Abstract
Stem/progenitor cell transplantation is a potential novel therapeutic strategy to induce angiogenesis in ischemic tissue, which can prevent major amputation in patients with advanced peripheral artery disease (PAD). Thus, clinicians can use cell therapies worldwide to treat PAD. However, some cell therapy studies did not report beneficial outcomes. Clinical researchers have suggested that classical risk factors and comorbidities may adversely affect the efficacy of cell therapy. Some studies have indicated that the response to stem cell therapy varies among patients, even in those harboring limited risk factors. This suggests the role of undetermined risk factors, including genetic alterations, somatic mutations, and clonal hematopoiesis. Personalized stem cell-based therapy can be developed by analyzing individual risk factors. These approaches must consider several clinical biomarkers and perform studies (such as genome-wide association studies (GWAS)) on disease-related genetic traits and integrate the findings with those of transcriptome-wide association studies (TWAS) and whole-genome sequencing in PAD. Additional unbiased analyses with state-of-the-art computational methods, such as machine learning-based patient stratification, are suited for predictions in clinical investigations. The integration of these complex approaches into a unified analysis procedure for the identification of responders and non-responders before stem cell therapy, which can decrease treatment expenditure, is a major challenge for increasing the efficacy of therapies.
Keywords:
cell therapy; chronic limb-threating ischemia; peripheral artery disease; diabetes; atherosclerosis obliterans; thromboangiitis obliterans; personalized medicine; artificial intelligence; machine learning; genome-wide association studies; transcriptome-wide association studies; clonal hematopoiesis of indeterminate potential 1. Introduction
Peripheral arterial disease (PAD) is the second leading cause of mortality and morbidity among cardiovascular diseases (CVDs) [1,2,3]. According to the global disease-burden data, the incidence of PAD in low-income and middle-income countries increased by 28.7%, while that in high-income countries increased by 13.1% compared to the preceding decade [1,3]. Chronic limb-threatening ischemia (CLTI) is clinically defined as chronic and severe limb-perfusion insufficiency that leads to tissue ulceration and gangrene [2]. The poor prognostic outcomes and high mortality rate of PAD are attributed to the following three main “conventional” risk factors: lifestyle (lifestyle risk factors subcategory, such as western diet, cigarette smoking, sedentary lifestyle, and alcohol consumption); comorbidities (comorbidities risk factors subcategories, such as hypertension, dyslipidemia (including all atherogenic lipid subsets or familial hypercholesterolemia), diabetes mellitus, obesity, homocysteine, high-sensitive C-reactive protein (hsCRP) and fibrinogen, and chronic kidney disease; and the genetic background risk factors subcategory, such as race and ethnicity [1,2,3]. Conventional cardiovascular risk factors contribute to the development of PAD. In the presence of four of the subcategory risk factors, the incidence rate of PAD can increase up to 140 cases per 100,000 individuals [4]. Clinical evidence suggests that additional or unidentified genetic risk factors may contribute to CVD development [5,6]. For example, the accumulation of somatic mutations in hematopoietic cells throughout life, which is referred to as clonal hematopoiesis of indeterminate potential (CHIP) (see Box 1), contributes to the development of CVD [5]. Several studies have demonstrated that the frequency of JAK2 V617F mutations in patients with atherosclerosis obliterans (ASO) of the lower extremities is five-fold higher than that in healthy controls [7]. Moreover, CHIP carriers are associated with a significantly higher risk of CVD events than non-carriers, such as atherosclerosis, PAD, and myocardial infarction [5,7]. Recent studies have demonstrated that 20% of the elderly population (60 to 90 years old) harbors mutated DNMT3A, TET2, ASXL1, JAK2, and NOTCH1 in hematopoietic stem cells (HSCs), such as CD34+ and CD133+ cells. CD133+ cells have applications in advanced treatment for patients with terminal-stage PAD [5,8]. Thus, there is a need to reassess the stem/progenitor cell genetic background to develop improved therapeutic strategies for PAD [9,10].
Box 1. Definition of terms.
Clonal hematopoiesis is the expansion of a clonal population of hematopoietic cells with one or more somatic mutations [6,11].
CHIP is defined by the absence of definitive morphological evidence of hematological neoplasms and the presence of a somatic mutation with a variant allele frequency of at least 2% to 4% [11].
Somatic mutations occur in somatic cells and cannot be inherited (only tissues derived from the mutated cell are affected) [6,12].
Germline mutations occur in gametes and can be passed on to offspring (every cell in the entire organism will be affected).
Variant allele frequency (VAF) is the percentage of sequence reads matching a specific DNA variant divided by the overall coverage at that locus [13].
Therapeutic angiogenesis with stem/progenitor cells is a promising strategy for treating ischemic tissue and preventing major amputation. Stem cell application accelerates angiogenesis (formation of new vessels from pre-existing vessels), vasculogenesis, and de novo synthesis of new vessels from transplanted and circulating precursor cells [14]. Previously, intra-arterial or intramuscular transplantation of stem cells has been performed in PAD patients with Rutherford grades 3–6. Some studies have obtained promising results using this strategy, including improved limb-blood perfusion, increased major amputation-free periods, and enhanced quality of life (Table 1). However, the response rate for stem cell-based therapy in patients with PAD exhibiting comorbidities (such as diabetes, hypertension, and dyslipidemia) and with risk factors (such as cigarette smoking) is lower or poorer than that in placebo-treated groups (Table 2). Hence, patients who respond or do not respond to stem/progenitor cell therapy must be determined before starting a clinical trial in cases of advanced CLTI. Previously, we determined cases responding to stem/progenitor cell therapy among patients with heart failure [15].

Table 1.
Positive stem cell-based trial outcomes in patients with CLI.

Table 2.
Negative stem/cell-based trial outcomes in patients with CLTI.
Personalized stem cell-based therapy approaches employing several clinical biomarkers, disease-related genetic-trait evaluation methods, such as the analysis of the findings of transcriptome-wide association studies (TWAS)/genome-wide association studies (GWAS)) of PAD, and advanced analyses with state-of-the-art computational methods (such as machine learning- (ML) based prediction) can contribute to clinical investigations [10]. The integration of these complex approaches is a major challenge for increasing the efficacy of therapies and decreasing treatment costs. Here, the pros and cons of the transplantation of granulocyte colony-stimulating factor- (G-CSF) mobilized CD34+ or endothelial progenitor cells (EPCs) were compared with those of bone marrow-derived mononuclear cells (BMMCs) and peripheral blood-derived mononuclear cells (PBMCs) by analyzing the results of randomized, placebo-controlled clinical trials with a focus on clonal hematopoiesis (CH). Additionally, the possibility of increasing the efficacy and safety of stem cell transplantation for different cell types based on somatic mutations in HSCs and studies on predictors that can identify the responder (R) and non-responder (NR) groups before stem cell therapy have been highlighted.
2. Personalized Stem Cell Therapy for Patients with PAD Based on Phenotype and Genotype Findings
Clinical trials of CLTI/PAD have identified several clinical risk factors and biomarkers associated with poor cell transplantation outcomes. Precise evaluation techniques are needed to achieve beneficial clinical outcomes and reduce the financial burden of advanced medical treatment technologies for patients with CLTI/PAD.
2.1. Characteristic Features of the R and NR Groups
Various clinical trials of stem/progenitor cell transplantation for PAD or CLTI have identified several predictors and biomarkers of response to cellular therapy (Table 3). Klepanec et al. [25] reported several parameters that delineated the R group from the NR group. Compared with the NR group, the R group exhibited a two-fold higher absolute CD34+ cell count but a similar number of total bone marrow mononuclear cells (BMNCs). Additionally, the CRP levels and total leucocyte counts in the R group were lower than those in the NR group. The general inflammation process and the number of CD34+ cells are directly associated with the cellular therapy outcomes [25,26]. For example, three independent clinical trials reported that increased levels of IL-6, CRP, serum leucocyte count, fibrinogen, and basic fibroblast growth factor are associated with a weak cellular therapy response (Table 3). Two studies have reported that age < 50 years was an independent predictor of improved cellular therapy response in patients with CLTI [26,27]. Consistently, Jaiswal et al. reported that the frequency of somatic mutations markedly increased in patients aged > 50 years [5,28]. Moreover, patients with myelodysplastic syndrome (MDS) aged > 65 years and harboring TET2, DNMT3A, and AXSL1 mutations were susceptible to PAD and systemic inflammation [29]. These findings indicate a correlation between inflammation and CHIP, as well as CVDs, including PAD. PAD and coronary artery disease (CAD) are associated with decreased HSC proliferation and inhibition of angiogenesis, which lead to tissue ischemia and inflammation [15,30]. In the PERFECT trial, ML determined that the response of bone marrow stem cell transplantation-based myocardial regeneration was correlated with PLCG1, LPCAT2, AP1B1, AFAP1, GRB2, KLF8, and MARK3 expression levels and serum EPO and VEGF levels but was not correlated with the expression of CHIP-related genes (DNMT3a, TET2, and ASXL1) (Table 3). Sequencing data revealed that the R group exhibited 161 differentially expressed genes when compared with the NR group. Mutation analysis revealed that the number of specific variants in the R group (48 genes) was lower than that in the NR group (224 genes). Additionally, R-related and NR-related genes determined using ML were correlated with SH2B3, as well as with other regulatory genes, such as NOTCH2, PDCD1/PD-1, and CD133 [10].

Table 3.
Clinical biomarkers of R and NR groups among patients with PAD.
2.2. BMNC Transplantation Alleviates Thromboangiitis Obliterans (TAO), and Purified CD34/CD133+ Stem Cell Therapy Alleviates ASO
Clinical studies have revealed that autologous G-CSF-mobilized peripheral blood EPCs exert higher therapeutic effects than BMMCs or PBMCs in patients with TAO (Table 4). Arai et al. [32] demonstrated that the legs injected with CD34+ exhibited a significantly higher ankle-brachial index (ABI) (0.56 ± 0.04) than those injected with BMMCs (0.53 ± 0.06) at week 4 after cell therapy. Similar improvements in transcutaneous oxygen pressure (TcPO2) were observed in both CD34+-transplanted (from 27 ± 4 to 37 ± 3; p < 0.05) and BMT-transplanted groups (from 24 ± 6 to 32 ± 8; p < 0.05) at week 4 post-implantation. One clinical trial demonstrated that CD34+ cells dose-dependently prevented major or minor amputation when compared with the placebo. At month 6 post-injection, 67%, 43%, and 22% of the control, low-dose-administered, and high-dose-administered groups underwent a major or minor amputation (p > 0.137), respectively. This trend continued at month 12 post-injection, with 75%, 45%, and 22% of the control, low-dose-administered, and high-dose-administered groups undergoing amputation (p > 0.058), respectively. The amputation rate in the combined cell-treated groups was lower than in the control group (month 6, p > 0.125; month 12, p > 0.054). The low-dose-administered and high-dose-administered groups exhibited improved amputation-free survival at months 6 and 12 post-injection [24].

