MicroRNA-214 in Health and Disease
Abstract
1. Introduction
2. MicroRNAs
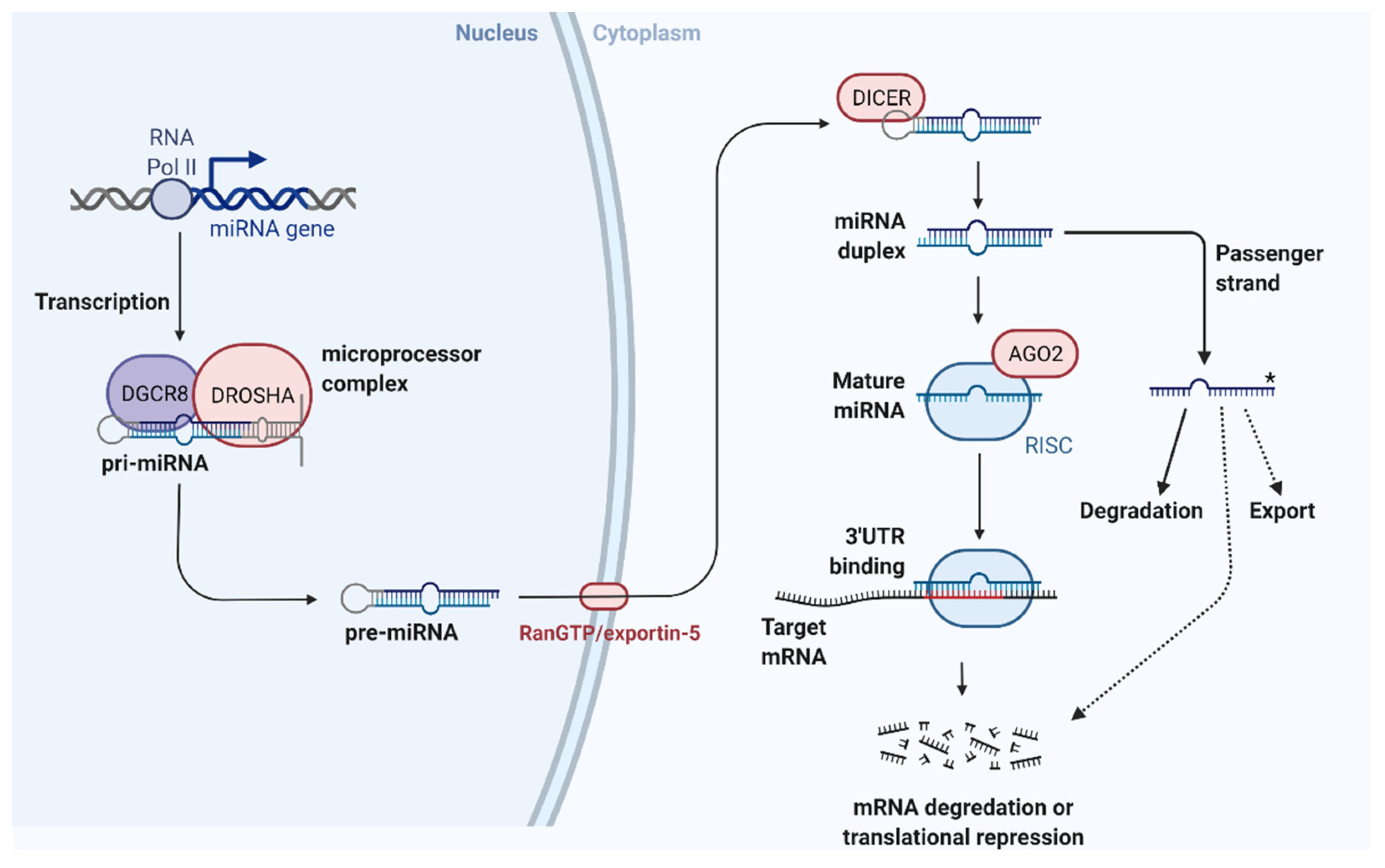
2.1. Biogenesis and Action of MicroRNAs
2.2. Investigating MicroRNAs
2.3. MicroRNA-214
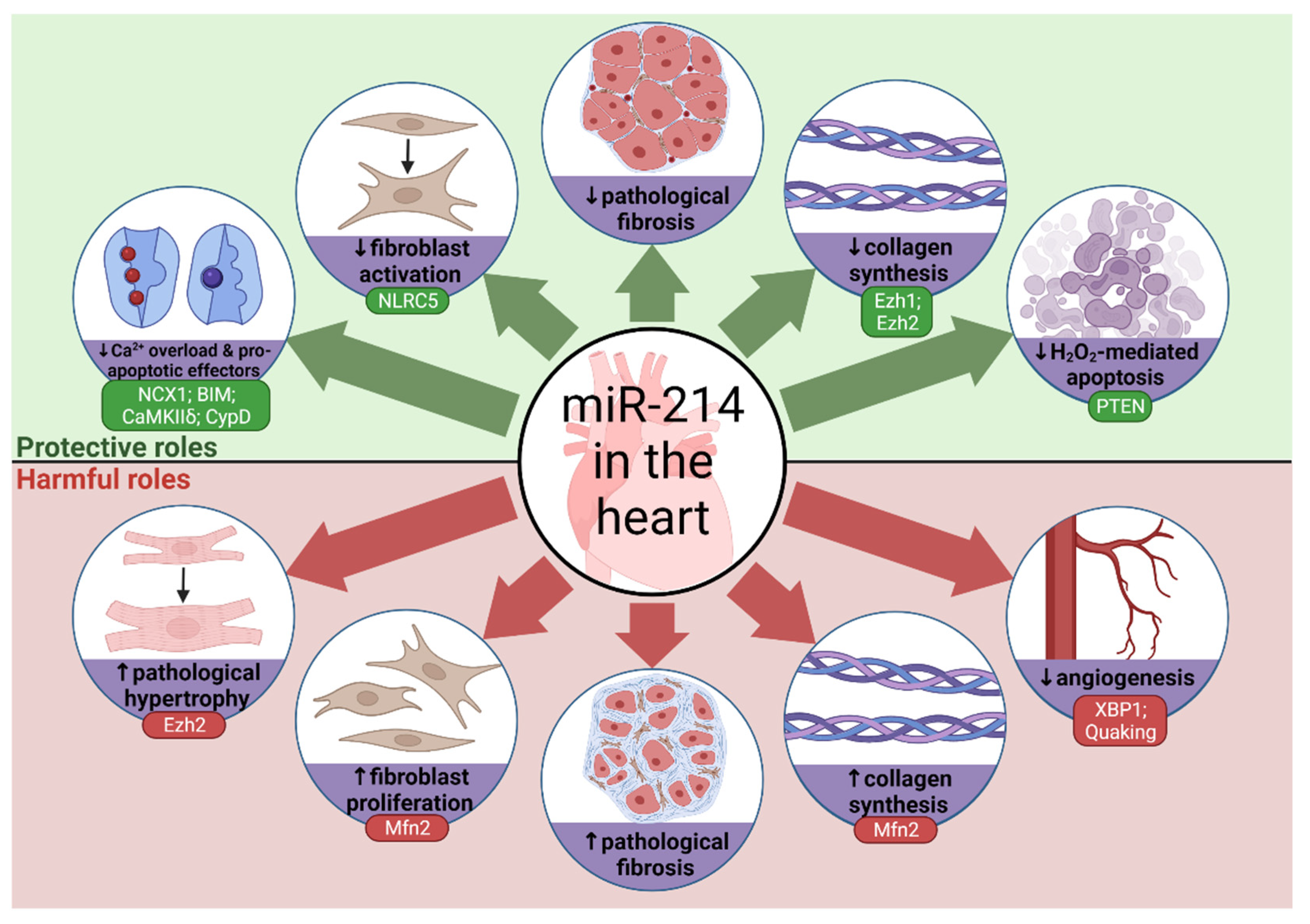
3. MicroRNA-214 in Cardiovascular Physiology and Pathophysiology
3.1. Calcium Overload
3.2. Reactive Oxygen Species
3.3. Hypertrophy and Angiogenesis
3.4. Fibrosis
4. MicroRNA-214 in Cancer Progression
4.1. Gastric Cancer
4.2. Liver Cancer
4.3. Ovarian Cancer
4.4. Cervical Cancer
4.5. Skin Cancer
4.6. Lung Cancer
4.7. Breast Cancer
5. MicroRNA-214 in Bone Formation
5.1. Osteoblasts
5.2. Osteoclasts
5.3. Therapeutics
6. MicroRNA-214 in Cell Differentiation
6.1. Muscle Cells
6.2. Neuronal Cells
6.3. T Cells
7. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Consortium, E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Beermann, J.; Piccoli, M.T.; Viereck, J.; Thum, T. Non-coding RNAs in Development and Disease: Background, Mechanisms, and Therapeutic Approaches. Physiol. Rev. 2016, 96, 1297–1325. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402. [Google Scholar] [CrossRef]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Alles, J.; Fehlmann, T.; Fischer, U.; Backes, C.; Galata, V.; Minet, M.; Hart, M.; Abu-Halima, M.; Grässer, F.A.; Lenhof, H.P.; et al. An estimate of the total number of true human miRNAs. Nucleic Acids Res. 2019, 47, 3353–3364. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–d162. [Google Scholar] [CrossRef] [PubMed]
- Baek, D.; Villén, J.; Shin, C.; Camargo, F.D.; Gygi, S.P.; Bartel, D.P. The impact of microRNAs on protein output. Nature 2008, 455, 64–71. [Google Scholar] [CrossRef] [PubMed]
- Tang, F.; Kaneda, M.; O’Carroll, D.; Hajkova, P.; Barton, S.C.; Sun, Y.A.; Lee, C.; Tarakhovsky, A.; Lao, K.; Surani, M.A. Maternal microRNAs are essential for mouse zygotic development. Genes Dev. 2007, 21, 644–648. [Google Scholar] [CrossRef]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.M.; Byrom, M.W.; Shelton, J.; Ford, L.P. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005, 33, 1290–1297. [Google Scholar] [CrossRef]
- Zhao, Y.; Ponnusamy, M.; Zhang, L.; Zhang, Y.; Liu, C.; Yu, W.; Wang, K.; Li, P. The role of miR-214 in cardiovascular diseases. Eur. J. Pharm. 2017, 816, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Penna, E.; Orso, F.; Taverna, D. miR-214 as a key hub that controls cancer networks: Small player, multiple functions. J. Investig. Dermatol. 2015, 135, 960–969. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Kuek, V.; Liu, Y.; Tickner, J.; Yuan, Y.; Chen, L.; Zeng, Z.; Shao, M.; He, W.; Xu, J. MiR-214 is an important regulator of the musculoskeletal metabolism and disease. J. Cell. Physiol. 2018, 234, 231–245. [Google Scholar] [CrossRef]
- Aurora, A.B.; Mahmoud, A.I.; Luo, X.; Johnson, B.A.; van Rooij, E.; Matsuzaki, S.; Humphries, K.M.; Hill, J.A.; Bassel-Duby, R.; Sadek, H.A.; et al. MicroRNA-214 protects the mouse heart from ischemic injury by controlling Ca2+ overload and cell death. J. Clin. Investig. 2012, 122, 1222–1232. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Shao, S.; Dong, H.; Bian, X.; Yang, X.; Dong, S. MicroRNA-214 protects cardiac myocytes against H2O2-induced injury. J. Cell. Biochem. 2014, 115, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Duan, Q.; Yang, L.; Gong, W.; Chaugai, S.; Wang, F.; Chen, C.; Wang, P.; Zou, M.H.; Wang, D.W. MicroRNA-214 Is Upregulated in Heart Failure Patients and Suppresses XBP1-Mediated Endothelial Cells Angiogenesis. J. Cell. Physiol. 2015, 230, 1964–1973. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Yu, H.; Zhang, Y.; Li, Z.; Gao, W. MicroRNA-214 mediates isoproterenol-induced proliferation and collagen synthesis in cardiac fibroblasts. Sci. Rep. 2015, 5, 18351. [Google Scholar] [CrossRef] [PubMed]
- Bageghni, S.A.; Hemmings, K.E.; Zava, N.; Denton, C.P.; Porter, K.E.; Ainscough, J.F.X.; Drinkhill, M.J.; Turner, N.A. Cardiac fibroblast-specific p38α MAP kinase promotes cardiac hypertrophy via a putative paracrine interleukin-6 signaling mechanism. FASEB J. 2018, 32, 4941–4954. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.S.; Tang, C.M.; Xiao, Z.; Zhu, J.N.; Lin, Q.X.; Fu, Y.H.; Hu, Z.Q.; Zhang, Z.; Yang, M.; Zheng, X.L.; et al. Targeting EZH1 and EZH2 contributes to the suppression of fibrosis-associated genes by miR-214-3p in cardiac myofibroblasts. Oncotarget 2016, 7, 78331–78342. [Google Scholar] [CrossRef] [PubMed]
- Dong, H.; Dong, S.; Zhang, L.; Gao, X.; Lv, G.; Chen, W.; Shao, S. MicroRNA-214 exerts a Cardio-protective effect by inhibition of fibrosis. Anat. Rec. (Hoboken N. J. 2007) 2016, 299, 1348–1357. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Shi, J.; Hu, Z.; Hu, X. The deficiency of miR-214-3p exacerbates cardiac fibrosis via miR-214-3p/NLRC5 axis. Clin. Sci. (Lond. Engl. 1979) 2019, 133, 1845–1856. [Google Scholar] [CrossRef]
- Yang, T.S.; Yang, X.H.; Wang, X.D.; Wang, Y.L.; Zhou, B.; Song, Z.S. MiR-214 regulate gastric cancer cell proliferation, migration and invasion by targeting PTEN. Cancer Cell Int. 2013, 13, 68. [Google Scholar] [CrossRef]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef]
- Penna, E.; Orso, F.; Cimino, D.; Tenaglia, E.; Lembo, A.; Quaglino, E.; Poliseno, L.; Haimovic, A.; Osella-Abate, S.; De Pittà, C.; et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011, 30, 1990–2007. [Google Scholar] [CrossRef]
- Wang, Y.S.; Wang, Y.H.; Xia, H.P.; Zhou, S.W.; Schmid-Bindert, G.; Zhou, C.C. MicroRNA-214 regulates the acquired resistance to gefitinib via the PTEN/AKT pathway in EGFR-mutant cell lines. Asian Pac. J. Cancer Prev. 2012, 13, 255–260. [Google Scholar] [CrossRef]
- Yang, Z.; Chen, S.; Luan, X.; Li, Y.; Liu, M.; Li, X.; Liu, T.; Tang, H. MicroRNA-214 is aberrantly expressed in cervical cancers and inhibits the growth of HeLa cells. IUBMB Life 2009, 61, 1075–1082. [Google Scholar] [CrossRef]
- Zhang, X.J.; Ye, H.; Zeng, C.W.; He, B.; Zhang, H.; Chen, Y.Q. Dysregulation of miR-15a and miR-214 in human pancreatic cancer. J. Hematol. Oncol. 2010, 3, 46. [Google Scholar] [CrossRef]
