. Phytochrome and light signaling in marchantia. Methods Mol. Biol. 2019, 2026, 215–223. [Google Scholar] [PubMed]
- Ma, L.; Li, J.; Qu, L.; Hager, J.; Chen, Z.; Zhao, H.; Deng, X.W. Light control of arabidopsis development entails coordinated regulation of genome expression and cellular pathways. Plant Cell 2001, 13, 2589–2607. [Google Scholar] [CrossRef]
- Sellaro, R.; Hoecker, U.; Yanovsky, M.; Chory, J.; Casal, J.J. Synergism of red and blue light in the control of arabidopsis gene expression and development. Curr. Biol. CB 2009, 19, 1216–1220. [Google Scholar] [CrossRef]
- Wang, Q.; Zuo, Z.; Wang, X.; Gu, L.; Yoshizumi, T.; Yang, Z.; Yang, L.; Liu, Q.; Liu, W.; Han, Y.J.; et al. Photoactivation and inactivation of arabidopsis cryptochrome 2. Science 2016, 354, 343–347. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zuo, Z.; Liu, H.; Liu, X.; Lin, C. Arabidopsis cryptochrome 1 interacts with spa1 to suppress cop1 activity in response to blue light. Genes Dev. 2011, 25, 1029–1034. [Google Scholar] [CrossRef] [PubMed]
- He, Z.; Zhao, X.; Kong, F.; Zuo, Z.; Liu, X. Tcp2 positively regulates hy5/hyh and photomorphogenesis in arabidopsis. J. Exp. Bot. 2016, 67, 775–785. [Google Scholar] [CrossRef]
- Pierik, R.; Ballaré, C.L. Control of plant growth and defense by photoreceptors: From mechanisms to opportunities in agriculture. Mol. Plant 2021, 14, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Fortunato, A.E.; Annunziata, R.; Jaubert, M.; Bouly, J.P.; Falciatore, A. Dealing with light: The widespread and multitasking cryptochrome/photolyase family in photosynthetic organisms. J. Plant Physiol. 2015, 172, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Boccalandro, H.E.; Giordano, C.V.; Ploschuk, E.L.; Piccoli, P.N.; Bottini, R.; Casal, J.J. Phototropins but not cryptochromes mediate the blue light-specific promotion of stomatal conductance, while both enhance photosynthesis and transpiration under full sunlight. Plant Physiol. 2012, 158, 1475–1484. [Google Scholar] [CrossRef]
- Tsunoyama, Y.; Morikawa, K.; Shiina, T.; Toyoshima, Y. Blue light specific and differential expression of a plastid sigma factor, sig5 in arabidopsis thaliana. FEBS Lett. 2002, 516, 225–228. [Google Scholar] [CrossRef]
- Thum, K.E.; Kim, M.; Christopher, D.A.; Mullet, J.E. Cryptochrome 1, cryptochrome 2, and phytochrome a co-activate the chloroplast psbd blue light-responsive promoter. Plant Cell 2001, 13, 2747–2760. [Google Scholar] [PubMed]
- Mochizuki, T.; Onda, Y.; Fujiwara, E.; Wada, M.; Toyoshima, Y. Two independent light signals cooperate in the activation of the plastid psbd blue light-responsive promoter in arabidopsis. FEBS Lett. 2004, 571, 26–30. [Google Scholar] [CrossRef]
- Barneche, F.; Winter, V.; Crèvecoeur, M.; Rochaix, J.-D. Atab2 is a novel factor in the signalling pathway of light-controlled synthesis of photosystem proteins. EMBO J. 2006, 25, 5907–5918. [Google Scholar] [CrossRef]
- Lopez, L.; Carbone, F.; Bianco, L.; Giuliano, G.; Facella, P.; Perrotta, G. Tomato plants overexpressing cryptochrome 2 reveal altered expression of energy and stress-related gene products in response to diurnal cues. Plant Cell Environ. 2012, 35, 994–1012. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Hu, C.; Liu, C.; Wang, J.; Zhou, Y.; Yu, J. Elongated hypocotyl 5 mediates blue light-induced starch degradation in tomato. J. Exp. Bot. 2021, 72, 2627–2641. [Google Scholar] [CrossRef]
- Koide, E.; Suetsugu, N.; Iwano, M.; Gotoh, E.; Nomura, Y.; Stolze, S.C.; Nakagami, H.; Kohchi, T.; Nishihama, R. Regulation of photosynthetic carbohydrate metabolism by a raf-like kinase in the liverwort marchantia polymorpha. Plant Cell Physiol. 2019, 61, 631–643. [Google Scholar] [CrossRef]