Table 4.
Comparative features of TAO and ASO.
The long-term clinical outcomes of CD34+ transplantation revealed that compared with those at the baseline, toe-brachial pressure index (TBPI) and TcPO2 significantly improved at week 12 post-transplantation in both CLI and TAO cases but improved only at week 52 in TAO cases. The improvements in TBPI and TcPO2 were sustained until weeks 156 and 208, respectively. The ulcers completely healed in all patients with TAO and two patients with atherosclerotic PAD at week 52 [19]. A recent randomized, single-blinded trial evaluated the 12-month treatment outcomes of purified CD34+ cell transplantation and PBMC transplantation for advanced TAO. The total amputation rates at month 6 post-transplantation in the CD34+-transplanted and PBMC-transplanted groups were 28.0% and 16.0%, respectively (p > 0.343), which remained unchanged at month 12 post-transplantation. Furthermore, the complete wound-healing rates did not significantly differ between the groups at months 3, 6, and 12. The two groups exhibited significantly increased ABI, toe-brachial index (TBI), TcPO2, and pain-free walking time (PFWT) values over time when compared with baseline values (ΔABI, ΔTBI, ΔTcPO2, and ΔPFWT, respectively) [33]. Another clinical trial evaluated the long-term efficacy of autologous BMMC transplantation in ASO and TAO [34]. The four-year amputation-free rates in the control ASO, BMMC-transplanted ASO, control TAO, and BMMC-transplanted groups were 0%, 48%, 6%, and 95%, respectively. ABI and TcPO2 significantly increased after one month in the BMMC-transplanted TAO group and remained high during the three-year follow-up visit. In contrast, ABI and TcO2 significantly increased only in the first month in the BMMC-transplanted ASO group and gradually decreased during the three-year follow-up before finally returning to baseline levels [34]. The 10-year amputation-free survival rates in the autologous BMMC-transplanted and aspirin-treated groups were 85.3% (29/34) and 40% (6/15) (p < 0.0019), respectively. Autologous BMMC transplantation significantly decreased the ulcer area (p < 0.0001), TBI (p < 0.0001), TcPO2 (p < 0.0001), and pain score (p < 0.0001) [35]. Moreover, transplantation of autologous stem cells harboring mutations is reported to promote the expansion of mutant blood cell clones, leading to increased risks of CHIP-associated complications [36]. However, patients with TAO are younger than those with ASO. Thus, TAO cases exhibited decreased somatic mutations and inflammation in the peripheral blood (Table 4 and Table 5).

Table 5.
Evidence-based information on peripheral artery disease (PAD) and somatic mutations.
These findings indicate that BMMC or PBMC transplantation is safe and feasible for patients with TAO. The parameters, such as the amputation-free rate (86%–95% in TAO and 40%–50% in ASO) and the enhanced ABI, TBI, TcPO2, and PFWT values compared with baseline values, indicated the effectiveness of cell therapies. BMMC or PBMC transplantation can exhibit similar efficacy as G-CSF-mobilized CD34+ cell transplantation in patients with TAO. However, purified or enriched transplantation of EPCs with a decreased somatic mutation profile must be performed for ASO. Thus, optimal stem cell-based therapy must be determined depending on the type of disease to reduce treatment expenditure.
3. CLTI/PAD in Patients with Diabetes Mellitus
Approximately one third of patients with diabetes mellitus develop PAD, with various grades of disease severity and type [43]. Hence, personalized stem cell therapy can increase the response rate, ameliorate patient-specific clinical parameters, enhance quality of life, and decrease therapy cost. The mobilization of CD34+ stem cells is impaired in patients with diabetes mellitus, while patients who previously received G-CSF have a decreased probability of achieving a CD34+ stem cell count of >50/μL [46,47]. This may be associated with diabetic bone marrow (BM) autonomic neuropathy, which impairs Lin− cKit+ Sca1+ cells and EPC mobilization through the upregulation of 66-kDa protein from the src homology and collagen homology domains, as well as the downregulation of sirtulin 1 in mice and humans [46,48] (Figure 1). Teraa et al., reported that CLTI induces alterations in bone marrow vasculature and sympathetic nerve innervation in patients with and without diabetes mellitus [49]. Bonnefond et al. [50] examined the correlation between type two diabetes mellitus (T2DM) and CHIP events in the blood. The occurrence of CHIP events was correlated with T2DM (odds ratio (OR) = 5.3; p = 5.1 × 10−5), and the correlation was strong when non-obese individuals with T2DM (OR = 5.6; p = 4.9 × 10−5) were included. Previous studies have revealed that advanced age and a long history of T2DM, atherosclerosis, vessel formation, and revascularization are associated with persistent glucotoxicity, oxidative, and inflammatory damages, which may impair vascular regenerative cell lineages [48,51].

Figure 1.
Negative epigenetic regulation of gene expression. The figure was reproduced from Reactome ver. 72 (URL.: https://reactome.org/PathwayBrowser/#/R-HSA-5250941, accessed on 18 January 2021. Stable ID: R-HAS-5250941. Abbreviations: NoRC, nucleolar remodeling complex.
Moreover, chronic inflammation and enhanced reactive oxygen species (ROS) production can lead to dysfunctional HSC self-renewal in the endosteal niche, which leads to persistent premature mobilization of HSCs into the peripheral vessels and, consequently, the exhaustion of the reservoir of early myeloid progenitor cells with proangiogenic secretory function [52,53]. This can be attributed to the genetic background or CH events in T2DM, which impair vascular regeneration and promote inflammation through the above-mentioned mechanisms. For example, clonal mosaic event carriers (71.4%) with T2DM exhibited a higher prevalence of vascular complications, such as microvascular and macrovascular lesions, than non-carriers with T2DM (37.1%) p = 7.7 × 10−4) [50]. This indicates that multiple risk factors contribute to T2DM-related stem/progenitor cell-quality impairments, including somatic mutations (Figure 1).
Diabetes affects the BM niche, decreases the availability of circulating stem/progenitor cells, impairs the function of vascular progenitors, and prevents mobilization. Hence, ex vivo incubation of BM-derived stem/progenitor cells of diabetic animals with growth factor cocktails can restore vasculogenic and anti-inflammatory potential in vitro and in vivo [54,55]. Furthermore, enriched cell transplantation in acute myocardial ischemia [56,57] and diabetic animal wounding models [48,55] effectively restored the vasculogenic and anti-inflammatory potential in a small number of regeneration-associated cells, such as EPCs and T regulatory cells, and alternatively activated macrophages. This may indicate that “exhausted” or dysfunctional cells in diabetes mellitus gain several regenerative functions after incubation with growth factor cocktails.
In summary, genetic alterations are high in patients with diabetes mellitus and PAD. Chronic inflammation caused by increased blood glucose levels and the resulting processes (ROS production and oxidative stress) may impair the function of HSCs and endothelial cell lineages through the introduction of additional mutations.
4. Findings of GWAS on Variations in PAD-Associated Gene Loci Involved in Inflammation, Thrombosis, and Lipid Imbalance
Findings of GWAS on ASO
High-throughput genotypic platforms, such as hybridization-based single nucleotide polymorphism (SNP) arrays and various next-generation sequencing (NGS) technologies, are widely applied for gene-variant-disease association studies worldwide. In GWAS, the correlation between the phenotypes and the genetic variants across the genome within, around, and between genes is analyzed. Early GWAS on PAD involving a small Japanese population reported that PAD was strongly correlated with rs1902341 or OSBPL10 gene locus. OSBPL10 variation may enhance triglyceride levels in patients with dyslipidemia, which indicates that it regulates cellular lipid metabolism [58,59]. Kullo et al. performed a two-stage genomic association study. In the first stage, 537 and 872 SNPs were examined in 1641 PAD cases, as well as 1604 control subjects of European ancestry. Next, the top 48 SNPs associated with PAD were genotyped. The SNP rs653178 in the ATXN2-SH2B3 locus was significantly associated with PAD in the discovery (OR = 1.23; p = 5.59 × 10−5), replication (OR = 1.22; 8.9 × 10−4), and combined (OR = 1.22; p = 6.46 × 10−7) cohorts. SH2B3 encodes an adapter protein that plays a key role in immune- and inflammatory-response pathways, hematopoietic cell regulation in the BM niche, and vascular homeostasis [15,60]. Other GWAS on CVDs (e.g., stroke, PAD, hypertension, and acute coronary syndrome (ACS)) have indicated that the SH2B3 locus is a key risk factor for the development of CVDs (Figure 2).
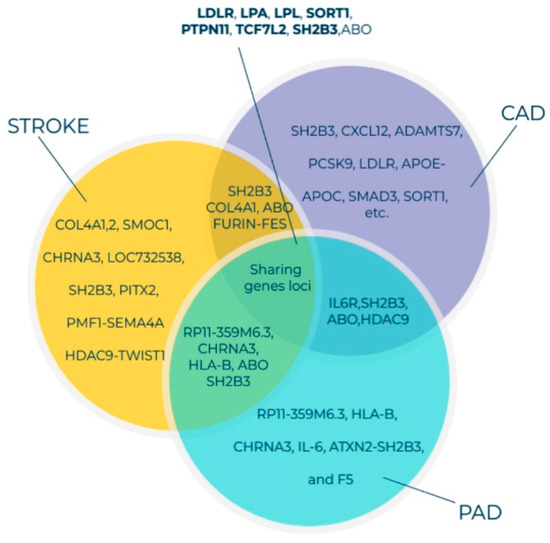
Figure 2.
Venn diagram of genes associated with susceptibility to PAD, CAD, and stroke identified through GWAS and the shared gene loci. All listed genes shown here were adapted from the GWAS catalog database for PAD, CAD, and stroke [61,62,63]. Currently, more than 190, 1060, and 327 gene loci have been identified for PAD, CAD, and stroke, respectively. The GWAS 71 catalog accession date is 18 January 2021 Abbreviations: PAD, peripheral arterial disease; CAD, coronary artery disease; GWAS, genome-wide association studies.
