Immunological Characterization of HIV and SARS-CoV-2 Coinfected Young Individuals
Abstract
:1. Introduction
2. Materials and Methods
2.1. Enrollment and Clinical Evaluation
2.2. SARS-CoV-2 Genome Quantification
2.3. SARS-CoV-2 Specific Antibodies
2.4. Quantigene Plex Gene Expression Analysis
2.5. Multiplex Analysis
2.6. In Vitro Cell Culture, Co-Culture, and Infection Assay
2.7. Gene Expression Analyses by RT-QPCR
2.8. Statistical Analysis
3. Results
3.1. Clinical Aspects
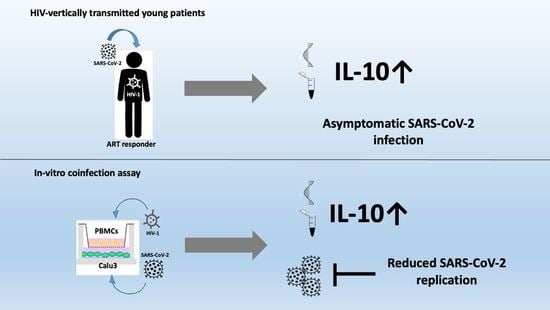
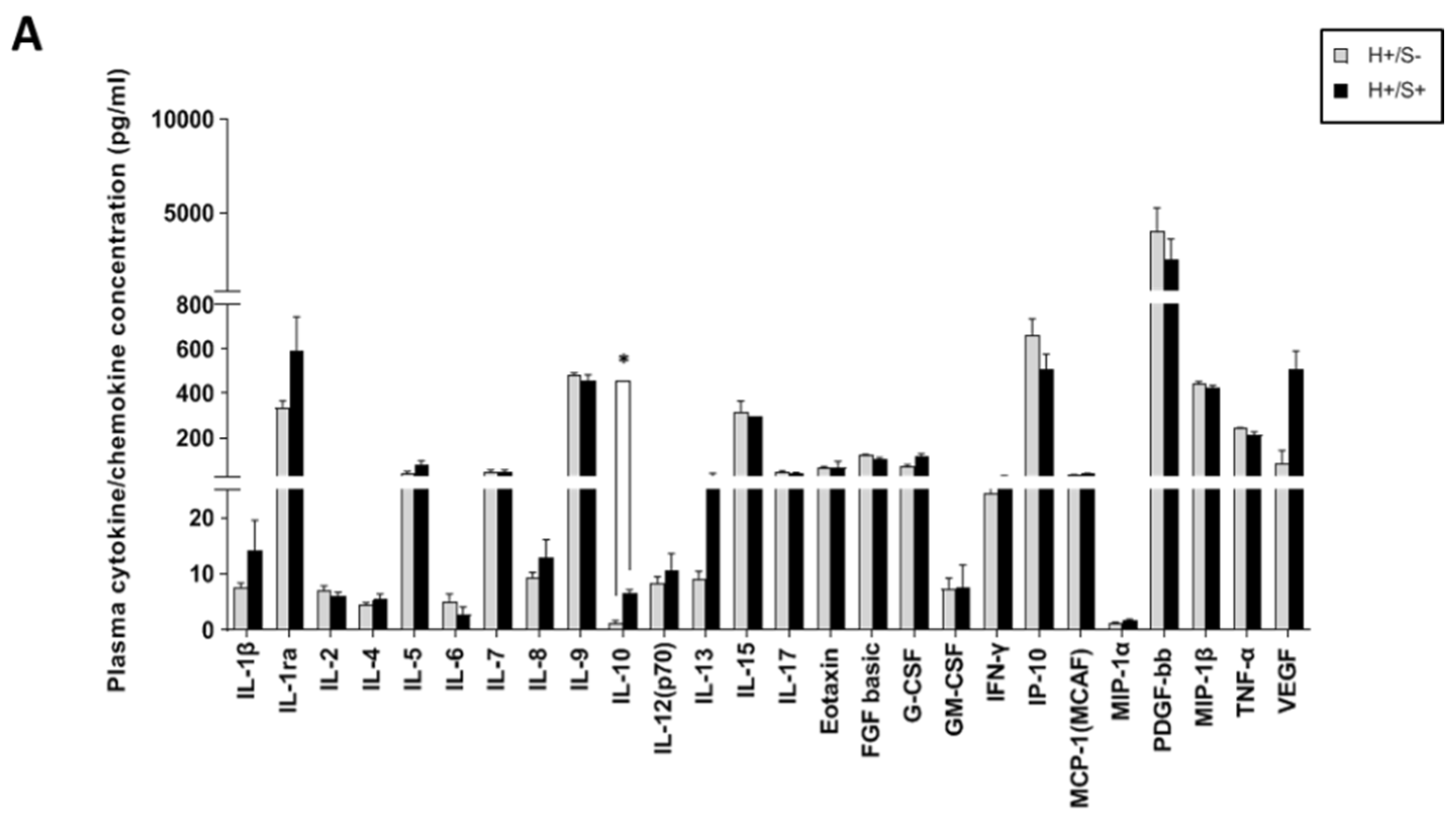
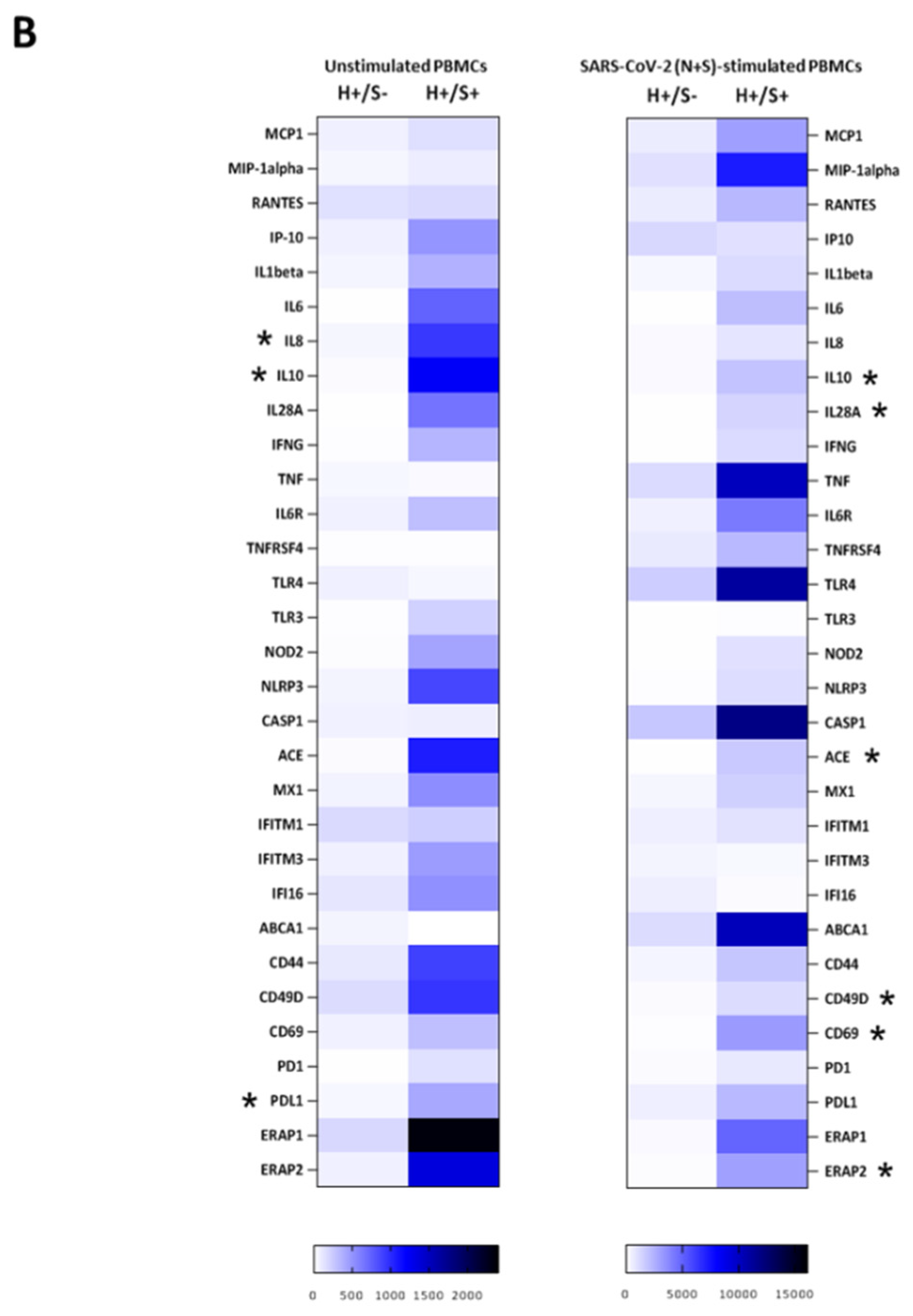
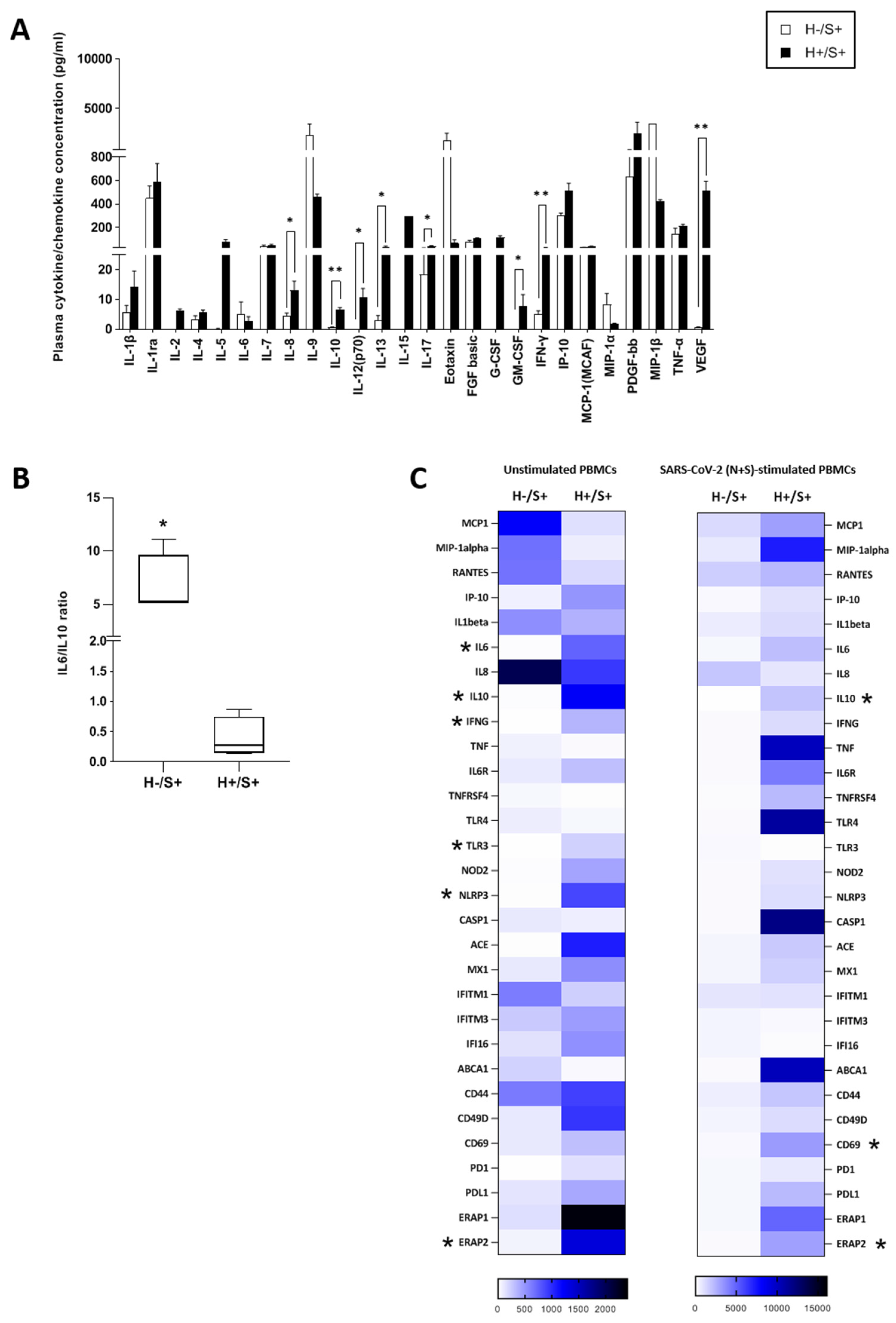
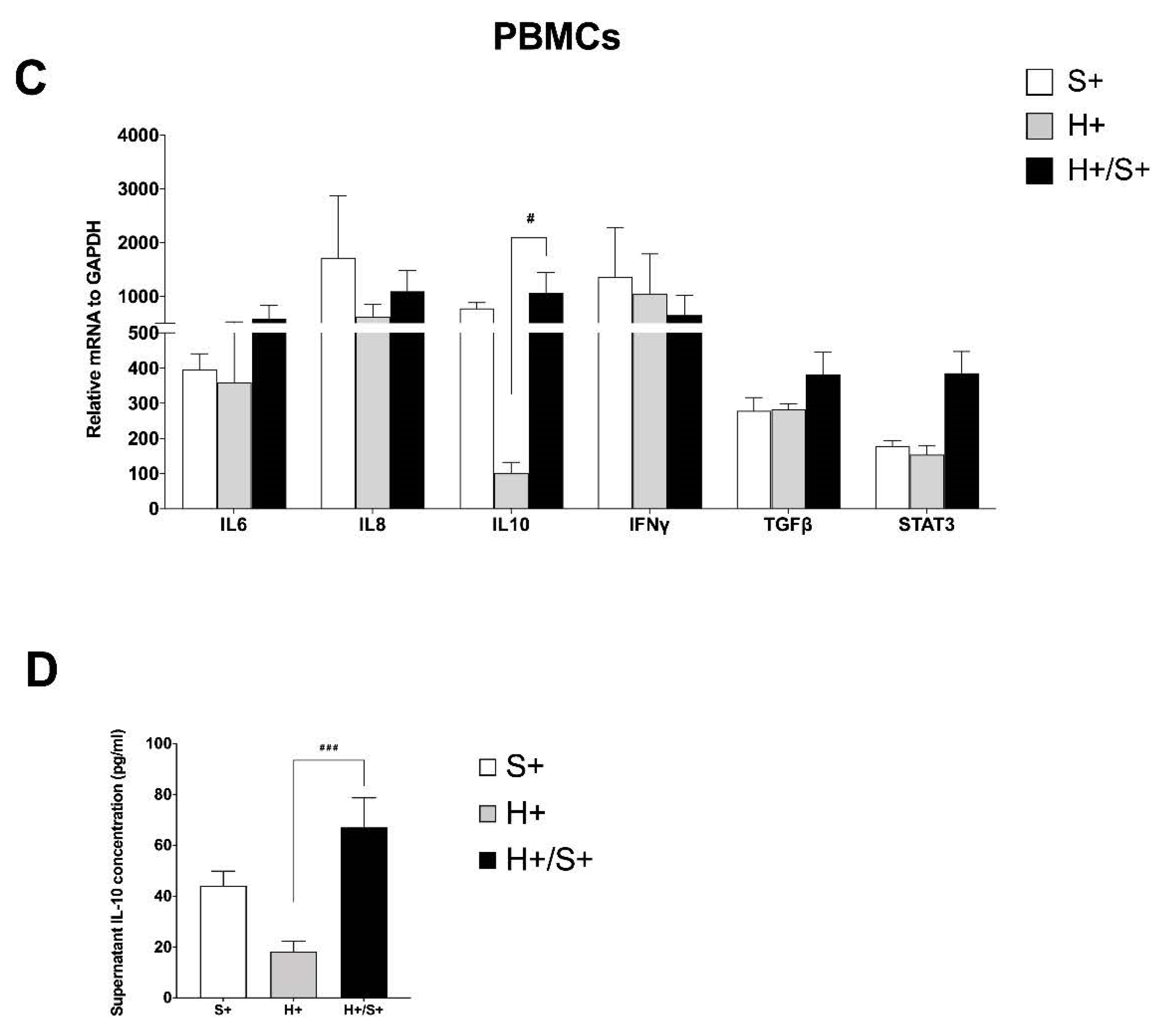
3.2. H+/+ Subjects Reported Higher Production and mRNA Expression of IL-10 When Compared to H+/S− Individuals
3.3. H+/S+ Patients Showed a Peculiar Inflammatory Profile When Compared to H−/S+ Individuals
3.4. In Vitro HIV/SARS-CoV-2 Coinfection Assays Confirmed the Upregulation of IL-10