- Misiewicz-Krzeminska, I.; Sarasquete, M.E.; Quwaider, D.; Krzeminski, P.; Ticona, F.V.; Paíno, T.; Delgado, M.; Aires, A.; Ocio, E.M.; García-Sanz, R.; et al. Restoration of microRNA-214 expression reduces growth of myeloma cells through positive regulation of P53 and inhibition of DNA replication. Haematologica 2013, 98, 640–648. [Google Scholar] [CrossRef]
- Deng, M.; Ye, Q.; Qin, Z.; Zheng, Y.; He, W.; Tang, H.; Zhou, Y.; Xiong, W.; Zhou, M.; Li, X.; et al. miR-214 promotes tumorigenesis by targeting lactotransferrin in nasopharyngeal carcinoma. Tumour Biol. 2013, 34, 1793–1800. [Google Scholar] [CrossRef]
- Wang, X.; Guo, B.; Li, Q.; Peng, J.; Yang, Z.; Wang, A.; Li, D.; Hou, Z.; Lv, K.; Kan, G.; et al. miR-214 targets ATF4 to inhibit bone formation. Nat. Med. 2013, 19, 93–100. [Google Scholar] [CrossRef]
- Juan, A.H.; Kumar, R.M.; Marx, J.G.; Young, R.A.; Sartorelli, V. Mir-214-dependent regulation of the polycomb protein Ezh2 in skeletal muscle and embryonic stem cells. Mol. Cell 2009, 36, 61–74. [Google Scholar] [CrossRef]
- Wu, Y.; Li, Z.; Yang, M.; Dai, B.; Hu, F.; Yang, F.; Zhu, J.; Chen, T.; Zhang, L. MicroRNA-214 regulates smooth muscle cell differentiation from stem cells by targeting RNA-binding protein QKI. Oncotarget 2017, 8, 19866–19878. [Google Scholar] [CrossRef]
- Yin, Y.; Cai, X.; Chen, X.; Liang, H.; Zhang, Y.; Li, J.; Wang, Z.; Chen, X.; Zhang, W.; Yokoyama, S.; et al. Tumor-secreted miR-214 induces regulatory T cells: A major link between immune evasion and tumor growth. Cell Res. 2014, 24, 1164–1180. [Google Scholar] [CrossRef]
- Derfoul, A.; Juan, A.H.; Difilippantonio, M.J.; Palanisamy, N.; Ried, T.; Sartorelli, V. Decreased microRNA-214 levels in breast cancer cells coincides with increased cell proliferation, invasion and accumulation of the Polycomb Ezh2 methyltransferase. Carcinogenesis 2011, 32, 1607–1614. [Google Scholar] [CrossRef]
- Wang, F.; Li, L.; Chen, Z.; Zhu, M.; Gu, Y. MicroRNA-214 acts as a potential oncogene in breast cancer by targeting the PTEN-PI3K/Akt signaling pathway. Int. J. Mol. Med. 2016, 37, 1421–1428. [Google Scholar] [CrossRef]
- Dangwal, S.; Bang, C.; Thum, T. Novel techniques and targets in cardiovascular microRNA research. Cardiovasc. Res. 2012, 93, 545–554. [Google Scholar] [CrossRef]
- Yang, T.; Gu, H.; Chen, X.; Fu, S.; Wang, C.; Xu, H.; Feng, Q.; Ni, Y. Cardiac hypertrophy and dysfunction induced by overexpression of miR-214 in vivo. J. Surg. Res 2014, 192, 317–325. [Google Scholar] [CrossRef]
- Orso, F.; Quirico, L.; Virga, F.; Penna, E.; Dettori, D.; Cimino, D.; Coppo, R.; Grassi, E.; Elia, A.R.; Brusa, D.; et al. miR-214 and miR-148b Targeting Inhibits Dissemination of Melanoma and Breast Cancer. Cancer Res. 2016, 76, 5151–5162. [Google Scholar] [CrossRef]
- Li, K.C.; Chang, Y.H.; Yeh, C.L.; Hu, Y.C. Healing of osteoporotic bone defects by baculovirus-engineered bone marrow-derived MSCs expressing MicroRNA sponges. Biomaterials 2016, 74, 155–166. [Google Scholar] [CrossRef]
- Schwarzenbach, H.; Milde-Langosch, K.; Steinbach, B.; Müller, V.; Pantel, K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res. Treat. 2012, 134, 933–941. [Google Scholar] [CrossRef]
- Lu, H.Q.; Liang, C.; He, Z.Q.; Fan, M.; Wu, Z.G. Circulating miR-214 is associated with the severity of coronary artery disease. J. Geriatr. Cardiol. 2013, 10, 34–38. [Google Scholar]
- Ruby, J.G.; Jan, C.H.; Bartel, D.P. Intronic microRNA precursors that bypass Drosha processing. Nature 2007, 448, 83–86. [Google Scholar] [CrossRef]
- Yang, J.S.; Maurin, T.; Robine, N.; Rasmussen, K.D.; Jeffrey, K.L.; Chandwani, R.; Papapetrou, E.P.; Sadelain, M.; O’Carroll, D.; Lai, E.C. Conserved vertebrate mir-451 provides a platform for Dicer-independent, Ago2-mediated microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2010, 107, 15163–15168. [Google Scholar] [CrossRef]
- Lee, I.; Ajay, S.S.; Yook, J.I.; Kim, H.S.; Hong, S.H.; Kim, N.H.; Dhanasekaran, S.M.; Chinnaiyan, A.M.; Athey, B.D. New class of microRNA targets containing simultaneous 5’-UTR and 3’-UTR interaction sites. Genome Res. 2009, 19, 1175–1183. [Google Scholar] [CrossRef]
- van Rooij, E. The art of microRNA research. Circ. Res. 2011, 108, 219–234. [Google Scholar] [CrossRef]
- Huang, X.Z.; Huang, J.; Li, W.Z.; Wang, J.J.; Song, D.Y.; Ni, J.D. LncRNA-MALAT1 promotes osteogenic differentiation through regulating ATF4 by sponging miR-214: Implication of steroid-induced avascular necrosis of the femoral head. Steroids 2020, 154, 108533. [Google Scholar] [CrossRef]
- Feng, Y.; Wan, P.; Yin, L. Long Noncoding RNA X-Inactive Specific Transcript (XIST) Promotes Osteogenic Differentiation of Periodontal Ligament Stem Cells by Sponging MicroRNA-214-3p. Med. Sci. Monit. 2020, 26, e918932. [Google Scholar] [CrossRef]
- Loebel, D.A.; Tsoi, B.; Wong, N.; Tam, P.P. A conserved noncoding intronic transcript at the mouse Dnm3 locus. Genomics 2005, 85, 782–789. [Google Scholar] [CrossRef]
- Watanabe, T.; Sato, T.; Amano, T.; Kawamura, Y.; Kawamura, N.; Kawaguchi, H.; Yamashita, N.; Kurihara, H.; Nakaoka, T. Dnm3os, a non-coding RNA, is required for normal growth and skeletal development in mice. Dev. Dyn. 2008, 237, 3738–3748. [Google Scholar] [CrossRef]
- Yin, G.; Chen, R.; Alvero, A.B.; Fu, H.H.; Holmberg, J.; Glackin, C.; Rutherford, T.; Mor, G. TWISTing stemness, inflammation and proliferation of epithelial ovarian cancer cells through MIR199A2/214. Oncogene 2010, 29, 3545–3553. [Google Scholar] [CrossRef]
- Lee, Y.B.; Bantounas, I.; Lee, D.Y.; Phylactou, L.; Caldwell, M.A.; Uney, J.B. Twist-1 regulates the miR-199a/214 cluster during development. Nucleic Acids Res. 2009, 37, 123–128. [Google Scholar] [CrossRef]
- Li, H.; Wang, H.; Ren, Z. MicroRNA-214-5p Inhibits the Invasion and Migration of Hepatocellular Carcinoma Cells by Targeting Wiskott-Aldrich Syndrome Like. Cell. Physiol. Biochem. 2018, 46, 757–764. [Google Scholar] [CrossRef]
- Cao, T.H.; Ling, X.; Chen, C.; Tang, W.; Hu, D.M.; Yin, G.J. Role of miR-214-5p in the migration and invasion of pancreatic cancer cells. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 7214–7221. [Google Scholar]