Previously, we had reported that SH2B3/LNK mutations in the NR group were higher than those in the R group and that altered SH2B3/LNK expression, along with regeneration-associated pathways, contributes to cardiac healing [13,15]. One Asian population-based GWAS identified the following three novel PAD susceptibility loci with genome-wide significance: IPO5/RAP2A (a member of the importin beta family that promotes apolipoprotein A-1 excretion; p = 6.8 × 10−14), EDNRA (p = 5.3 × 10−9), and HDAC9 (p = 8.8 × 10−8). For example, the HDAC9 locus was identified in GWAS on stroke, ACS, and PAD [64,65]. HDAC9 increases the risk of disease development by enhancing the atherosclerosis process in large- and small-caliber vessels [64,65]. GWAS involving Hispanic/Latino ethnic subgroups have demonstrated ethnic diversity of the PAD-related gene locus among the European and Asian populations. Two genome-wide significant associations were identified between ABI and the SNP rs4466200 at the COMMD10 locus in the Puerto Rican population and the SNP rs12520838 at the SYBU locus in the Caribbean population, which suggested ethnic genetic diversity of PAD on different continents [66]. Klarin et al. [62] recently identified 19 novel PAD loci, among which 18 were not reported previously (Figure 3). A large cohort study included 31,307 PAD cases and 211,753 controls and examined approximately 32 million DNA sequence variants in veterans of European, African, and Hispanic ancestry. A phenome-wide association study was applied to examine the association of risk variants across various phenotypes, which detected 11 PAD risk variants. A significant association was demonstrated between all three (coronary, cerebral, and peripheral) vascular beds, lipids (LDLR, LPA, LPL, and SORT1), hypertension (PTPN11), and diabetes (TCF7L2). Variants in the RP11-359M6.3, HLA-B, CHRNA3, and F5 loci were uniquely associated with PAD. This indicated that a tobacco-smoking-related gene (CHRNA3) and a thrombosis-related gene (F5) might play an essential role in PAD but not in other arterial diseases (Figure 3). Cyclin-dependent kinase inhibitor 2 B antisense (CDKN2B-AS1), which enhances lipid uptake and accumulation, is another risk variant that is upregulated in atherosclerotic plaque tissues and human primary macrophage-derived foam cells. Long, non-coding RNA CDKN2B-AS1 recruits EZH2 and CTCF in the CDKN2B promoter region and consequently inhibits CDKN2B transcription by promoting histone methylation [67].
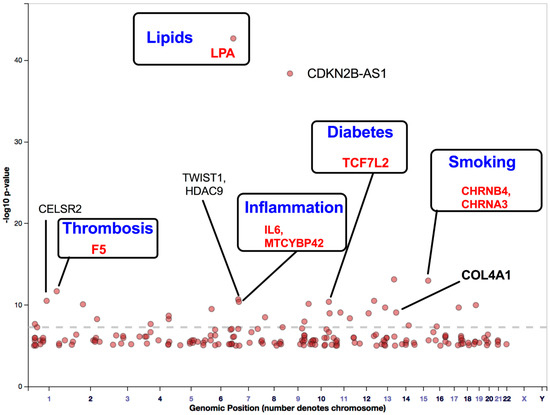
Figure 3.
Gene loci associated with PAD identified through GWAS. This figure was modified from the GWAS catalog database with 261,600 patients with PAD. The seven GWAS have been registered as PAD traits (study accession # GCST000720, GCST002504, GCST003154, GCST009133, GCST009134, and GCST008474 downloaded on 18 January 2021). Here, we show a possible improvement of five genes that may reduce disease-development risk and enhance stem cell therapy efficacy. Abbreviations: PAD, peripheral arterial disease; GWAS, genome-wide association studies.
GWAS have identified several gene loci that significantly contribute to PAD development. Most of the identified gene loci share similar traits with CADs and stroke (Figure 2 and Figure 3). Several genes are directly associated with traditional PAD risk factors (e.g., hyperlipidemia, hypertension, aging, and diabetes). These classical risk factors impair stem/progenitor cell-dependent therapeutic effects (Figure 3). Hence, loci identified using GWAS were integrated into the findings of TWAS to detect gene-trait associations. TWAS leverage expression reference panels (expression quantitative trait loci (eQTL) cohorts with expression and genotype data) to identify gene-trait associations from GWAS datasets [68]. This methodology may help to understand transcription loss in the tissues or cells during PAD development. Moreover, high-throughput deep-sequencing methods enable the discovery of somatic mutations and epigenetic regulation of genes, proving valuable insights into single-nucleotide variants, SNPs, and mutation load that are not currently available in GWAS.
5. Genetic Alterations in PAD
5.1. Somatic Mutation in PAD
In 2019, 240 million cases of PAD were reported worldwide. The prevalence of PAD markedly increases with age (from 6% in individuals aged 40 years to approximately 27% in individuals at an advanced age [42] https://www.cdc.gov/heartdisease/PAD.htm. Accessed on 18 October 2021) These statistical data are positively correlated with the CHIP prevalence in patients with CVD. CHIP rarely occurs in the young population. However, the prevalence of CHIP increases to 10%–20% in the aging population [5] (see Table 5).
Muendlein et al. revealed that the prevalence of JAK2 V617F mutation frequency in a cohort of 287 patients with sonographically confirmed PAD was higher than that in 997 healthy subjects. The acquired JAK2 V617F mutation frequency in patients with PAD was five-fold higher than that in healthy individuals (p < 0.001). Interestingly, the frequency of mutations in patients with PAD markedly decreased in patients who received aspirin (p < 0.003) [7]. Moreover, JAK2 mutations are reported to play an essential role in systemic inflammation, coagulation, decreased proliferation and angiogenesis, and thrombosis through the activation of the downstream STAT1,6, MAPK, and PI3K/AKT signaling pathways [73]. Mechanistically, patients with JAK2 V617F mutation exhibit decreased blood endothelial-cell outgrowth and enhanced expression of interferon-related genes, including serine protease inhibitor B2, early growth response protein 1, and chemokine ligand 2 [30]. This indicated that individuals with JAK2 V617F mutation exhibit sustained inflammation/permeability, limited cell growth (or angiogenesis), and EPC senescence when compared with healthy controls [30]. Walsh et al. [74] recently demonstrated that JAK2 V617F-mediated CH promotes pathological cardiac remodeling by enhancing the pro-inflammatory properties of circulating myeloid cells, such as secretion of IL-1b, IL-6, and TNFα. Clinical trials on previously transplanted CD34+ or cultured “early EPC” and (modestly) “late EPC” did not reveal beneficial effects on ischemic diseases (Table 4) [24,75]. This may suggest that the cell product is impaired due to CHIP. Additionally, a comparative study of somatic mutation profiling of CD34+, HSCs, and circulating endothelial cells (CECs) revealed that in 70% of patients with MDS—both CECs and HSCs—harbor the most frequently mutated genes, such as JAK2, ASXL1, TET2, NOTCH1, and SRSF2 [76].
5.2. Inflammaging and PAD
Inflammation promotes several pathologies associated with aging, which is called “inflammaging” [77]. Recent studies have reported the downstream regulation of mutated genes involved in the inflammatory process. For example, TET2 deficiency in hematopoietic cells promotes an aberrant production of inflammatory cytokines/chemokines, such as IL-6 and IL-1b in macrophages. This provides a direct link between TET2 loss and microenvironmental changes within the BM niche [78,79] (Figure 4). Moreover, a recent large cross-sectional analysis of (n = 750, 249) patients with MDS across the United States revealed that patients with TET2, DNMT3A, and AXSL1 mutations were associated with a high prevalence of PAD (prevalence among individuals aged <65 and >65 years was 14.5% and 43.2%, respectively; p > 0.0001) and CAD (prevalence among individuals aged <65 and >65 years was 9.1% and 17.3%, respectively; p < 0.04) [29]. These findings indicate that CHIP carrier patients with PAD aged >65 were at three- to four-fold higher risk of developing PAD than non-carriers. Patients with PAD who have previously undergone transplantation with cells, such as BMNCs or PBMCs or with enriched CD34 cells may exhibit a pro-inflammatory cell phenotype due to somatic mutations in these cells. Transplantation of enriched CD34 cells exacerbates cell therapy outcomes rather than regenerating ischemic tissues [10,15,80]. Transplanted mutant clones increase clone size after auto-transplantation of cells and enhance the secretion of inflammatory cytokines and CRP (Figure 4) [36,81]. The enhanced levels of pro-inflammatory cytokines maintain mutant hematopoietic stem/progenitor cell survival and growth as they inhibit the HDAC2-mediated suppression of IL-6 transcription or upregulate a novel anti-apoptotic long, non-coding RNA (Figure 1) [82,83].
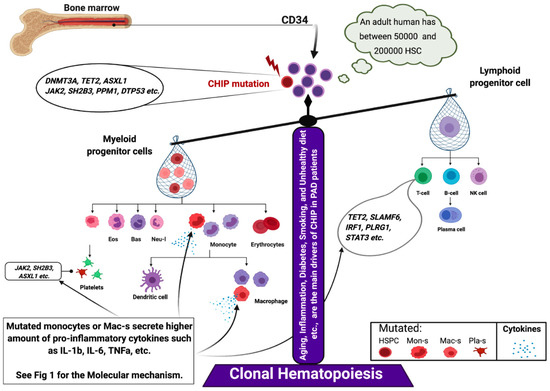
Figure 4.
CHIP-induced hematopoiesis. Abbreviation: CHIP, clonal hematopoiesis of indeterminate potential.
5.3. SH2B3/Lnk Mutations Interfere with Key Regeneration Pathways
A key inhibitor of HSC proliferation and inflammation is the lymphocyte adaptor protein Lnk, which is encoded by SH2B3, a member of the SH2B (Src homology 2-B) adaptor protein family [84]. Aberrant SH2B3/Lnk expression is associated with multiple HSC proliferation-related pathologies [85]. In the PERFECT trial, patients with CAD expressed SNP rs3184504 variant of SH2B3 and exhibited altered SH2B3 expression levels in the peripheral blood [10,15]. Compared with SH2B3 knockout mouse models, the downregulated SH2B3 expression levels promoted CH in various human cells, including myeloid and lymphocytic subsets. In addition to downregulated SH2B3 expression levels, mutations, such as rs3184504, may enhance inflammation in precursor/immune cells and endothelial cells. The regulatory role of SH2B3 in hematopoietic and immune regulation, which involves the inhibition of stem cell proliferation (PLCG1 a.o.) and inflammation by promoting lipid oxidation (LPCAT2 a.o.), can be classified as a stem cell switch [10,86]. Therefore, SH2B3/Lnk mutation-mediated interference of proliferation or inflammatory pathway downregulation may affect PAD/CAD progression based on pleiotropic involvement of the Lnk protein in stem/precursor proliferation, integrin signaling, platelet thrombus formation, and endothelial activation [5,86,87]. Therefore, SH2B3 is a potential therapeutic target for PAD/CAD.
Thus, recent data indicate that somatic mutations in blood cells significantly increase with age. The mutated blood cells (mainly myeloid cells) secrete increased levels of inflammatory cytokines, such as IL-6, TNFα, and IL-1b, and consequently impair organ function rather than promote tissue regeneration.
6. Patient Screening
Recent findings on genetic risk factors that are directly associated with CVD complications (including PAD, CAD, stroke, and hypertension) must be considered, along with comorbid diseases and inflammation, to evaluate the health of individuals with CH, as well as to treat and predict five-year or ten-year mortality. Screening of the population and identification of at least three or more of the following parameters may increase identification of CHIP carriers:
- (1)
- Age: Older adults > 60 years of age [6,28,88].
- (2)
- Blood status: Patients with unexplained anemia or aberrant laboratory parameters (altered myeloid cell/erythroid cell ratio or myeloid/lymphoid ratio) may be associated with an underlying stem cell disorder [89,90].
- (3)
- Pre-existing comorbidities: CAD, PAD, stroke, atherosclerosis, hypertension, diabetes, etc. [5,91].