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zimmermann, P.; Curtis, N. Why Is COVID-19 Less Severe in Children? A Review of the Proposed Mechanisms Underlying the Age-Related Difference in Severity of SARS-CoV-2 Infections. Arch. Dis. Child. 2021, 106, 429–439. [Google Scholar] [CrossRef]
- Zhu, F.; Cao, Y.; Xu, S.; Zhou, M. Co-Infection of SARS-CoV-2 and HIV in a Patient in Wuhan City, China. J. Med. Virol. 2020, 92, 529–530. [Google Scholar] [CrossRef] [Green Version]
- Vizcarra, P.; Pérez-Elías, M.J.; Quereda, C.; Moreno, A.; Vivancos, M.J.; Dronda, F.; Casado, J.L.; Moreno, S.; Pérez-Elías, M.J.; Fortún, J.; et al. Description of COVID-19 in HIV-Infected Individuals: A Single-Centre, Prospective Cohort. Lancet HIV 2020, 7, e554–e564. [Google Scholar] [CrossRef]
- Gervasoni, C.; Meraviglia, P.; Riva, A.; Giacomelli, A.; Oreni, L.; Minisci, D.; Atzori, C.; Ridolfo, A.; Cattaneo, D. Clinical Features and Outcomes of Patients with Human Immunodeficiency Virus with COVID-19. Clin. Infect. Dis. 2020, 71, 2276–2278. [Google Scholar] [CrossRef]