- Iizuka, M.; Ogawa, T.; Enomoto, M.; Motoyama, H.; Yoshizato, K.; Ikeda, K.; Kawada, N. Induction of microRNA-214-5p in human and rodent liver fibrosis. Fibrogenesis Tissue Repair 2012, 5, 12. [Google Scholar] [CrossRef]
- Li, D.; Liu, J.; Guo, B.; Liang, C.; Dang, L.; Lu, C.; He, X.; Cheung, H.Y.; Xu, L.; Lu, C.; et al. Osteoclast-derived exosomal miR-214-3p inhibits osteoblastic bone formation. Nat. Commun. 2016, 7, 10872. [Google Scholar] [CrossRef]
- Feng, Y.; Cao, J.H.; Li, X.Y.; Zhao, S.H. Inhibition of miR-214 expression represses proliferation and differentiation of C2C12 myoblasts. Cell Biochem. Funct. 2011, 29, 378–383. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Li, S.; Dong, L.; Li, Y.; Mao, Y.; Liang, Y.; Tao, Y.; Ma, J. Inhibition of miR-214 attenuates the migration and invasion of triple-negative breast cancer cells. Mol. Med. Rep. 2019, 19, 4035–4042. [Google Scholar] [CrossRef]
- Yao, S.; Zhao, W.; Ou, Q.; Liang, L.; Lin, X.; Wang, Y. MicroRNA-214 Suppresses Osteogenic Differentiation of Human Periodontal Ligament Stem Cells by Targeting ATF4. Stem Cells Int. 2017, 2017, 3028647. [Google Scholar] [CrossRef]
- Wang, X.; Chen, J.; Li, F.; Lin, Y.; Zhang, X.; Lv, Z.; Jiang, J. MiR-214 inhibits cell growth in hepatocellular carcinoma through suppression of β-catenin. Biochem. Biophys. Res. Commun. 2012, 428, 525–531. [Google Scholar] [CrossRef]
- Yi, S.J.; Li, L.L.; Tu, W.B. MiR-214 negatively regulates proliferation and WNT/β-catenin signaling in breast cancer. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 5148–5154. [Google Scholar]
- Li, J.P.; Zhuang, H.T.; Xin, M.Y.; Zhou, Y.L. MiR-214 inhibits human mesenchymal stem cells differentiating into osteoblasts through targeting β-catenin. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 4777–4783. [Google Scholar]
- Wang, F.; Liu, M.; Li, X.; Tang, H. MiR-214 reduces cell survival and enhances cisplatin-induced cytotoxicity via down-regulation of Bcl2l2 in cervical cancer cells. FEBS Lett. 2013, 587, 488–495. [Google Scholar] [CrossRef]
- Yang, Y.; Liu, Y.; Li, G.; Li, L.; Geng, P.; Song, H. microRNA-214 suppresses the growth of cervical cancer cells by targeting EZH2. Oncol. Lett. 2018, 16, 5679–5686. [Google Scholar] [CrossRef]
- Wang, J.; Li, J.; Wang, X.; Zheng, C.; Ma, W. Downregulation of microRNA-214 and overexpression of FGFR-1 contribute to hepatocellular carcinoma metastasis. Biochem. Biophys. Res. Commun. 2013, 439, 47–53. [Google Scholar] [CrossRef]
- Yang, L.; Ge, D.; Cao, X.; Ge, Y.; Chen, H.; Wang, W.; Zhang, H. MiR-214 Attenuates Osteogenic Differentiation of Mesenchymal Stem Cells via Targeting FGFR1. Cell. Physiol. Biochem. 2016, 38, 809–820. [Google Scholar] [CrossRef]
- Wang, J.M.; Ju, B.H.; Pan, C.J.; Gu, Y.; Li, M.Q.; Sun, L.; Xu, Y.Y.; Yin, L.R. MiR-214 inhibits cell migration, invasion and promotes the drug sensitivity in human cervical cancer by targeting FOXM1. Am. J. Transl Res. 2017, 9, 3541–3557. [Google Scholar]
- Shih, T.C.; Tien, Y.J.; Wen, C.J.; Yeh, T.S.; Yu, M.C.; Huang, C.H.; Lee, Y.S.; Yen, T.C.; Hsieh, S.Y. MicroRNA-214 downregulation contributes to tumor angiogenesis by inducing secretion of the hepatoma-derived growth factor in human hepatoma. J. Hepatol. 2012, 57, 584–591. [Google Scholar] [CrossRef]
- Chandrasekaran, K.S.; Sathyanarayanan, A.; Karunagaran, D. MicroRNA-214 suppresses growth, migration and invasion through a novel target, high mobility group AT-hook 1, in human cervical and colorectal cancer cells. Br. J. Cancer 2016, 115, 741–751. [Google Scholar] [CrossRef]
- Liu, J.; Luo, X.J.; Xiong, A.W.; Zhang, Z.D.; Yue, S.; Zhu, M.S.; Cheng, S.Y. MicroRNA-214 promotes myogenic differentiation by facilitating exit from mitosis via down-regulation of proto-oncogene N-ras. J. Biol. Chem. 2010, 285, 26599–26607. [Google Scholar] [CrossRef]
- Shi, K.; Lu, J.; Zhao, Y.; Wang, L.; Li, J.; Qi, B.; Li, H.; Ma, C. MicroRNA-214 suppresses osteogenic differentiation of C2C12 myoblast cells by targeting Osterix. Bone 2013, 55, 487–494. [Google Scholar] [CrossRef]
- Wang, F.; Lv, P.; Liu, X.; Zhu, M.; Qiu, X. microRNA-214 enhances the invasion ability of breast cancer cells by targeting p53. Int. J. Mol. Med. 2015, 35, 1395–1402. [Google Scholar] [CrossRef]
- Qiang, R.; Wang, F.; Shi, L.Y.; Liu, M.; Chen, S.; Wan, H.Y.; Li, Y.X.; Li, X.; Gao, S.Y.; Sun, B.C.; et al. Plexin-B1 is a target of miR-214 in cervical cancer and promotes the growth and invasion of HeLa cells. Int. J. Biochem. Cell Biol. 2011, 43, 632–641. [Google Scholar] [CrossRef] [PubMed]
- Xin, R.; Bai, F.; Feng, Y.; Jiu, M.; Liu, X.; Bai, F.; Nie, Y.; Fan, D. MicroRNA-214 promotes peritoneal metastasis through regulating PTEN negatively in gastric cancer. Clin. Res. Hepatol. Gastroenterol. 2016, 40, 748–754. [Google Scholar] [CrossRef]
- Zhang, K.; Zhang, M.; Jiang, H.; Liu, F.; Liu, H.; Li, Y. Down-regulation of miR-214 inhibits proliferation and glycolysis in non-small-cell lung cancer cells via down-regulating the expression of hexokinase 2 and pyruvate kinase isozyme M2. Biomed. Pharmacother. 2018, 105, 545–552. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Sun, W.; Zhang, P.; Ling, S.; Li, Y.; Zhao, D.; Peng, J.; Wang, A.; Li, Q.; Song, J.; et al. miR-214 promotes osteoclastogenesis by targeting Pten/PI3k/Akt pathway. RNA Biol. 2015, 12, 343–353. [Google Scholar] [CrossRef]
- Jindra, P.T.; Bagley, J.; Godwin, J.G.; Iacomini, J. Costimulation-dependent expression of microRNA-214 increases the ability of T cells to proliferate by targeting Pten. J. Immunol. 2010, 185, 990–997. [Google Scholar] [CrossRef] [PubMed]
- van Mil, A.; Grundmann, S.; Goumans, M.J.; Lei, Z.; Oerlemans, M.I.; Jaksani, S.; Doevendans, P.A.; Sluijter, J.P. MicroRNA-214 inhibits angiogenesis by targeting Quaking and reducing angiogenic growth factor release. Cardiovasc. Res. 2012, 93, 655–665. [Google Scholar] [CrossRef] [PubMed]