- (4)
- Inflammatory status: elevated peripheral blood cytokines, such as IL-6, TNF-α, IL-1b, and CRP [78,79,81,91].
- (5)
- Patients with pre-oncology-related factors in the blood may need additional screening for CVD diseases, such as CAD, PAD, and stroke [29].
Thus, experimental and clinical data demonstrate that decreasing or controlling somatic mutation-inducing risk factors, such as inflammation, lipid metabolism, glucose level, diet, and smoking. significantly reduces major adverse cardiovascular events (MACE) and improves stem cell therapy outcomes. State-of-the-art artificial intelligence (AI) algorithms must be utilized to handle these vast amounts of diverse, large medical datasets that include individual genetic features, clinical findings, laboratory biomarkers, and connective computational analyses in stem cell therapy.
7. AI-Based R and NR Stratification of Patients with PAD Undergoing Stem Cell Transplantation
AI has contributed to the analysis of clinical data in biomedical fields, including cardiovascular medicine. Additionally, AI assists and supports clinical decisions within a short duration [92].
7.1. High Data Quality Is Required for Accurate Prediction Models
The major challenges associated with clinical research on diseases (including PAD) include accessing and retrieving high-quality datasets [15]. The integrated analysis of high-throughput deep-sequencing data, patient phenotypes, images, and SNP loci enables the identification of robust biomarker candidates if controlled, manually curated, and high-quality data are used [93]. Data quality for advanced analysis is critical because it determines the outcomes of subsequent computational analysis [94]. Most studies utilize structured data for predictive modeling and ignore potentially valuable information in unstructured clinical notes, such as doctor reports. This is because of the challenges associated with integrating diverse reports, which are mostly handwritten text documents, into common AI algorithms. The integration of heterogeneous data types across electronic health records (EHRs) through deep learning (DL) techniques is reported to improve the performance of AI prediction models. Zhang et al. demonstrated that the models constructed based on the integration of unstructured clinical notes with structured data outperformed other models that utilize only unstructured notes or structured data [95]. Other DL methods have recently been reviewed for heterogeneous medical data [96] and image analysis [97]. However, AI-assisted clinical decisions should mainly be obtained from structured data to avoid overgeneralization from sparse datasets. Unstructured data allow adequate comprehensibility of the findings. Personalized therapy data, such as individual medical documents obtained from unstructured data, can be integrated. Thus, all data under investigation should be obtained under good practice (GxP) [98]. However, GxP cannot ensure the usage of appropriate or scientifically relevant methods nor the scientific significance of analyses or examinations. The data integrated into an AI study should be carefully analyzed, as using more data may decrease the signal-to-noise ratio [99].
7.2. Supporting Decision Making through AI-Based Complex Data Analysis
AI models are currently developed to mitigate the black-box effect, which leads to a lack of interpretability and transparency. Previously, this was the most important reason for the skeptical view of this technology held by patients and clinicians. This is due to the lack of trust in unfamiliar interfaces and hesitancy to rely on a machine or mathematical algorithm for making critical life decisions [100,101].
However, AI-based data analysis algorithms can be an independent extension of previously established statistical approaches for disease risk assessment, such as the recommendations by the American Heart Association/American College of Cardiology (ACC/AHA), which can predict the prognostic risk of CVD based on common risk factors, such as cholesterol, age, smoking, and diabetes [102]. However, several patients are not identified through the classical linear prediction models, and some patients are unnecessarily treated due to false-positive classifications [103,104]. Classical models may thus oversimplify complex, high-dimensional datasets using insufficient parameters. However, the integration of several parameters can result in overfitting the model. This so-called bias-variance dilemma occurs while generating an AI model and has a direct effect on the prediction accuracy, interpretability, and robustness to interpret new data. Complex models with many parameters and a high variance can often lead to overfitting. In these cases, the model adapts itself too closely to the training data and exhibits limited performance on new patients. In contrast, high-biased models that are not complex tend to ignore data points and important features, which ultimately leads to underfitting and decreased model accuracy [105]. This tradeoff must be specifically evaluated and taken into consideration for each dataset and newly trained model by testing different parametrizations. The increased availability of highly efficient AI algorithms has enabled the development of alternative approaches to classical linear prediction models. These models can utilize large, integrative datasets for improved prognosis and diagnosis [94]. In 2014, Dilsizian and Siegel reported that the vast amount of information obtained from patients and pre-clinical studies is too complex and heterogeneous for humans to comprehensively interpret without any technological support [106].
7.3. Identification of Responsive Patients for Cell Therapy
The number of patients undergoing genetic testing for various diseases, ranging from cancer to cardiomyopathy, has steadily increased. Additionally, RNA-seq can reveal specific mutational status and utilize the same guidelines used for DNA sequencing [107]. The ethical principles and history underlying clinical genetics will provide clinicians with improved tools to guide their practice and help patients navigate through complex medical-psychosocial terrain [108].
Recent biomedical studies have aimed to identify patient-specific biomarker signatures from high-throughput data to effectively predict postoperative results by stratifying patients into the R and NR groups before therapy. Wolfien et al. proposed a diagnostic strategy to predict the response of patients undergoing coronary artery bypass grafting to BMSC-mediated myocardial repair [10]. Predictive analyses in this case would provide useful insights to identify individuals who are most likely to benefit from BMSC treatment with patient-specific diagnostic characteristics.
This focused panel of molecular targets is consistent with the current comparison of AI approaches and traditional models for using administrative claims with EHR to predict heart-failure outcomes [109]. In this case, approaches with traditional logistic regression were compared in order to predict key outcomes in patients with heart failure, and the added value of predictive models was evaluated using EHR data. In total, 9502 patients (aged ≥ 65 years) with at least one heart-failure diagnosis were identified. Of these, 6113 were included in the training set, while 3389 were used as the testing set. The study comprised a large dataset with standard clinical parameters that do not have increased predictive value for ML-based stratification. The authors initially observed limited predictive capacity in the PERFECT study. Hence, they used additional molecular data, which improved the prediction accuracy of the ML model from 64% to 82% [15]. Further, the authors used specific molecular data, such as RNA-seq data. The stratification accuracy and sensitivity for a larger cohort increased by more than 10% [10]. Firouzi and Sussman suggested that this novel strategic approach of combining transcriptome profiling with precise patient phenotyping and AI-guided feature selection is a potentially valuable tool for advancing personalized medicine and cell-based therapies [110].
7.4. A New Diagnostic Tool Supporting Individual Disease Characterization
Generally, all studies utilize different ML methods to identify decision boundaries or specific patterns within patient data to individually characterize patients, identify similar groups or subgroups (referred to as clustering), or assign them to a certain disease, disease stage, or treatment option (referred to as stratification). The underlying “learning step” from data input to prediction involves a test of all available features for their capability of separating patients or subgroups in a supervised or unsupervised manner. The algorithm knows the correct label (e.g., control, treatment, and diagnosis) (the so-called ground-truth) in supervised ML. Each data point consists of manually selected features and the corresponding label (e.g., a data point represents a patient, the features are the clinical data, and the label is the stage of a specific disease). The algorithms attempt to identify a suitable relation between the features and the known label during the learning process, which is usually performed retrospectively on previously generated data. A trained ML model, which serves as a decision boundary, can now be applied to new patient data points to predict a label on previously unlabeled patient data. Here, a well-chosen patient parameter set without high imbalance ratios between the investigated groups and a well-defined ground truth is the key for accurate model performance. However, this can often not be assessed in medical data. New ML-based algorithms for dimensional reduction and visualization, such as t-SNE or UMAP, may enable new possibilities for classifying or reconsidering disease subgroups among patients with unknown labels. These unsupervised technologies for medical cases are predominantly used with heterogeneous data, especially for activated or aberrant signaling pathways (e.g., measured in peripheral blood or single-cell RNA-seq), genetic background (GWAS/TWAS), acquired SNPs, or other comorbidities [111,112]. Based on the integrative data, unique combinations and hybrid forms of diseases will be observed, as has been previously demonstrated on a smaller scale, in which AI will support their identification and characterization [10]. These investigations at this specific individual level would have been impossible without the use of AI. Therefore, these investigations assist in routine clinical decisions, especially concerning personalized cell therapy for PAD.
As previously mentioned, the analysis of medical images is one of the greatest success stories of AI, spanning the analysis of histopathological images [113], electrocardiograms [114], radiographs [115], magnetic resonance imaging slices [116], and many more [108]. Since imaging is very much an emerging field in PAD, this bears high potential because several already-established biomarkers could already be used for an unbiased patient stratification [117]. Similarly, a current review of Flores et al. points towards the infancy of AI in PAD but also foresees a broad spectrum of potential applications [118]. One of the first proofs of concept was implemented by Kim et al. [119] who used a deep convolutional neural network for the detection and assessment of the severity of PAD based on the analysis of brachial and ankle arterial pulse waveforms. These findings were compared with the state-of-the-art ankle-brachial index (ABI) using the virtual patients and showed, according to the authors, that DL may diagnose PAD more accurately and robustly than ABI. This work demonstrates a DL-based arterial pulse waveform analysis for affordable and convenient PAD screening, as well as the open challenges that need to be addressed for real-world clinical applications. However, one current limitation of imaging in PAD is the profound correlation of biomarkers with genetic alterations, which might be addressed with the help of AI, as already demonstrated in other fields, [120] as these ensemble models allow for complex data integration.
Nevertheless, a simple in silico identification of novel subgroups or biomarkers without a specific explanation for the choice a treatment option is insufficient. Therefore, AI algorithms in personalized PAD treatment also must utilize heterogeneous information at the gene level (e.g., GWAS, gene expression, pathway activity, identification of mutations) and phenotype level (e.g., cardiac functionality, angiogenesis potential, and inflammation status) to optimize predictions for cell therapy applications in the future. To achieve this, large patient cohorts are needed to utilize the full potential of personalized medicine therapies assisted by AI approaches.
8. Conclusions and Future Perspectives
Cell therapy can improve ABI, TCPO2, rest pain, pain-free walking distance, ulcer healing, and limb salvage markedly in some patients and moderately in other patients. However, cell therapy outcomes may be improved using state-of-the-art AI-based prediction methods before cell therapy to define the R and NR groups. Previous predictor findings and other studies have reported that up to 40% of CLTI patients are not candidates for revascularization [121]. Hence, autologous personalized cell therapy may be considered as a new standard of care for these patients [10,122]. Growing evidence indicates that genetic screening of patients before stem cell transplantation for ischemic CVD facilitates the assessment of regenerative potential. The average treatment effect of randomized cell therapy trials and precision medicine must be reconsidered based on genetic and clinical laboratory findings by employing an unbiased AI approach to identify the most effective treatment option for patients and decrease treatment expenditure.