- Calza, L.; Bon, I.; Tadolini, M.; Borderi, M.; Colangeli, V.; Badia, L.; Verucchi, G.; Rossini, G.; Vocale, C.; Gaibani, P.; et al. COVID-19 in Patients with HIV-1 Infection: A Single-Centre Experience in Northern Italy. Infection 2021, 49, 333–337. [Google Scholar] [CrossRef]
- Costenaro, P.; Minotti, C.; Barbieri, E.; Giaquinto, C.; Donà, D. SARS-CoV-2 Infection in People Living with HIV: A Systematic Review. Rev. Med. Virol. 2021, 31, 1–12. [Google Scholar] [CrossRef]
- Maggiolo, F.; Zoboli, F.; Arosio, M.; Valenti, D.; Guarneri, D.; Sangiorgio, L.; Ripamonti, D.; Callegaro, A. SARS-CoV-2 Infection in Persons Living with HIV: A Single Center Prospective Cohort. J. Med. Virol. 2021, 93, 1145–1149. [Google Scholar] [CrossRef]
- Wyllie, A.L.; Fournier, J.; Casanovas-Massana, A.; Campbell, M.; Tokuyama, M.; Vijayakumar, P.; Warren, J.L.; Geng, B.; Muenker, M.C.; Moore, A.J.; et al. Saliva or Nasopharyngeal Swab Specimens for Detection of SARS-CoV-2. N. Engl. J. Med. 2020, 383, 1283–1286. [Google Scholar] [CrossRef]