- Shu, P.; Fu, H.; Zhao, X.; Wu, C.; Ruan, X.; Zeng, Y.; Liu, W.; Wang, M.; Hou, L.; Chen, P.; et al. MicroRNA-214 modulates neural progenitor cell differentiation by targeting Quaking during cerebral cortex development. Sci. Rep. 2017, 7, 8014. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Su, B.; Gong, C.; Xi, Q.; Chao, T. miR-214 promotes apoptosis and sensitizes breast cancer cells to doxorubicin by targeting the RFWD2-p53 cascade. Biochem. Biophys. Res. Commun. 2016, 478, 337–342. [Google Scholar] [CrossRef] [PubMed]
- Min, L.; Liu, C.; Kuang, J.; Wu, X.; Zhu, L. miR-214 inhibits epithelial-mesenchymal transition of breast cancer cells via downregulation of RNF8. Acta Biochim. Biophys. Sin. 2019, 51, 791–798. [Google Scholar] [CrossRef]
- Wang, Z.; Yin, H.; Zhang, Y.; Feng, Y.; Yan, Z.; Jiang, X.; Bukhari, I.; Iqbal, F.; Cooke, H.J.; Shi, Q. miR-214-mediated downregulation of RNF8 induces chromosomal instability in ovarian cancer cells. Cell Cycle 2014, 13, 3519–3528. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Qiu, Y.Y.; Zhang, Y.W.; Qian, X.F.; Bian, T. miR-371, miR-138, miR-544, miR-145, and miR-214 could modulate Th1/Th2 balance in asthma through the combinatorial regulation of Runx3. Am. J. Transl. Res. 2017, 9, 3184–3199. [Google Scholar] [PubMed]
- Tao, Y.; Zhao, Z.; Ma, J.; Dong, L.; Liang, Y.; Li, S.; Mao, Y.; Li, Y.; Zhang, Y. MiR-214-3p regulates the viability, invasion, migration and EMT of TNBC cells by targeting ST6GAL1. Cytotechnology 2019, 71, 1155–1165. [Google Scholar] [CrossRef] [PubMed]
- Flynt, A.S.; Li, N.; Thatcher, E.J.; Solnica-Krezel, L.; Patton, J.G. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 2007, 39, 259–263. [Google Scholar] [CrossRef]
- He, S.; Yang, F.; Yang, M.; An, W.; Maguire, E.M.; Chen, Q.; Xiao, R.; Wu, W.; Zhang, L.; Wang, W.; et al. miR-214-3p-Sufu-GLI1 is a novel regulatory axis controlling inflammatory smooth muscle cell differentiation from stem cells and neointimal hyperplasia. Stem Cell Res. Ther. 2020, 11, 465. [Google Scholar] [CrossRef]
- Han, L.C.; Wang, H.; Niu, F.L.; Yan, J.Y.; Cai, H.F. Effect miR-214-3p on proliferation and apoptosis of breast cancer cells by targeting survivin protein. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 7469–7474. [Google Scholar]
- Liu, J.; Li, D.; Dang, L.; Liang, C.; Guo, B.; Lu, C.; He, X.; Cheung, H.Y.; He, B.; Liu, B.; et al. Osteoclastic miR-214 targets TRAF3 to contribute to osteolytic bone metastasis of breast cancer. Sci. Rep. 2017, 7, 40487. [Google Scholar] [CrossRef]
- Yu, X.; Luo, A.; Liu, Y.; Wang, S.; Li, Y.; Shi, W.; Liu, Z.; Qu, X. MiR-214 increases the sensitivity of breast cancer cells to tamoxifen and fulvestrant through inhibition of autophagy. Mol. Cancer 2015, 14, 208. [Google Scholar] [CrossRef]
- Duan, Q.; Wang, X.; Gong, W.; Ni, L.; Chen, C.; He, X.; Chen, F.; Yang, L.; Wang, P.; Wang, D.W. ER stress negatively modulates the expression of the miR-199a/214 cluster to regulates tumor survival and progression in human hepatocellular cancer. PLoS ONE 2012, 7, e31518. [Google Scholar] [CrossRef]
- Decembrini, S.; Bressan, D.; Vignali, R.; Pitto, L.; Mariotti, S.; Rainaldi, G.; Wang, X.; Evangelista, M.; Barsacchi, G.; Cremisi, F. MicroRNAs couple cell fate and developmental timing in retina. Proc. Natl. Acad. Sci. USA 2009, 106, 21179–21184. [Google Scholar] [CrossRef]
- Murphy, E.; Steenbergen, C. Mechanisms underlying acute protection from cardiac ischemia-reperfusion injury. Physiol. Rev. 2008, 88, 581–609. [Google Scholar] [CrossRef]
- Imahashi, K.; Pott, C.; Goldhaber, J.I.; Steenbergen, C.; Philipson, K.D.; Murphy, E. Cardiac-specific ablation of the Na+-Ca2+ exchanger confers protection against ischemia/reperfusion injury. Circ. Res. 2005, 97, 916–921. [Google Scholar] [CrossRef]
- Tani, M.; Neely, J.R. Role of intracellular Na+ in Ca2+ overload and depressed recovery of ventricular function of reperfused ischemic rat hearts. Possible involvement of H+-Na+ and Na+-Ca2+ exchange. Circ. Res. 1989, 65, 1045–1056. [Google Scholar] [CrossRef]
- Zhang, Z.C.; Li, Y.Y.; Wang, H.Y.; Fu, S.; Wang, X.P.; Zeng, M.S.; Zeng, Y.X.; Shao, J.Y. Knockdown of miR-214 promotes apoptosis and inhibits cell proliferation in nasopharyngeal carcinoma. PLoS ONE 2014, 9, e86149. [Google Scholar] [CrossRef]
- Vila-Petroff, M.; Salas, M.A.; Said, M.; Valverde, C.A.; Sapia, L.; Portiansky, E.; Hajjar, R.J.; Kranias, E.G.; Mundiña-Weilenmann, C.; Mattiazzi, A. CaMKII inhibition protects against necrosis and apoptosis in irreversible ischemia-reperfusion injury. Cardiovasc. Res. 2007, 73, 689–698. [Google Scholar] [CrossRef]
- Hampton, T.G.; Wang, J.F.; DeAngelis, J.; Amende, I.; Philipson, K.D.; Morgan, J.P. Enhanced gene expression of Na(+)/Ca(2+) exchanger attenuates ischemic and hypoxic contractile dysfunction. Am. J. Physiol. Heart Circ. Physiol. 2000, 279, H2846–H2854. [Google Scholar] [CrossRef]
- Studer, R.; Reinecke, H.; Bilger, J.; Eschenhagen, T.; Böhm, M.; Hasenfuss, G.; Just, H.; Holtz, J.; Drexler, H. Gene expression of the cardiac Na(+)-Ca2+ exchanger in end-stage human heart failure. Circ. Res. 1994, 75, 443–453. [Google Scholar] [CrossRef]
- Zhou, T.; Chuang, C.C.; Zuo, L. Molecular Characterization of Reactive Oxygen Species in Myocardial Ischemia-Reperfusion Injury. BioMed Res. Int. 2015, 2015, 864946. [Google Scholar] [CrossRef]
- Halestrap, A.P. What is the mitochondrial permeability transition pore? J. Mol. Cell. Cardiol. 2009, 46, 821–831. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Vazquez, E.J.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Production of reactive oxygen species by mitochondria: Central role of complex III. J. Biol. Chem. 2003, 278, 36027–36031. [Google Scholar] [CrossRef]
- Chen, Q.; Moghaddas, S.; Hoppel, C.L.; Lesnefsky, E.J. Ischemic defects in the electron transport chain increase the production of reactive oxygen species from isolated rat heart mitochondria. Am. J. Physiol. Cell Physiol. 2008, 294, C460–C466. [Google Scholar] [CrossRef]