Possible next steps should involve larger patient groups and an integration of the AI supportive systems directly in clinical care [123]. This way, all involved stakeholders, including clinicians, patients, and IT practitioners, will become more familiar with the new technologies and the final applications will likewise grow based on the given feedback [124]. However, multimodal data integration, security, federated learning, model performance, understanding, and bias still pose challenges and hurdles to the use of AI in health care in general, which, of course, also applies to PAD [125]. In turn, this is why a mechanistic understanding of genetic alterations and mutational profiles is crucial, in addition to sole AI predictions, as it was recently demonstrated for SH2B3/Lnk mutations in mice. A well-suited clinical AI model should not only simply select responding patients but should give indications of the decision being made to get more transparent and accepted.
GWAS and broad cross-sectional studies have demonstrated that race, age, and sex variations must be examined accurately due to the distribution of diseases and genotype variations between ethnic groups, such as Asian, Western, Hispanic, and Afro-American populations. Allogeneic cell and extracellular vesicle (EV) transplantation for CVD is safe and feasible (decreased mutations when transplanting young donor cells/EVs to patients with PAD) and may be optimal for the NR group [57,126]. Taken together, future studies must elucidate strong predictive biomarkers obtained from genotype and phenotype findings of the R and NR groups using ML-based methods to support clinical decisions for managing PAD.
Author Contributions
Conceptualization, A.A.S., M.W., T.A. and G.S.; software, A.A.S. and M.W.; writing—original draft preparation, A.A.S. and M.W.; writing—review and editing, T.A. and G.S.; visualization, A.A.S.; funding acquisition, S.K. and A.A.S. All authors have read and agreed to the published version of the manuscript.
Funding
Salybekov is supported by JSPS KAKENHI (grant no. 20K17163), and M. Wolfien was supported by the EU Social Fund (ESF/14-BM-A55-0027/18).
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Michael, H.; Criqui, V.A. Epidemiology of Peripheral Artery Disease. Circ. Res. 2015, 116, 1509–1526. [Google Scholar]
- Howard, D.P.; Banerjee, A.; Fairhead, J.F.; Hands, L.; Silver, L.E.; Rothwell, P.M.; Oxford Vascular, S. Population-Based Study of Incidence, Risk Factors, Outcome, and Prognosis of Ischemic Peripheral Arterial Events: Implications for Prevention. Circulation 2015, 132, 1805–1815. [Google Scholar] [CrossRef]
- Fowkes, F.G.; Rudan, D.; Rudan, I.; Aboyans, V.; Denenberg, J.O.; McDermott, M.M.; Norman, P.E.; Sampson, U.K.; Williams, L.J.; Mensah, G.A.; et al. Comparison of global estimates of prevalence and risk factors for peripheral artery disease in 2000 and 2010: A systematic review and analysis. Lancet 2013, 382, 1329–1340. [Google Scholar] [CrossRef]
- Joosten, M.M.; Pai, J.K.; Bertoia, M.L.; Rimm, E.B.; Spiegelman, D.; Mittleman, M.A.; Mukamal, K.J. Associations between conventional cardiovascular risk factors and risk of peripheral artery disease in men. JAMA 2012, 308, 1660–1667. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [CrossRef] [PubMed]
- Fuster, J.J.; Walsh, K. Somatic Mutations and Clonal Hematopoiesis: Unexpected Potential New Drivers of Age-Related Cardiovascular Disease. Circ. Res. 2018, 122, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Muendlein, A.; Kinz, E.; Gasser, K.; Leiherer, A.; Rein, P.; Saely, C.H.; Grallert, H.; Peters, A.; Fraunberger, P.; Drexel, H.; et al. Occurrence of the JAK2 V617F mutation in patients with peripheral arterial disease. Am. J. Hematol. 2015, 90, E17–E21. [Google Scholar] [CrossRef]
- Jaiswal, S.; Ebert, B.L. Clonal Hematopoiesis in Human Aging and Disease. Science 2019, 366. [Google Scholar] [CrossRef] [PubMed]
- Gibson, C.J.; Lindsley, R.C.; Tchekmedyian, V.; Mar, B.G.; Shi, J.; Jaiswal, S.; Bosworth, A.; Francisco, L.; He, J.; Bansal, A.; et al. Clonal Hematopoiesis Associated With Adverse Outcomes After Autologous Stem-Cell Transplantation for Lymphoma. J. Clin. Oncol. 2017, 35, 1598–1605. [Google Scholar] [CrossRef]
- Wolfien, M.; Klatt, D.; Salybekov, A.A.; Ii, M.; Komatsu-Horii, M.; Gaebel, R.; Philippou-Massier, J.; Schrinner, E.; Akimaru, H.; Akimaru, E.; et al. Hematopoietic stem-cell senescence and myocardial repair—Coronary artery disease genotype/phenotype analysis of post-MI myocardial regeneration response induced by CABG/CD133+ bone marrow hematopoietic stem cell treatment in RCT PERFECT Phase 3. EBioMedicine 2020, 57, 102862. [Google Scholar] [CrossRef]
- Steensma, D.P.; Bejar, R.; Jaiswal, S.; Lindsley, R.C.; Sekeres, M.A.; Hasserjian, R.P.; Ebert, B.L. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood 2015, 126, 9–16. [Google Scholar] [CrossRef]
- Falconi, G.; Fabiani, E.; Piciocchi, A.; Criscuolo, M.; Fianchi, L.; Rossi, E.L.L.; Finelli, C.; Cerqui, E.; Ottone, T.; Molteni, A.; et al. Somatic mutations as markers of outcome after azacitidine and allogeneic stem cell transplantation in higher-risk myelodysplastic syndromes. Leukemia 2019, 33, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Young, A.L.; Challen, G.A.; Birmann, B.M.; Druley, T.E. Clonal haematopoiesis harbouring AML-associated mutations is ubiquitous in healthy adults. Nat. Commun. 2016, 7, 12484. [Google Scholar] [CrossRef]
- Asahara, T.; Masuda, H.; Takahashi, T.; Kalka, C.; Pastore, C.; Silver, M.; Kearne, M.; Magner, M.; Isner, J.M. Bone marrow origin of endothelial progenitor cells responsible for postnatal vasculogenesis in physiological and pathological neovascularization. Circ. Res. 1999, 85, 221–228. [Google Scholar] [CrossRef]
- Steinhoff, G.; Nesteruk, J.; Wolfien, M.; Kundt, G.; Börgermann, J.; David, R.; Garbade, J.; Große, J.; Haverich, A.; Hennig, H.; et al. Cardiac Function Improvement and Bone Marrow Response -: Outcome Analysis of the Randomized PERFECT Phase III Clinical Trial of Intramyocardial CD133(+) Application After Myocardial Infarction. EBioMedicine 2017, 22, 208–224. [Google Scholar] [CrossRef]
- Tateishi-Yuyama, E.; Matsubara, H.; Murohara, T.; Ikeda, U.; Shintani, S.; Masaki, H.; Amano, K.; Kishimoto, Y.; Yoshimoto, K.; Akashi, H.; et al. Therapeutic angiogenesis for patients with limb ischaemia by autologous transplantation of bone-marrow cells: A pilot study and a randomised controlled trial. Lancet 2002, 360, 427–435. [Google Scholar] [CrossRef]
- Molavi, B.; Zafarghandi, M.R.; Aminizadeh, E.; Hosseini, S.E.; Mirzayi, H.; Arab, L.; Baharvand, H.; Aghdami, N. Safety and Efficacy of Repeated Bone Marrow Mononuclear Cell Therapy in Patients with Critical Limb Ischemia in a Pilot Randomized Controlled Trial. Arch. Iran. Med. 2016, 19, 388–396. [Google Scholar] [PubMed]
- Kawamoto, A.; Katayama, M.; Handa, N.; Kinoshita, M.; Takano, H.; Horii, M.; Sadamoto, K.; Yokoyama, A.; Yamanaka, T.; Onodera, R.; et al. Intramuscular transplantation of G-CSF-mobilized CD34(+) cells in patients with critical limb ischemia: A phase I/IIa, multicenter, single-blinded, dose-escalation clinical trial. Stem Cells 2009, 27, 2857–2864. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Kinoshita, M.; Furukawa, Y.; Nagano, T.; Hashimoto, H.; Hirami, Y.; Kurimoto, Y.; Arakawa, K.; Yamazaki, K.; Okada, Y.; et al. Phase II clinical trial of CD34+ cell therapy to explore endpoint selection and timing in patients with critical limb ischemia. Circ. J. 2014, 78, 490–501. [Google Scholar] [CrossRef]
- Barć, P.; Skóra, J.; Pupka, A.; Turkiewicz, D.; Dorobisz, A.T.; Garcarek, J.; Tomasiewicz, B.; Szyber, P. Bone-marrow cells in therapy of critical limb ischemia of lower extremities - own experience. Acta Angiol. 2006, 12, 155–166. Available online: https://journals.viamedica.pl/acta_angiologica/article/view/9862 (accessed on 13 May 2021).