- Saulle, I.; Vanetti, C.; Goglia, S.; Vicentini, C.; Tombetti, E.; Garziano, M.; Clerici, M.; Biasin, M. A New ERAP2/Iso3 Isoform Expression Is Triggered by Different Microbial Stimuli in Human Cells. Could It Play a Role in the Modulation of SARS-CoV-2 Infection? Cells 2020, 9, 1951. [Google Scholar] [CrossRef]
- Bandera, A.; Masetti, M.; Fabbiani, M.; Biasin, M.; Muscatello, A.; Squillace, N.; Clerici, M.; Gori, A.; Trabattoni, D. The NLRP3 Inflammasome Is Upregulated in HIV-Infected Antiretroviral Therapy-Treated Individuals with Defective Immune Recovery. Front. Immunol. 2018, 9, 214. [Google Scholar] [CrossRef] [Green Version]
- Mondi, A.; Cimini, E.; Colavita, F.; Cicalini, S.; Pinnetti, C.; Matusali, G.; Casetti, R.; Maeurer, M.; Vergori, A.; Mazzotta, V.; et al. COVID-19 in People Living with HIV: Clinical Implications of Dynamics of the Immune Response to SARS-CoV-2. J. Med. Virol. 2021, 93, 1796–1804. [Google Scholar] [CrossRef]