- Zhu, X.; Zuo, L. Characterization of oxygen radical formation mechanism at early cardiac ischemia. Cell Death Dis. 2013, 4, e787. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, X.; Zhang, S.; Lin, Y.; Yang, J.; Zhang, C. MicroRNA-21 protects against the H(2)O(2)-induced injury on cardiac myocytes via its target gene PDCD4. J. Mol. Cell. Cardiol. 2009, 47, 5–14. [Google Scholar] [CrossRef]
- Maes, O.C.; An, J.; Sarojini, H.; Wang, E. Murine microRNAs implicated in liver functions and aging process. Mech. Ageing Dev. 2008, 129, 534–541. [Google Scholar] [CrossRef]
- Li, J.; Yen, C.; Liaw, D.; Podsypanina, K.; Bose, S.; Wang, S.I.; Puc, J.; Miliaresis, C.; Rodgers, L.; McCombie, R.; et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science 1997, 275, 1943–1947. [Google Scholar] [CrossRef]
- Tamura, M.; Gu, J.; Matsumoto, K.; Aota, S.; Parsons, R.; Yamada, K.M. Inhibition of cell migration, spreading, and focal adhesions by tumor suppressor PTEN. Science 1998, 280, 1614–1617. [Google Scholar] [CrossRef]
- Zheng, H.C.; Li, Y.L.; Sun, J.M.; Yang, X.F.; Li, X.H.; Jiang, W.G.; Zhang, Y.C.; Xin, Y. Growth, invasion, metastasis, differentiation, angiogenesis and apoptosis of gastric cancer regulated by expression of PTEN encoding products. World J. Gastroenterol. 2003, 9, 1662–1666. [Google Scholar] [CrossRef]
- Sayed, D.; Hong, C.; Chen, I.Y.; Lypowy, J.; Abdellatif, M. MicroRNAs play an essential role in the development of cardiac hypertrophy. Circ. Res. 2007, 100, 416–424. [Google Scholar] [CrossRef]
- Cheng, Y.; Ji, R.; Yue, J.; Yang, J.; Liu, X.; Chen, H.; Dean, D.B.; Zhang, C. MicroRNAs are aberrantly expressed in hypertrophic heart: Do they play a role in cardiac hypertrophy? Am. J. Pathol. 2007, 170, 1831–1840. [Google Scholar] [CrossRef]
- Oka, T.; Akazawa, H.; Naito, A.T.; Komuro, I. Angiogenesis and cardiac hypertrophy: Maintenance of cardiac function and causative roles in heart failure. Circ. Res. 2014, 114, 565–571. [Google Scholar] [CrossRef]
- Chan, L.S.; Yue, P.Y.; Mak, N.K.; Wong, R.N. Role of microRNA-214 in ginsenoside-Rg1-induced angiogenesis. Eur. J. Pharm. Sci. 2009, 38, 370–377. [Google Scholar] [CrossRef] [PubMed]
- Souders, C.A.; Bowers, S.L.; Baudino, T.A. Cardiac fibroblast: The renaissance cell. Circ. Res. 2009, 105, 1164–1176. [Google Scholar] [CrossRef] [PubMed]
- Porter, K.E.; Turner, N.A. Cardiac fibroblasts: At the heart of myocardial remodeling. Pharmacol. Ther. 2009, 123, 255–278. [Google Scholar] [CrossRef]
- Cleutjens, J.P.; Verluyten, M.J.; Smiths, J.F.; Daemen, M.J. Collagen remodeling after myocardial infarction in the rat heart. Am. J. Pathol. 1995, 147, 325–338. [Google Scholar]
- Kong, P.; Christia, P.; Frangogiannis, N.G. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef]
- Iwanaga, Y.; Aoyama, T.; Kihara, Y.; Onozawa, Y.; Yoneda, T.; Sasayama, S. Excessive activation of matrix metalloproteinases coincides with left ventricular remodeling during transition from hypertrophy to heart failure in hypertensive rats. J. Am. Coll. Cardiol. 2002, 39, 1384–1391. [Google Scholar] [CrossRef]
- Turner, N.A.; Das, A.; Warburton, P.; O’Regan, D.J.; Ball, S.G.; Porter, K.E. Interleukin-1α stimulates pro-inflammatory cytokine expression in human cardiac myofibroblasts. Am. J. Physiol. Heart Circ. Physiol. 2009, 297, H1117–H1127. [Google Scholar] [CrossRef]
- Turner, N.A.; Mughal, R.S.; Warburton, P.; O’Regan, D.J.; Ball, S.G.; Porter, K.E. Mechanism of TNFα-induced IL-1α, IL-1β and IL-6 expression in human cardiac fibroblasts: Effects of statins and thiazolidinediones. Cardiovasc. Res. 2007, 76, 81–90. [Google Scholar] [CrossRef] [PubMed]
- Siwik, D.A.; Chang, D.L.; Colucci, W.S. Interleukin-1β and tumor necrosis factor-α decrease collagen synthesis and increase matrix metalloproteinase activity in cardiac fibroblasts in vitro. Circ. Res. 2000, 86, 1259–1265. [Google Scholar] [CrossRef]
- Siwik, D.A.; Pagano, P.J.; Colucci, W.S. Oxidative stress regulates collagen synthesis and matrix metalloproteinase activity in cardiac fibroblasts. Am. J. Physiol. Cell Physiol. 2001, 280, C53–C60. [Google Scholar] [CrossRef]
- Hou, Y.; Sun, Y.; Shan, H.; Li, X.; Zhang, M.; Zhou, X.; Xing, S.; Sun, H.; Chu, W.; Qiao, G.; et al. β-adrenoceptor regulates miRNA expression in rat heart. Med. Sci. Monit. 2012, 18, Br309–Br314. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Xing, R.; Yang, C.; Tian, A.; Lv, Z.; Sun, N.; Gao, X.; Zhang, Y.; Li, Z. HIP-55/DBNL-dependent regulation of adrenergic receptor mediates the ERK1/2 proliferative pathway. Mol. Biosyst. 2014, 10, 1932–1939. [Google Scholar] [CrossRef] [PubMed]
- Bisognano, J.D.; Weinberger, H.D.; Bohlmeyer, T.J.; Pende, A.; Raynolds, M.V.; Sastravaha, A.; Roden, R.; Asano, K.; Blaxall, B.C.; Wu, S.C.; et al. Myocardial-directed overexpression of the human beta(1)-adrenergic receptor in transgenic mice. J. Mol. Cell. Cardiol. 2000, 32, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Denby, L.; Ramdas, V.; Lu, R.; Conway, B.R.; Grant, J.S.; Dickinson, B.; Aurora, A.B.; McClure, J.D.; Kipgen, D.; Delles, C.; et al. MicroRNA-214 antagonism protects against renal fibrosis. J. Am. Soc. Nephrol. 2014, 25, 65–80. [Google Scholar] [CrossRef]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef] [PubMed]
- Rawla, P.; Barsouk, A. Epidemiology of gastric cancer: Global trends, risk factors and prevention. Prz. Gastroenterol. 2019, 14, 26–38. [Google Scholar] [CrossRef]
- Ueda, T.; Volinia, S.; Okumura, H.; Shimizu, M.; Taccioli, C.; Rossi, S.; Alder, H.; Liu, C.G.; Oue, N.; Yasui, W.; et al. Relation between microRNA expression and progression and prognosis of gastric cancer: A microRNA expression analysis. Lancet Oncol. 2010, 11, 136–146. [Google Scholar] [CrossRef]
- Akyala, A.I.; Peppelenbosch, M.P. Gastric cancer and Hedgehog signaling pathway: Emerging new paradigms. Genes Cancer 2018, 9, 1–10. [Google Scholar] [CrossRef][Green Version]
- Marin, J.J.; Al-Abdulla, R.; Lozano, E.; Briz, O.; Bujanda, L.; Banales, J.M.; Macias, R.I. Mechanisms of Resistance to Chemotherapy in Gastric Cancer. Anti-Cancer Agents Med. Chem. 2016, 16, 318–334. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhang, H.; Bai, M.; Ning, T.; Ge, S.; Deng, T.; Liu, R.; Zhang, L.; Ying, G.; Ba, Y. Exosomes Serve as Nanoparticles to Deliver Anti-miR-214 to Reverse Chemoresistance to Cisplatin in Gastric Cancer. Mol. Ther. 2018, 26, 774–783. [Google Scholar] [CrossRef] [PubMed]