- Miyamoto, K.; Nishigami, K.; Nagaya, N.; Akutsu, K.; Chiku, M.; Kamei, M.; Soma, T.; Miyata, S.; Higashi, M.; Tanaka, R.; et al. Unblinded pilot study of autologous transplantation of bone marrow mononuclear cells in patients with thromboangiitis obliterans. Circulation 2006, 114, 2679–2684. [Google Scholar] [CrossRef]
- Benoit, E.; O’Donnell, T.F., Jr.; Iafrati, M.D.; Asher, E.; Bandyk, D.F.; Hallett, J.W.; Lumsden, A.B.; Pearl, G.J.; Roddy, S.P.; Vijayaraghavan, K.; et al. The role of amputation as an outcome measure in cellular therapy for critical limb ischemia: Implications for clinical trial design. J. Transl. Med. 2011, 9, 165. [Google Scholar] [CrossRef] [PubMed]
- Teraa, M.; Sprengers, R.W.; Schutgens, R.E.; Slaper-Cortenbach, I.C.; van der Graaf, Y.; Algra, A.; van der Tweel, I.; Doevendans, P.A.; Mali, W.P.; Moll, F.L.; et al. Effect of repetitive intra-arterial infusion of bone marrow mononuclear cells in patients with no-option limb ischemia: The randomized, double-blind, placebo-controlled Rejuvenating Endothelial Progenitor Cells via Transcutaneous Intra-arterial Supplementation (JUVENTAS) trial. Circulation 2015, 131, 851–860. [Google Scholar] [CrossRef]
- Losordo, D.W.; Kibbe, M.R.; Mendelsohn, F.; Marston, W.; Driver, V.R.; Sharafuddin, M.; Teodorescu, V.; Wiechmann, B.N.; Thompson, C.; Kraiss, L.; et al. A randomized, controlled pilot study of autologous CD34+ cell therapy for critical limb ischemia. Circ. Cardiovasc. Interv. 2012, 5, 821–830. [Google Scholar] [CrossRef] [PubMed]
- Klepanec, A.; Mistrik, M.; Altaner, C.; Valachovicova, M.; Olejarova, I.; Slysko, R.; Balazs, T.; Urlandova, T.; Hladikova, D.; Liska, B.; et al. No difference in intra-arterial and intramuscular delivery of autologous bone marrow cells in patients with advanced critical limb ischemia. Cell Transpl. 2012, 21, 1909–1918. [Google Scholar] [CrossRef] [PubMed]
- Madaric, J.; Klepanec, A.; Valachovicova, M.; Mistrik, M.; Bucova, M.; Olejarova, I.; Necpal, R.; Madaricova, T.; Paulis, L.; Vulev, I. Characteristics of responders to autologous bone marrow cell therapy for no-option critical limb ischemia. Stem Cell Res. Ther. 2016, 7, 116. [Google Scholar] [CrossRef] [PubMed]
- Pan, T.; Liu, H.; Fang, Y.; Wei, Z.; Gu, S.; Fang, G.; Liu, Y.; Luo, Y.; Guo, D.; Xu, X.; et al. Predictors of responders to mononuclear stem cell-based therapeutic angiogenesis for no-option critical limb ischemia. Stem Cell Res. Ther. 2019, 10, 15. [Google Scholar] [CrossRef]
- Jaiswal, S.; Fontanillas, P.; Flannick, J.; Manning, A.; Grauman, P.V.; Mar, B.G.; Lindsley, R.C.; Mermel, C.H.; Burtt, N.; Chavez, A.; et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 2014, 371, 2488–2498. [Google Scholar] [CrossRef]
- Rivero, G.A.; Perli, E.; Moreno, S.; Salemi, J.L. Excess in Atherosclerotic and Inflammametabolic Diseases Are Differentially Expressed in Myelodysplasia and Are Highly Dependent on Age, R-IPSS and Ethnicity. Blood 2018, 132. [Google Scholar] [CrossRef]
- Cosgrove, M.E.; Suman, R.; Harrison, H.J.; Jackson, G.E.; Howard, M.R.; Hitchcock, I.S. Endothelial JAK2V617F Expression Drives Inflammation and Cellular Senescence; New Evidence for the Roles of Endothelial Cells in MPN-Related Clotting Abnormalities? Blood 2016, 128. [Google Scholar] [CrossRef]
- Malyar, N.M.; Radtke, S.; Malyar, K.; Arjumand, J.; Horn, P.A.; Kroger, K.; Freisinger, E.; Reinecke, H.; Giebel, B.; Brock, F.E. Autologous bone marrow mononuclear cell therapy improves symptoms in patients with end-stage peripheral arterial disease and reduces inflammation-associated parameters. Cytotherapy 2014, 16, 1270–1279. [Google Scholar] [CrossRef]
- Arai, M.; Misao, Y.; Nagai, H.; Kawasaki, M.; Nagashima, K.; Suzuki, K.; Tsuchiya, K.; Otsuka, S.; Uno, Y.; Takemura, G.; et al. Granulocyte Colony-Stimulating Factor A Noninvasive Regeneration Therapy for Treating Atherosclerotic Peripheral Artery Disease. Circ. J. 2006, 70, 1093–1098. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Pan, T.; Fang, Y.; Wei, Z.; Gu, S.; Fang, G.; Liu, Y.; Luo, Y.; Liu, H.; Zhang, T.; et al. Purified CD34(+) cells versus peripheral blood mononuclear cells in the treatment of angiitis-induced no-option critical limb ischaemia: 12-Month results of a prospective randomised single-blinded non-inferiority trial. EBioMedicine 2018, 35, 46–57. [Google Scholar] [CrossRef]
- Idei, N.; Soga, J.; Hata, T.; Fujii, Y.; Fujimura, N.; Mikami, S.; Maruhashi, T.; Nishioka, K.; Hidaka, T.; Kihara, Y.; et al. Autologous bone-marrow mononuclear cell implantation reduces long-term major amputation risk in patients with critical limb ischemia: A comparison of atherosclerotic peripheral arterial disease and Buerger disease. Circ. Cardiovasc. Interv. 2011, 4, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Guo, L.; Cui, S.; Tong, Z.; Dardik, A.; Gu, Y. Autologous bone marrow-derived mononuclear cell therapy in Chinese patients with critical limb ischemia due to thromboangiitis obliterans: 10-year results. Stem Cell Res. Ther. 2018, 9, 43. [Google Scholar] [CrossRef] [PubMed]
- Ortmann, C.A.; Dorsheimer, L.; Abou-El-Ardat, K.; Hoffrichter, J.; Assmus, B.; Bonig, H.; Scholz, A.; Pfeifer, H.; Martin, H.; Schmid, T.; et al. Functional Dominance of CHIP-Mutated Hematopoietic Stem Cells in Patients Undergoing Autologous Transplantation. Cell Rep. 2019, 27, 2022–2028.e3. [Google Scholar] [CrossRef]
- Piazza, G.; Creager, M.A. Thromboangiitis obliterans. Circulation 2010, 121, 1858–1861. [Google Scholar] [CrossRef]
- Rivera-Chavarria, I.J.; Brenes-Gutierrez, J.D. Thromboangiitis obliterans (Buerger’s disease). Ann. Med. Surg. 2016, 7, 79–82. [Google Scholar] [CrossRef]
- Shu, J.; Santulli, G. Update on peripheral artery disease: Epidemiology and evidence-based facts. Atherosclerosis 2018, 275, 379–381. [Google Scholar] [CrossRef]
- Fazeli, B.; Dadgar Moghadam, M.; Niroumand, S. How to Treat a Patient with Thromboangiitis Obliterans: A Systematic Review. Ann. Vasc. Surg. 2018, 49, 219–228. [Google Scholar] [CrossRef]
- Cooper, L.T.; Henderson, S.S.; Ballman, K.V.; Offord, K.P.; Tse, T.S.; Holmes, D.R.; Hurt, R.D. A prospective, case-control study of tobacco dependence in thromboangiitis obliterans (Buerger’s Disease). Angiology 2006, 57, 73–78. [Google Scholar] [CrossRef]
- Song, P.; Rudan, D.; Zhu, Y.; Fowkes, F.J.I.; Rahimi, K.; Fowkes, F.G.R.; Rudan, I. Global, regional, and national prevalence and risk factors for peripheral artery disease in 2015: An updated systematic review and analysis. Lancet Glob. Health 2019, 7, e1020–e1030. [Google Scholar] [CrossRef]
- Thiruvoipati, T.; Kielhorn, C.E.; Armstrong, E.J. Peripheral artery disease in patients with diabetes: Epidemiology, mechanisms, and outcomes. World J. Diabetes 2015, 6, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Adar, R.; Papa, M.Z.; Halpern, Z.; Mozes, M.; Shoshan, S.; Sofer, B.; Zinger, H.; Dayan, M.; Mozes, E. Cellular sensitivity to collagen in thromboangiitis obliterans. N. Engl. J. Med. 1983, 308, 1113–1116. [Google Scholar] [CrossRef]
- Wei, Z.; Jiang, W.; Wang, H.; Li, H.; Tang, B.; Liu, B.; Jiang, H.; Sun, X. The IL-6/STAT3 pathway regulates adhesion molecules and cytoskeleton of endothelial cells in thromboangiitis obliterans. Cell. Signal. 2018, 44, 118–126. [Google Scholar] [CrossRef] [PubMed]
- Fadini, G.P.; Albiero, M.; Vigili de Kreutzenberg, S.; Boscaro, E.; Cappellari, R.; Marescotti, M.; Poncina, N.; Agostini, C.; Avogaro, A. Diabetes impairs stem cell and proangiogenic cell mobilization in humans. Diabetes Care 2013, 36, 943–949. [Google Scholar] [CrossRef]
- Fadini, G.P.; Fiala, M.; Cappellari, R.; Danna, M.; Park, S.; Poncina, N.; Menegazzo, L.; Albiero, M.; DiPersio, J.; Stockerl-Goldstein, K.; et al. Diabetes Limits Stem Cell Mobilization Following G-CSF but Not Plerixafor. Diabetes 2015, 64, 2969–2977. [Google Scholar] [CrossRef]
- Albiero, M.; Poncina, N.; Tjwa, M.; Ciciliot, S.; Menegazzo, L.; Ceolotto, G.; de Kreutzenberg, S.V.; Moura, R.; Giorgio, M.; Pelicci, P.; et al. Diabetes causes bone marrow autonomic neuropathy and impairs stem cell mobilization via dysregulated p66Shc and Sirt1. Diabetes 2014, 63, 1353–1365. [Google Scholar] [CrossRef]
- Teraa, M.; Fledderus, J.O.; Rozbeh, R.I.; Leguit, R.J.; Verhaar, M.C.; Juventas Study, G. Bone marrow microvascular and neuropathic alterations in patients with critical limb ischemia. Circ. Res. 2014, 114, 311–314. [Google Scholar] [CrossRef]
- Bonnefond, A.; Skrobek, B.; Lobbens, S.; Eury, E.; Thuillier, D.; Cauchi, S.; Lantieri, O.; Balkau, B.; Riboli, E.; Marre, M.; et al. Association between large detectable clonal mosaicism and type 2 diabetes with vascular complications. Nat. Genet. 2013, 45, 1040–1043. [Google Scholar] [CrossRef]
- Fadini, G.P.; Ciciliot, S.; Albiero, M. Concise Review: Perspectives and Clinical Implications of Bone Marrow and Circulating Stem Cell Defects in Diabetes. Stem Cells 2017, 35, 106–116. [Google Scholar] [CrossRef]
- Kojima, H.; Kim, J.; Chan, L. Emerging roles of hematopoietic cells in the pathobiology of diabetic complications. Trends Endocrinol. Metab. 2014, 25, 178–187. [Google Scholar] [CrossRef]