- Griffin, J.H.C.; Prado, K.; Sutton, P.; Toledo-Ortiz, G. Coordinating light responses between the nucleus and the chloroplast, a role for plant cryptochromes and phytochromes. Physiol. Plant. 2020, 169, 515–528. [Google Scholar] [CrossRef] [PubMed]
- Buccitelli, C.; Selbach, M. Mrnas, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.L.; Jin, Y.; Yang, Y.J.; Li, G.J.; Boyer, J.S. Sugar input, metabolism, and signaling mediated by invertase: Roles in development, yield potential, and response to drought and heat. Mol. Plant 2010, 3, 942–955. [Google Scholar] [CrossRef] [PubMed]

| TF | Background All | Background _Bind | Query_All | Query_Bind | p_Value | q_Value | Best Hit in Arabidopsis thaliana | Description |
|---|---|---|---|---|---|---|---|---|
| Mp7g09490 | 19,287 | 404 | 1086 | 43 | 2.19 × 10−5 | 1.78 × 10−3 | AT1G69690 | TCP family protein |
| Mp8g03820 | 19,287 | 490 | 1086 | 48 | 7.06 × 10−5 | 2.53 × 10−3 | AT5G28770 | bZIP family protein |
| Mp2g23600 | 19,287 | 604 | 1086 | 56 | 9.36 × 10−5 | 2.53 × 10−3 | AT1G49720 | abscisic acid responsive element-binding factor 1 |
| Mp6g03920 | 19,287 | 484 | 1086 | 41 | 3.64 × 10−3 | 7.38 × 10−2 | AT1G75390 | basic leucine-zipper 44 |
| Mp6g18650 | 19,287 | 571 | 1086 | 45 | 9.36 × 10−3 | 1.52 × 10−1 | AT5G19790 | related to AP2 11 |
| Mp1g13640 | 19,287 | 103 | 1086 | 11 | 1.30 × 10−2 | 1.57 × 10−1 | AT1G20980 | squamosa promoter binding protein-like 14 |
| Mp5g06970 | 19,287 | 436 | 1086 | 35 | 1.41 × 10−2 | 1.57 × 10−1 | AT5G13910 | ERF family protein |
| Mp2g07170 | 19,287 | 503 | 1086 | 39 | 1.77 × 10−2 | 1.57 × 10−1 | AT2G46270 | G-box binding factor 3 |
| Mp5g12480 | 19,287 | 520 | 1086 | 40 | 1.88 × 10−2 | 1.57 × 10−1 | AT1G72360 | ERF family protein |
| Mp4g14530 | 19,287 | 162 | 1086 | 15 | 2.06 × 10−2 | 1.57 × 10−1 | AT1G66810 | C3H family protein |
| Mp5g21080 | 19,287 | 1391 | 1086 | 95 | 2.13 × 10−2 | 1.57 × 10−1 | AT5G63090 | LBD family protein |
| Mp6g04830 | 19,287 | 239 | 1086 | 20 | 2.92 × 10−2 | 1.70 × 10−1 | AT3G49690 | myb domain protein 84 |
| Mp2g02230 | 19,287 | 415 | 1086 | 32 | 2.92 × 10−2 | 1.70 × 10−1 | AT2G40950 | bZIP family protein |
| Mp2g13020 | 19,287 | 225 | 1086 | 19 | 2.94 × 10−2 | 1.70 × 10−1 | AT5G08520 | MYB family protein |
| Mp3g17350 | 19,287 | 359 | 1086 | 28 | 3.27 × 10−2 | 1.77 × 10−1 | AT1G09530 | phytochrome interacting factor 3 |
| Mp7g02640 | 19,287 | 274 | 1086 | 22 | 3.67 × 10−2 | 1.86 × 10−1 | AT3G20770 | EIL family protein |
| Mp1g25020 | 19,287 | 3001 | 1086 | 189 | 4.00 × 10−2 | 1.91 × 10−1 | AT1G72050 | transcription factor IIIA |
| Mp1g16800 | 19,287 | 400 | 1086 | 30 | 4.51 × 10−2 | 2.03 × 10−1 | AT5G11260 | bZIP family protein |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, T.; Zhang, L.; Su, S.; Li, S.; Zhang, J.; Yang, Z.; Zuo, Z. Roles of a Cryptochrome in Carbon Fixation and Sucrose Metabolism in the Liverwort Marchantia polymorpha. Cells 2021, 10, 3387. https://doi.org/10.3390/cells10123387
Li T, Zhang L, Su S, Li S, Zhang J, Yang Z, Zuo Z. Roles of a Cryptochrome in Carbon Fixation and Sucrose Metabolism in the Liverwort Marchantia polymorpha. Cells. 2021; 10(12):3387. https://doi.org/10.3390/cells10123387
Chicago/Turabian StyleLi, Tianhong, Li Zhang, Shengzhong Su, Sudi Li, Junchuan Zhang, Zhenming Yang, and Zecheng Zuo. 2021. "Roles of a Cryptochrome in Carbon Fixation and Sucrose Metabolism in the Liverwort Marchantia polymorpha" Cells 10, no. 12: 3387. https://doi.org/10.3390/cells10123387
APA StyleLi, T., Zhang, L., Su, S., Li, S., Zhang, J., Yang, Z., & Zuo, Z. (2021). Roles of a Cryptochrome in Carbon Fixation and Sucrose Metabolism in the Liverwort Marchantia polymorpha. Cells, 10(12), 3387. https://doi.org/10.3390/cells10123387