- Romero-Ramirez, L.; Cao, H.; Nelson, D.; Hammond, E.; Lee, A.H.; Yoshida, H.; Mori, K.; Glimcher, L.H.; Denko, N.C.; Giaccia, A.J.; et al. XBP1 is essential for survival under hypoxic conditions and is required for tumor growth. Cancer Res. 2004, 64, 5943–5947. [Google Scholar] [CrossRef]
- Lee, A.H.; Iwakoshi, N.N.; Anderson, K.C.; Glimcher, L.H. Proteasome inhibitors disrupt the unfolded protein response in myeloma cells. Proc. Natl. Acad. Sci. USA 2003, 100, 9946–9951. [Google Scholar] [CrossRef]
- Qiao, L.; Zhang, H.; Yu, J.; Francisco, R.; Dent, P.; Ebert, M.P.; Röcken, C.; Farrell, G. Constitutive activation of NF-kappaB in human hepatocellular carcinoma: Evidence of a cytoprotective role. Hum. Gene Ther. 2006, 17, 280–290. [Google Scholar] [CrossRef]
- Koong, A.C.; Chen, E.Y.; Giaccia, A.J. Hypoxia causes the activation of nuclear factor kappa B through the phosphorylation of I kappa B alpha on tyrosine residues. Cancer Res. 1994, 54, 1425–1430. [Google Scholar] [PubMed]
- Nhieu, J.T.; Renard, C.A.; Wei, Y.; Cherqui, D.; Zafrani, E.S.; Buendia, M.A. Nuclear accumulation of mutated beta-catenin in hepatocellular carcinoma is associated with increased cell proliferation. Am. J. Pathol. 1999, 155, 703–710. [Google Scholar] [CrossRef]
- Momenimovahed, Z.; Tiznobaik, A.; Taheri, S.; Salehiniya, H. Ovarian cancer in the world: Epidemiology and risk factors. Int. J. Women’s Health 2019, 11, 287–299. [Google Scholar] [CrossRef]
- Iorio, M.V.; Visone, R.; Di Leva, G.; Donati, V.; Petrocca, F.; Casalini, P.; Taccioli, C.; Volinia, S.; Liu, C.G.; Alder, H.; et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007, 67, 8699–8707. [Google Scholar] [CrossRef]
- Martinez, I.; Gardiner, A.S.; Board, K.F.; Monzon, F.A.; Edwards, R.P.; Khan, S.A. Human papillomavirus type 16 reduces the expression of microRNA-218 in cervical carcinoma cells. Oncogene 2008, 27, 2575–2582. [Google Scholar] [CrossRef]
- Wang, X.; Tang, S.; Le, S.Y.; Lu, R.; Rader, J.S.; Meyers, C.; Zheng, Z.M. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS ONE 2008, 3, e2557. [Google Scholar] [CrossRef]
- Morgan, E.L.; Scarth, J.A.; Patterson, M.R.; Wasson, C.W.; Hemingway, G.C.; Barba-Moreno, D.; Macdonald, A. E6-mediated activation of JNK drives EGFR signalling to promote proliferation and viral oncoprotein expression in cervical cancer. Cell Death Differ. 2021, 28, 1669–1687. [Google Scholar] [CrossRef] [PubMed]
- Prickett, T.D.; Brautigan, D.L. Cytokine activation of p38 mitogen-activated protein kinase and apoptosis is opposed by alpha-4 targeting of protein phosphatase 2A for site-specific dephosphorylation of MEK3. Mol. Cell. Biol. 2007, 27, 4217–4227. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.W.; Choi, C.H.; Choi, J.J.; Park, Y.A.; Kim, S.J.; Hwang, S.Y.; Kim, W.Y.; Kim, T.J.; Lee, J.H.; Kim, B.G.; et al. Altered MicroRNA expression in cervical carcinomas. Clin. Cancer Res. 2008, 14, 2535–2542. [Google Scholar] [CrossRef] [PubMed]
- Yang, D.D.; Salciccioli, J.D.; Marshall, D.C.; Sheri, A.; Shalhoub, J. Trends in malignant melanoma mortality in 31 countries from 1985 to 2015. Br. J. Dermatol. 2020, 183, 1056–1064. [Google Scholar] [CrossRef]
- Leonardi, G.C.; Falzone, L.; Salemi, R.; Zanghì, A.; Spandidos, D.A.; McCubrey, J.A.; Candido, S.; Libra, M. Cutaneous melanoma: From pathogenesis to therapy. Int. J. Oncol. 2018, 52, 1071–1080. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef]
- Mueller, D.W.; Rehli, M.; Bosserhoff, A.K. miRNA expression profiling in melanocytes and melanoma cell lines reveals miRNAs associated with formation and progression of malignant melanoma. J. Investig. Dermatol. 2009, 129, 1740–1751. [Google Scholar] [CrossRef]
- Worley, L.A.; Long, M.D.; Onken, M.D.; Harbour, J.W. Micro-RNAs associated with metastasis in uveal melanoma identified by multiplexed microarray profiling. Melanoma Res. 2008, 18, 184–190. [Google Scholar] [CrossRef]
- Segura, M.F.; Belitskaya-Lévy, I.; Rose, A.E.; Zakrzewski, J.; Gaziel, A.; Hanniford, D.; Darvishian, F.; Berman, R.S.; Shapiro, R.L.; Pavlick, A.C.; et al. Melanoma MicroRNA signature predicts post-recurrence survival. Clin. Cancer Res. 2010, 16, 1577–1586. [Google Scholar] [CrossRef] [PubMed]
- Mitchell, K.; Svenson, K.B.; Longmate, W.M.; Gkirtzimanaki, K.; Sadej, R.; Wang, X.; Zhao, J.; Eliopoulos, A.G.; Berditchevski, F.; Dipersio, C.M. Suppression of integrin alpha3beta1 in breast cancer cells reduces cyclooxygenase-2 gene expression and inhibits tumorigenesis, invasion, and cross-talk to endothelial cells. Cancer Res. 2010, 70, 6359–6367. [Google Scholar] [CrossRef]
- Penna, E.; Orso, F.; Cimino, D.; Vercellino, I.; Grassi, E.; Quaglino, E.; Turco, E.; Taverna, D. miR-214 coordinates melanoma progression by upregulating ALCAM through TFAP2 and miR-148b downmodulation. Cancer Res. 2013, 73, 4098–4111. [Google Scholar] [CrossRef]
- Maemondo, M.; Inoue, A.; Kobayashi, K.; Sugawara, S.; Oizumi, S.; Isobe, H.; Gemma, A.; Harada, M.; Yoshizawa, H.; Kinoshita, I.; et al. Gefitinib or chemotherapy for non-small-cell lung cancer with mutated EGFR. N. Engl. J. Med. 2010, 362, 2380–2388. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Wu, Y.L.; Chen, G.; Feng, J.; Liu, X.Q.; Wang, C.; Zhang, S.; Wang, J.; Zhou, S.; Ren, S.; et al. Erlotinib versus chemotherapy as first-line treatment for patients with advanced EGFR mutation-positive non-small-cell lung cancer (OPTIMAL, CTONG-0802): A multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2011, 12, 735–742. [Google Scholar] [CrossRef]