- Orlandi, A.; Chavakis, E.; Seeger, F.; Tjwa, M.; Zeiher, A.M.; Dimmeler, S. Long-term diabetes impairs repopulation of hematopoietic progenitor cells and dysregulates the cytokine expression in the bone marrow microenvironment in mice. Basic Res. Cardiol. 2010, 105, 703–712. [Google Scholar] [CrossRef]
- Tanaka, R.; Vaynrub, M.; Masuda, H.; Ito, R.; Kobori, M.; Miyasaka, M.; Mizuno, H.; Warren, S.M.; Asahara, T. Quality-control culture system restores diabetic endothelial progenitor cell vasculogenesis and accelerates wound closure. Diabetes 2013, 62, 3207–3217. [Google Scholar] [CrossRef] [PubMed]
- Salybekov, A.A.; Masuda, H.; Miyazaki, K.; Sheng, Y.; Sato, A.; Shizuno, T.; Iida, Y.; Okada, Y.; Asahara, T. Dipeptidyl dipeptidase-4 inhibitor recovered ischemia through an increase in vasculogenic endothelial progenitor cells and regeneration-associated cells in diet-induced obese mice. PLoS ONE 2019, 14, e0205477. [Google Scholar] [CrossRef]
- Salybekov, A.A.; Kawaguchi, A.T.; Masuda, H.; Vorateera, K.; Okada, C.; Asahara, T. Regeneration-associated cells improve recovery from myocardial infarction through enhanced vasculogenesis, anti-inflammation, and cardiomyogenesis. PLoS ONE 2018, 13, e0203244. [Google Scholar] [CrossRef] [PubMed]
- Salybekov, A.A.; Salybekova, A.; Sheng, Y.; Shinozaki, Y.; Yokoyama, K.; Kobayashi, S.; Asahara, T. Extracellular Vesicles Derived From Regeneration Associated Cells Preserve Heart Function After Ischemia-Induced Injury. Front. Cardiovasc. Med. 2021, 8, 754254. [Google Scholar] [CrossRef] [PubMed]
- Perttila, J.; Merikanto, K.; Naukkarinen, J.; Surakka, I.; Martin, N.W.; Tanhuanpaa, K.; Grimard, V.; Taskinen, M.R.; Thiele, C.; Salomaa, V.; et al. OSBPL10, a novel candidate gene for high triglyceride trait in dyslipidemic Finnish subjects, regulates cellular lipid metabolism. J. Mol. Med. 2009, 87, 825–835. [Google Scholar] [CrossRef]
- Koriyama, H.; Nakagami, H.; Katsuya, T.; Sugimoto, K.; Yamashita, H.; Takami, Y.; Maeda, S.; Kubo, M.; Takahashi, A.; Nakamura, Y.; et al. Identification of evidence suggestive of an association with peripheral arterial disease at the OSBPL10 locus by genome-wide investigation in the Japanese population. J. Atheroscler. Thromb. 2010, 17. [Google Scholar] [CrossRef]
- Maslah, N.; Cassinat, B.; Verger, E.; Kiladjian, J.J.; Velazquez, L. The role of LNK/SH2B3 genetic alterations in myeloproliferative neoplasms and other hematological disorders. Leukemia 2017, 31, 1661–1670. [Google Scholar] [CrossRef]
- Klarin, D.; Lynch, J.; Aragam, K.; Chaffin, M.; Assimes, T.L.; Huang, J.; Lee, K.M.; Shao, Q.; Huffman, J.E.; Natarajan, P.; et al. Genome-wide Association Study of Peripheral Artery Disease in the Million Veteran Program. Nat. Med. 2019, 25, 1274–1279. [Google Scholar] [CrossRef]
- Nikpay, M.; Goel, A.; Won, H.H.; Hall, L.M.; Willenborg, C.; Kanoni, S.; Saleheen, D.; Kyriakou, T.; Nelson, C.P.; Hopewell, J.C.; et al. A comprehensive 1000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat. Genet. 2015, 47, 1121–1130. [Google Scholar] [CrossRef] [PubMed]
- Malik, R.; Chauhan, G.; Traylor, M.; Sargurupremraj, M.; Okada, Y.; Mishra, A.; Rutten-Jacobs, L.; Giese, A.K.; van der Laan, S.W.; Gretarsdottir, S.; et al. Multiancestry genome-wide association study of 520,000 subjects identifies 32 loci associated with stroke and stroke subtypes. Nat. Genet. 2018, 50, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Han, Z.; Dong, X.; Zhang, C.; Wu, Y.; Yuan, Z.; Wang, X. Polymorphism of HDAC9 Gene Is Associated with Increased Risk of Acute Coronary Syndrome in Chinese Han Population. Biomed. Res. Int. 2016, 2016, 3746276. [Google Scholar] [CrossRef]
- Markus, H.S.; Makela, K.M.; Bevan, S.; Raitoharju, E.; Oksala, N.; Bis, J.C.; O’Donnell, C.; Hainsworth, A.; Lehtimaki, T. Evidence HDAC9 genetic variant associated with ischemic stroke increases risk via promoting carotid atherosclerosis. Stroke 2013, 44, 1220–1225. [Google Scholar] [CrossRef] [PubMed]
- Sofer, T.; Emery, L.; Jain, D.; Ellis, A.M.; Laurie, C.C.; Allison, M.A.; Lee, J.; Kurniansyah, N.; Kerr, K.F.; Gonzalez, H.M.; et al. Variants Associated with the Ankle Brachial Index Differ by Hispanic/Latino Ethnic Group: A genome-wide association study in the Hispanic Community Health Study/Study of Latinos. Sci. Rep. 2019, 9, 11410. [Google Scholar] [CrossRef] [PubMed]
- Ou, M.; Li, X.; Zhao, S.; Cui, S.; Tu, J. Long non-coding RNA CDKN2B-AS1 contributes to atherosclerotic plaque formation by forming RNA-DNA triplex in the CDKN2B promoter. EBioMedicine 2020, 55. [Google Scholar] [CrossRef]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K.; et al. Opportunities and challenges for transcriptome-wide association studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef]
- Savji, N.; Rockman, C.B.; Skolnick, A.H.; Guo, Y.; Adelman, M.A.; Riles, T.; Berger, J.S. Association between advanced age and vascular disease in different arterial territories: A population database of over 3.6 million subjects. J. Am. Coll. Cardiol. 2013, 61, 1736–1743. [Google Scholar] [CrossRef]
- Fuster, J.J.; MacLauchlan, S.; Zuriaga, M.A.; Polackal, M.N.; Ostriker, A.C.; Chakraborty, R.; Wu, C.L.; Sano, S.; Muralidharan, S.; Rius, C.; et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 2017, 355, 842–847. [Google Scholar] [CrossRef]
- Shah, A.D.; Langenberg, C.; Rapsomaniki, E.; Denaxas, S.; Pujades-Rodriguez, M.; Gale, C.P.; Deanfield, J.; Smeeth, L.; Timmis, A.; Hemingway, H. Type 2 diabetes and incidence of cardiovascular diseases: A cohort study in 1.9 million people. Lancet Diabetes Endocrinol. 2015, 3, 105–113. [Google Scholar] [CrossRef]
- Yoshida, K.; Gowers, K.H.C.; Lee-Six, H.; Chandrasekharan, D.P.; Coorens, T.; Maughan, E.F.; Beal, K.; Menzies, A.; Millar, F.R.; Anderson, E.; et al. Tobacco smoking and somatic mutations in human bronchial epithelium. Nature 2020, 578, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Perner, F.; Perner, C.; Ernst, T.; Heidel, F.H. Roles of JAK2 in Aging, Inflammation, Hematopoiesis and Malignant Transformation. Cells 2019, 8, 854. [Google Scholar] [CrossRef]
- Sano, S.; Wang, Y.; Yura, Y.; Sano, M.; Oshima, K.; Yang, Y.; Katanasaka, Y.; Min, K.D.; Matsuura, S.; Ravid, K.; et al. JAK2 (V617F) -Mediated Clonal Hematopoiesis Accelerates Pathological Remodeling in Murine Heart Failure. JACC Basic Transl. Sci. 2019, 4, 684–697. [Google Scholar] [CrossRef] [PubMed]
- Losordo, D.W.; Henry, T.D.; Davidson, C.; Sup Lee, J.; Costa, M.A.; Bass, T.; Mendelsohn, F.; Fortuin, F.D.; Pepine, C.J.; Traverse, J.H.; et al. Intramyocardial, autologous CD34+ cell therapy for refractory angina. Circ. Res. 2011, 109, 428–436. [Google Scholar] [CrossRef]
- Farina, M.; Bernardi, S.; Polverelli, N.; Zanaglio, C.; Malagola, M.; Re, F.; Cattina, F.; D’Adda, M.; Rossi, G.; Zollner, T.; et al. Comparative Somatic Mutational Profiling of CD34+ Hematopoietic Precursors (HSC) and Circulating Endothelial Cells (CEC) in Patients with Primary Myelofibrosis (PMF). Blood 2019, 134, 1684. [Google Scholar] [CrossRef]
- Franceschi, C.; Capri, M.; Monti, D.; Giunta, S.; Olivieri, F.; Sevini, F.; Panourgia, M.P.; Invidia, L.; Celani, L.; Scurti, M.; et al. Inflammaging and anti-inflammaging: A systemic perspective on aging and longevity emerged from studies in humans. Mech. Ageing Dev. 2007, 128, 92–105. [Google Scholar] [CrossRef]
- Cull, A.H.; Snetsinger, B.; Buckstein, R.; Wells, R.A.; Rauh, M.J. Tet2 restrains inflammatory gene expression in macrophages. Exp. Hematol. 2017, 55, 56–70. [Google Scholar] [CrossRef] [PubMed]
- Abegunde, S.O.; Buckstein, R.; Wells, R.A.; Rauh, M.J. An inflammatory environment containing TNFalpha favors Tet2-mutant clonal hematopoiesis. Exp. Hematol. 2018, 59, 60–65. [Google Scholar] [CrossRef]
- Dorsheimer, L.; Assmus, B.; Rasper, T.; Ortmann, C.A.; Ecke, A.; Abou-El-Ardat, K.; Schmid, T.; Brüne, B.; Wagner, S.; Hoffmann, J.; et al. Association of Mutations Contributing to Clonal Hematopoiesis With Prognosis in Chronic Ischemic Heart Failure. JAMA Cardiol. 2020, 4, 25–33. [Google Scholar] [CrossRef] [PubMed]
- Busque, L.; Sun, M.; Buscarlet, M.; Ayachi, S.; Feroz Zada, Y.; Provost, S.; Bourgoin, V.; Mollica, L.; Meisel, M.; Hinterleitner, R.; et al. High-sensitivity C-reactive protein is associated with clonal hematopoiesis of indeterminate potential. Blood Adv. 2020, 4, 2430–2438. [Google Scholar] [CrossRef]
- King, K.Y.; Huang, Y.; Nakada, D.; Goodell, M.A. Environmental influences on clonal hematopoiesis. Exp. Hematol. 2020, 83, 66–73. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhao, K.; Shen, Q.; Han, Y.; Gu, Y.; Li, X.; Zhao, D.; Liu, Y.; Wang, C.; Zhang, X.; et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature 2015, 525, 389–393. [Google Scholar] [CrossRef] [PubMed]
- Elias, H.K.; Bryder, D.; Park, C.Y. Molecular mechanisms underlying lineage bias in aging hematopoiesis. Semin. Hematol. 2017, 54, 4–11. [Google Scholar] [CrossRef]
- Baughn, L.B.; Meredith, M.M.; Oseth, L.; Smolarek, T.A.; Hirsch, B. SH2B3 aberrations enriched in iAMP21 B lymphoblastic leukemia. Cancer Genet. 2018, 226-227. [Google Scholar] [CrossRef] [PubMed]
- Takaki, S.; Morita, H.; Tezuka, Y.; Takatsu, K. Enhanced hematopoiesis by hematopoietic progenitor cells lacking intracellular adaptor protein, Lnk. J. Exp. Med. 2002, 195. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Tang, Y.; Wang, Y.; Tascau, L.; Balcerek, J.; Tong, W.; Levine, R.L.; Welch, C.; Tall, A.R.; Wang, N. LNK/SH2B3 Loss of Function Promotes Atherosclerosis and Thrombosis. Circ. Res. 2016, 119. [Google Scholar] [CrossRef]
- Snetsinger, B.; Ferrone, C.K.; Rauh, M.J. Targeted, Amplicon-Based, Next-Generation Sequencing to Detect Age-Related Clonal Hematopoiesis. Methods Mol. Biol. 2019, 2045, 167–180. [Google Scholar] [CrossRef]
- Buckstein, R.; Jang, K.; Friedlich, J.; Zhang, L.; Reis, M.; Chesney, A.; Wells, R.A. Estimating the prevalence of myelodysplastic syndromes in patients with unexplained cytopenias: A retrospective study of 322 bone marrows. Leuk. Res. 2009, 33. [Google Scholar] [CrossRef]
- Rauw, J.; Wells, R.A.; Chesney, A.; Reis, M.; Zhang, L.; Buckstein, R. Validation of a scoring system to establish the probability of myelodysplastic syndrome in patients with unexplained cytopenias or macrocytosis. Leuk. Res. 2011, 35. [Google Scholar] [CrossRef]
- Svensson, E.C.; Madar, A.; Campbell, C.D.; He, Y.; Sultan, M.; Healey, M.L.; D’Aco, K.; Fernandez, A.; Wache-Mainier, C.; Ridker, P.M.; et al. TET2-Driven Clonal Hematopoiesis Predicts Enhanced Response to Canakinumab in the CANTOS Trial: An Exploratory Analysis. Circulation 2018, 138, A15111. [Google Scholar]
- Obermeyer, Z.; Emanuel, E.J. Predicting the Future—Big Data, Machine Learning, and Clinical Medicine. N. Engl. J. Med. 2016, 375, 1216–1219. [Google Scholar] [CrossRef] [PubMed]
- Müller, R.U.; Skorska, A.; Lemcke, H.; Steinhoff, G.; David, R. GLP: A requirement in cell therapies—Perspectives for the cardiovascular field. Adv. Drug Deliv. Rev. 2020. [Google Scholar] [CrossRef]
- Uellendahl-Werth, F.; Wolfien, M.; Franke, A.; Wolkenhauer, O.; Ellinghaus, D. A benchmark of hemoglobin blocking during library preparation for mRNA-Sequencing of human blood samples. Sci. Rep. 2020, 10, 5630. [Google Scholar] [CrossRef] [PubMed]
- Yue, L.; Tian, D.; Chen, W.; Han, X.; Yin, M. Deep learning for heterogeneous medical data analysis. World Wide Web 2020, 23, 2715–2737. [Google Scholar] [CrossRef]
- Zhang, D.; Yin, C.; Zeng, J.; Yuan, X.; Zhang, P. Combining structured and unstructured data for predictive models: A deep learning approach. BMC Med. Inform. Decis. Mak. 2020, 20, 280. [Google Scholar] [CrossRef]
- Lambin, P.; Leijenaar, R.T.H.; Deist, T.M.; Peerlings, J.; de Jong, E.E.C.; van Timmeren, J.; Sanduleanu, S.; Larue, R.; Even, A.J.G.; Jochems, A.; et al. Radiomics: The bridge between medical imaging and personalized medicine. Nat. Rev. Clin. Oncol. 2017, 14, 749–762. [Google Scholar] [CrossRef]
- Del Mazo-Barbara, A.; Nieto, V.; Mirabel, C.; Reyes, B.; García-López, J.; Oliver-Vila, I.; Vives, J. Streamlining the qualification of computerized systems in GxP-compliant academic cell therapy facilities. Cytotherapy 2016, 18, 1237–1239. [Google Scholar] [CrossRef]
- Shah, R.U. We Don’t Need More Data, We Need the Right Data. Circulation 2020, 142, 197–198. [Google Scholar] [CrossRef]
- Begoli, E.; Bhattacharya, T.; Kusnezov, D. The need for uncertainty quantification in machine-assisted medical decision making. Nat. Mach. Intell. 2019, 1, 20–23. [Google Scholar] [CrossRef]
- Rudin, C. Stop explaining black box machine learning models for high stakes decisions and use interpretable models instead. Nat. Mach. Intell. 2019, 1, 206–215. [Google Scholar] [CrossRef]
- Benjamin, E.J.; Virani, S.S.; Callaway, C.W.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Chiuve, S.E.; Cushman, M.; Delling, F.N.; Deo, R.; et al. Heart Disease and Stroke Statistics—2018 Update: A Report From the American Heart Association. Circulation 2018, 137, e67–e492. [Google Scholar] [CrossRef] [PubMed]
- Kwon, J.M.; Lee, Y.; Lee, Y.; Lee, S.; Park, J. An Algorithm Based on Deep Learning for Predicting In-Hospital Cardiac Arrest. J. Am. Hear. Assoc. 2018, 7, e008678. [Google Scholar] [CrossRef]
- Weng, S.F.; Reps, J.; Kai, J.; Garibaldi, J.M.; Qureshi, N. Can machine-learning improve cardiovascular risk prediction using routine clinical data? PLoS ONE 2017, 12, e0174944. [Google Scholar] [CrossRef] [PubMed]
- Hastie, T.; Tibshirani, R.; Friedman, J. Random Forests; Springer: New York, NY, USA, 2009; pp. 587–624. [Google Scholar]
- Dilsizian, S.E.; Siegel, E.L. Artificial intelligence in medicine and cardiac imaging: Harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr. Cardiol. Rep. 2014, 16, 441. [Google Scholar] [CrossRef]
- Karamperis, K.; Wadge, S.; Koromina, M.; Patrinos, G.P. Applied Genomics and Public Health; Academic Press: London, UK, 2020; pp. 189–207. Available online: https://www.sciencedirect.com/book/9780128136959/applied-genomics-and-public-health (accessed on 13 May 2021).
- Braverman, G.; Shapiro, Z.E.; Bernstein, J.A. Ethical Issues in Contemporary Clinical Genetics. Mayo Clin. Proc. Innov. Qual. Outcomes 2018, 2, e1918962. [Google Scholar] [CrossRef][Green Version]
- Desai, R.J.; Wang, S.V.; Vaduganathan, M.; Evers, T.; Schneeweiss, S. Comparison of Machine Learning Methods With Traditional Models for Use of Administrative Claims With Electronic Medical Records to Predict Heart Failure Outcomes. JAMA Netw. Open 2020, 3, e1918962. [Google Scholar] [CrossRef]
- Firouzi, F.; Sussman, M.A. Blood speaks: Personalised medicine profiling for heart failure patients. EBioMedicine 2020, 58, 102900. [Google Scholar] [CrossRef]
- Zhao, M.T.; Shao, N.Y.; Garg, V. Subtype-specific cardiomyocytes for precision medicine: Where are we now? Stem Cells 2020, 38, 822–833. [Google Scholar] [CrossRef]
- Sánchez-Rico, M.; Alvarado, J.M. A Machine Learning Approach for Studying the Comorbidities of Complex Diagnoses. Behav. Sci. 2019, 9, 122. [Google Scholar] [CrossRef]
- Komura, D.; Ishikawa, S. Machine Learning Methods for Histopathological Image Analysis. Comput. Struct. Biotechnol. J. 2018, 16, 34–42. [Google Scholar] [CrossRef]
- Siontis, K.C.; Noseworthy, P.A.; Attia, Z.I.; Friedman, P.A. Artificial intelligence-enhanced electrocardiography in cardiovascular disease management. Nat. Rev. Cardiol. 2021, 18, 465–478. [Google Scholar] [CrossRef]
- Dai, L.; Zhou, Q.; Zhou, H.; Zhang, H.; Cheng, P.; Ding, M.; Xu, X.; Zhang, X. Deep learning-based classification of lower extremity arterial stenosis in computed tomography angiography. Eur. J. Radiol. 2021, 136, 109528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.L.; Conlin, C.C.; Li, X.; Layec, G.; Chang, K.; Kalpathy-Cramer, J.; Lee, V.S. Exercise-induced calf muscle hyperemia: Rapid mapping of magnetic resonance imaging using deep learning approach. Physiol. Rep. 2020, 8, e14563. [Google Scholar] [CrossRef]
- Veit-Haibach, P.; Huellner, M.W.; Banyai, M.; Mafeld, S.; Heverhagen, J.; Strobel, K.; Sah, B.R. CT perfusion in peripheral arterial disease-hemodynamic differences before and after revascularisation. Eur. Radiol. 2021, 31, 5507–5513. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.M.; Demsas, F.; Leeper, N.J.; Ross, E.G. Leveraging Machine Learning and Artificial Intelligence to Improve Peripheral Artery Disease Detection, Treatment, and Outcomes. Circ. Res. 2021, 128, 1833–1850. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Hahn, J.-O.; Youn, B.D. Detection and Severity Assessment of Peripheral Occlusive Artery Disease via Deep Learning Analysis of Arterial Pulse Waveforms: Proof-of-Concept and Potential Challenges. Front. Bioeng. Biotechnol. 2020, 8, 720. [Google Scholar] [CrossRef]
- Fu, Y.; Jung, A.W.; Torne, R.V.; Gonzalez, S.; Vöhringer, H.; Shmatko, A.; Yates, L.R.; Jimenez-Linan, M.; Moore, L.; Gerstung, M. Pan-cancer computational histopathology reveals mutations, tumor composition and prognosis. Nat. Rev. Cancer 2020, 1, 800–810. [Google Scholar] [CrossRef]
- Gao, W.; Chen, D.; Liu, G.; Ran, X. Autologous stem cell therapy for peripheral arterial disease: A systematic review and meta-analysis of randomized controlled trials. Stem Cell Res. Ther. 2019, 10, 140. [Google Scholar] [CrossRef]
- Rigato, M.; Monami, M.; Fadini, G.P. Autologous Cell Therapy for Peripheral Arterial Disease: Systematic Review and Meta-Analysis of Randomized, Nonrandomized, and Noncontrolled Studies. Circ. Res. 2017, 120, 1326–1340. [Google Scholar] [CrossRef]
- Johnson, K.B.; Wei, W.; Weeraratne, D.; Frisse, M.E.; Misulis, K.; Rhee, K.; Zhao, J.; Snowdon, J.L. Precision Medicine, AI, and the Future of Personalized Health Care. Care. Clin. Transl. Sci. 2021, 14, 86–93. [Google Scholar] [CrossRef] [PubMed]
- Kelly, C.J.; Karthikesalingam, A.; Suleyman, M.; Corrado, G.; King, D. Key challenges for delivering clinical impact with artificial intelligence. BMC Med. 2019, 17, 195. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Preininger, A. Artificial intelligence in Health: State of the Art, Challenges, and Future Directions. Yearb. Med. Inform. 2019, 28, 16–26. [Google Scholar] [CrossRef]
- Salybekov, A.A.; Kunikeyev, A.D.; Kobayashi, S.; Asahara, T. Latest Advances in Endothelial Progenitor Cell-Derived Extracellular Vesicles Translation to the Clinic. Front. Cardiovasc. Med. 2021, 8, 734562. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).