- Yanaihara, N.; Caplen, N.; Bowman, E.; Seike, M.; Kumamoto, K.; Yi, M.; Stephens, R.M.; Okamoto, A.; Yokota, J.; Tanaka, T.; et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell 2006, 9, 189–198. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, M.; Hu, C. Exosomal transfer of miR-214 mediates gefitinib resistance in non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2018, 507, 457–464. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Chu, Z.; Mao, L. Antibodies targeting hepatoma-derived growth factor as a novel strategy in treating lung cancer. Mol. Cancer Ther. 2009, 8, 1106–1112. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Luo, J.; Zhao, Y.T.; Wang, Z.Y.; Zhou, J.; Huang, S.; Huang, J.N.; Long, H.X.; Zhu, B. TWIST1 upregulates miR-214 to promote epithelial-to-mesenchymal transition and metastasis in lung adenocarcinoma. Int. J. Mol. Med. 2018, 42, 461–470. [Google Scholar] [CrossRef] [PubMed]
- Sadighparvar, S.; Targhazeh, N.; Karimian, A.; Shafiei-Irannejad, V.; Farsad-Akhtar, N.; Rafieian, S.; Salamati, A.; Bastami, M.; Kafil, H.S.; Yousefi, M.; et al. Downregulation of microRNA-214 and PTEN in tissue samples of patients with breast cancer. Meta Gene 2020, 24, 100668. [Google Scholar] [CrossRef]
- Liu, B.; Tian, Y.; Li, F.; Zhao, Z.; Jiang, X.; Zhai, C.; Han, X.; Zhang, L. Tumor-suppressing roles of miR-214 and miR-218 in breast cancer. Oncol. Rep. 2016, 35, 3178–3184. [Google Scholar] [CrossRef]
- Kalniete, D.; Nakazawa-Miklaševiča, M.; Štrumfa, I.; Āboliņš, A.; Irmejs, A.; Gardovskis, J.; Miklaševičs, E. High expression of miR-214 is associated with a worse disease-specific survival of the triple-negative breast cancer patients. Hered. Cancer Clin. Prac. 2015, 13, 7. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dettori, D.; Orso, F.; Penna, E.; Baruffaldi, D.; Brundu, S.; Maione, F.; Turco, E.; Giraudo, E.; Taverna, D. Therapeutic Silencing of miR-214 Inhibits Tumor Progression in Multiple Mouse Models. Mol. Ther. 2018, 26, 2008–2018. [Google Scholar] [CrossRef]
- Chen, X.; Wang, Z.; Duan, N.; Zhu, G.; Schwarz, E.M.; Xie, C. Osteoblast-osteoclast interactions. Connect. Tissue Res. 2018, 59, 99–107. [Google Scholar] [CrossRef]
- Li, Z.; Hassan, M.Q.; Volinia, S.; van Wijnen, A.J.; Stein, J.L.; Croce, C.M.; Lian, J.B.; Stein, G.S. A microRNA signature for a BMP2-induced osteoblast lineage commitment program. Proc. Natl. Acad. Sci. USA 2008, 105, 13906–13911. [Google Scholar] [CrossRef]
- Mizuno, Y.; Yagi, K.; Tokuzawa, Y.; Kanesaki-Yatsuka, Y.; Suda, T.; Katagiri, T.; Fukuda, T.; Maruyama, M.; Okuda, A.; Amemiya, T.; et al. miR-125b inhibits osteoblastic differentiation by down-regulation of cell proliferation. Biochem. Biophys. Res. Commun. 2008, 368, 267–272. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Xie, H.; Liu, W.; Hu, R.; Huang, B.; Tan, Y.F.; Xu, K.; Sheng, Z.F.; Zhou, H.D.; Wu, X.P.; et al. A novel microRNA targeting HDAC5 regulates osteoblast differentiation in mice and contributes to primary osteoporosis in humans. J. Clin. Investig. 2009, 119, 3666–3677. [Google Scholar] [CrossRef]
- Tohmonda, T.; Miyauchi, Y.; Ghosh, R.; Yoda, M.; Uchikawa, S.; Takito, J.; Morioka, H.; Nakamura, M.; Iwawaki, T.; Chiba, K.; et al. The IRE1α-XBP1 pathway is essential for osteoblast differentiation through promoting transcription of Osterix. EMBO Rep. 2011, 12, 451–457. [Google Scholar] [CrossRef]
- Bonewald, L.F. The amazing osteocyte. J. Bone Miner. Res. 2011, 26, 229–238. [Google Scholar] [CrossRef]
- Yuan, Y.; Guo, J.; Zhang, L.; Tong, X.; Zhang, S.; Zhou, X.; Zhang, M.; Chen, X.; Lei, L.; Li, H.; et al. MiR-214 Attenuates the Osteogenic Effects of Mechanical Loading on Osteoblasts. Int. J. Sports Med. 2019, 40, 931–940. [Google Scholar] [PubMed]
- Wang, X.; Li, X.; Li, J.; Zhai, L.; Liu, D.; Abdurahman, A.; Zhang, Y.; Yokota, H.; Zhang, P. Mechanical loading stimulates bone angiogenesis through enhancing type H vessel formation and downregulating exosomal miR-214-3p from bone marrow-derived mesenchymal stem cells. FASEB J. 2021, 35, e21150. [Google Scholar] [CrossRef] [PubMed]
- Li, K.C.; Chang, Y.H.; Hsu, M.N.; Lo, S.C.; Li, W.H.; Hu, Y.C. Baculovirus-Mediated miR-214 Knockdown Shifts Osteoporotic ASCs Differentiation and Improves Osteoporotic Bone Defects Repair. Sci. Rep. 2017, 7, 16225. [Google Scholar] [CrossRef]
- Wang, C.G.; Wang, L.; Yang, T.; Su, S.L.; Hu, Y.H.; Zhong, D. Pseudogene PTENP1 sponges miR-214 to regulate the expression of PTEN to modulate osteoclast differentiation and attenuate osteoporosis. Cytotherapy 2020, 22, 412–423. [Google Scholar] [CrossRef] [PubMed]
- Cai, M.; Yang, L.; Zhang, S.; Liu, J.; Sun, Y.; Wang, X. A bone-resorption surface-targeting nanoparticle to deliver anti-miR214 for osteoporosis therapy. Int. J. Nanomed. 2017, 12, 7469–7482. [Google Scholar] [CrossRef] [PubMed]
- Li, N.; Bai, Y.; Zhou, G.; Ma, Y.; Tan, M.; Qiao, F.; Li, X.; Shen, M.; Song, X.; Zhao, X.; et al. miR-214 Attenuates Aortic Valve Calcification by Regulating Osteogenic Differentiation of Valvular Interstitial Cells. Mol. Ther. Nucleic Acids 2020, 22, 971–980. [Google Scholar] [CrossRef]
- Horak, M.; Novak, J.; Bienertova-Vasku, J. Muscle-specific microRNAs in skeletal muscle development. Dev. Biol. 2016, 410, 1–13. [Google Scholar] [CrossRef]
- O’Rourke, J.R.; Georges, S.A.; Seay, H.R.; Tapscott, S.J.; McManus, M.T.; Goldhamer, D.J.; Swanson, M.S.; Harfe, B.D. Essential role for Dicer during skeletal muscle development. Dev. Biol. 2007, 311, 359–368. [Google Scholar] [CrossRef] [PubMed]
- Huang, T.H.; Zhu, M.J.; Li, X.Y.; Zhao, S.H. Discovery of porcine microRNAs and profiling from skeletal muscle tissues during development. PLoS ONE 2008, 3, e3225. [Google Scholar] [CrossRef] [PubMed]
- Honardoost, M.; Soleimani, M.; Arefian, E.; Sarookhani, M.R. Expression Change of miR-214 and miR-135 during Muscle Differentiation. Cell J. 2015, 17, 461–470. [Google Scholar]
- Chen, H.; Shalom-Feuerstein, R.; Riley, J.; Zhang, S.D.; Tucci, P.; Agostini, M.; Aberdam, D.; Knight, R.A.; Genchi, G.; Nicotera, P.; et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem. Biophys. Res. Commun. 2010, 394, 921–927. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Githinji, J.; McLaughlin, B.; Wilczek, K.; Nolta, J. Role of miRNAs in neuronal differentiation from human embryonic stem cell-derived neural stem cells. Stem Cell Rev. Rep. 2012, 8, 1129–1137. [Google Scholar] [CrossRef]
- Ahmadian-Elmi, M.; Bidmeshki Pour, A.; Naghavian, R.; Ghaedi, K.; Tanhaei, S.; Izadi, T.; Nasr-Esfahani, M.H. miR-27a and miR-214 exert opposite regulatory roles in Th17 differentiation via mediating different signaling pathways in peripheral blood CD4+ T lymphocytes of patients with relapsing-remitting multiple sclerosis. Immunogenetics 2016, 68, 43–54. [Google Scholar] [CrossRef]
- Bonneau, E.; Neveu, B.; Kostantin, E.; Tsongalis, G.J.; De Guire, V. How close are miRNAs from clinical practice? A perspective on the diagnostic and therapeutic market. Electron. J. Int. Fed. Clin. Chem. Lab. Med. 2019, 30, 114–127. [Google Scholar]
- Ramalingam, H.; Yheskel, M.; Patel, V. Modulation of polycystic kidney disease by non-coding RNAs. Cell Signal. 2020, 71, 109548. [Google Scholar] [CrossRef] [PubMed]
- Gallant-Behm, C.L.; Piper, J.; Lynch, J.M.; Seto, A.G.; Hong, S.J.; Mustoe, T.A.; Maari, C.; Pestano, L.A.; Dalby, C.M.; Jackson, A.L.; et al. A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular Matrix Expression and Fibroplasia in the Skin. J. Investig. Dermatol. 2019, 139, 1073–1081. [Google Scholar] [CrossRef] [PubMed]





| Target mRNA/Protein | Effect Mediated by miR-214 Downregulation | Reference |
|---|---|---|
| α1-AT | Promotes cell viability, invasion, and migration in triple-negative breast cancer. | [57] |
| ATF4 | Inhibits osteoblast activity; inhibits osteoblast differentiation in hPDLSCs. | [30,58] |
| β-catenin | Reduced activation of pro-proliferative downstream effectors c-myc, TCF-1 and cyclinD1 in hepatocellular carcinoma; inhibits breast cancer proliferation; inhibits osteoblast differentiation. | [59,60,61] |
| Bcl2l2 | Inhibits cervical cancer growth. | [62] |
| BIM | Inhibits mitochondrial-dependent apoptosis. | [14] |
| CaMKIIδ | Impairs the regulation of excitation–contraction coupling in the heart. | [14] |
| CypD | Impairs the opening of the mitochondrial permeability transition pore. | [14] |
| Ezh1 | Inhibits Col1α1 and Col1α3 expression in myofibroblasts. | [19] |
| Ezh2 | Promotes cardiac hypertrophy; inhibits Col1α1 and Col1α3 expression in myofibroblasts; inhibits cervical cancer growth; inhibits breast cancer proliferation and cell invasion; establishes a feedback loop to promote skeletal muscle cell differentiation. | [19,31,34,37,63] |
| FGFR-1 | Inhibits cell invasion in hepatocellular carcinoma; inhibits osteoblast differentiation. | [64,65] |
| FOXM1 | Inhibits cervical cancer growth, invasion and promotes cisplatin sensitivity. | [66] |
| HDGF | Impairs angiogenic signalling in hepatocellular carcinoma. | [67] |
| HMGA1 | Inhibits cervical cancer growth and invasion. | [68] |
| ITGA3 | Increases melanoma cell migration. | [24] |
| JNK1 | Inhibits proliferation and metastasis in cervical cancer and affects EGFR signaling. | [26] |
| MEK3 | Inhibits cervical cancer progression. | [26] |
| Mitofusin2 (Mfn2) | Promotes ERK1/2-MAPK activation, cardiac fibroblast proliferation and collagen synthesis. | [17] |
| NCX1 | Attenuates calcium ion overload in the heart. | [14] |
| NLRC5 | Inhibits cardiac fibroblast activation and fibroblast to myofibroblast transition. | [21] |
| N-Ras | Promotes myogenic differentiation. | [69] |
| Osterix | Inhibits osteoblast differentiation. | [70] |
| p53 | Promotes breast cancer cell invasion. | [71] |
| Plexin-B1 | Inhibits cervical cancer growth and invasion. | [72] |
| PTEN | Inhibits PTEN signalling and promotes the PI3K/Akt pathway. Protects against H2O2-mediated apoptosis in cardiomyocytes; promotes gastric cancer progression; promotes peritoneal metastasis in gastric cancer; promotes ovarian cancer chemoresistance to cisplatin; promotes lung cancer resistance to gefitinib; promotes glycolysis in NSCLC; promotes cell growth and protects against apoptosis in breast cancer; promotes osteoclast differentiation; promotes regulatory T cell differentiation; increases T cell proliferation. | [15,22,23,25,33,35,73,74,75,76] |
| Quaking | Impairs angiogenic signalling; promotes vascular smooth muscle cell differentiation; promotes neurogenesis during cerebral cortex development. | [32,77,78] |
| RFWD2 | Promotes apoptosis and sensitises breast cancer to doxorubicin. | [79] |
| RNF8 | Inhibits metastatic epithelial–mesenchymal transition in breast cancer; encourages chromosomal instability in ovarian cancer. | [80,81] |
| Runx3 | Impairs the Th1/Th2 cell balance in asthmatic patients when simultaneously targeted by miR-214, miR-371, miR-138, miR-544, and miR-145. | [82] |
| ST6GAL1 | Promotes cell viability, invasion, migration, and epithelial–mesenchymal transition in triple-negative breast cancer. | [83] |
| Su(fu) | Activates hedgehog signalling and promotes muscle cell differentiation in zebrafish; inhibits inflammatory smooth muscle cell differentiation. | [84,85] |
| Survivin | Inhibits breast cancer proliferation and increases apoptosis. | [86] |
| TFAP2C | Increases melanoma cell migration and metastasis, increases progrowth VEGFA and suppresses ERBB2, and regulates many more factors. | [24] |
| TRAF3 | Promotes osteoclast activity and osteolytic bone metastasis in breast cancer patients. | [87] |
| UCP2 | Sensitises breast cancer cells to tamoxifen and fulvestrant. | [88] |
| XBP1 | Impairs angiogenic signalling; impairs hepatocellular carcinoma survival. | [16,89] |
| Xotx2 | Inhibits retinal bipolar neuron differentiation. | [90] |
| Xvsx1 | Inhibits retinal bipolar neuron differentiation. | [90] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amin, M.M.J.; Trevelyan, C.J.; Turner, N.A. MicroRNA-214 in Health and Disease. Cells 2021, 10, 3274. https://doi.org/10.3390/cells10123274
Amin MMJ, Trevelyan CJ, Turner NA. MicroRNA-214 in Health and Disease. Cells. 2021; 10(12):3274. https://doi.org/10.3390/cells10123274
Chicago/Turabian StyleAmin, Meer M. J., Christopher J. Trevelyan, and Neil A. Turner. 2021. "MicroRNA-214 in Health and Disease" Cells 10, no. 12: 3274. https://doi.org/10.3390/cells10123274
APA StyleAmin, M. M. J., Trevelyan, C. J., & Turner, N. A. (2021). MicroRNA-214 in Health and Disease. Cells, 10(12), 3274. https://doi.org/10.3390/cells10123274