- Peng, X.; Ouyang, J.; Isnard, S.; Lin, J.; Fombuena, B.; Zhu, B.; Routy, J.-P. Sharing CD4+ T Cell Loss: When COVID-19 and HIV Collide on Immune System. Front. Immunol. 2020, 11, 596631. [Google Scholar] [CrossRef]
- Istituto Superiore di Sanità (ISS). Epidemia COVID-19. Aggiornamento Nazionale 29 Dicembre 2020–Ore 12:00. Available online: https://www.epicentro.iss.it/coronavirus/bollettino/Bollettino-sorveglianza-integrata-COVID-19_29-dicembre-2020.pdf (accessed on 15 October 2021).
- Karmen-Tuohy, S.; Carlucci, P.M.; Zervou, F.N.; Zacharioudakis, I.M.; Rebick, G.; Klein, E.; Reich, J.; Jones, S.; Rahimian, J. Outcomes Among HIV-Positive Patients Hospitalized with COVID-19. JAIDS J. Acquir. Immune Defic. Syndr. 2020, 85, 6–10. [Google Scholar] [CrossRef]
- Van der Hoek, L.; Pyrc, K.; Berkhout, B. Human Coronavirus NL63, a New Respiratory Virus. FEMS Microbiol. Rev. 2006, 30, 760–773. [Google Scholar] [CrossRef] [Green Version]
- Makoti, P.; Fielding, B.C. HIV and Human Coronavirus Coinfections: A Historical Perspective. Viruses 2020, 12, 937. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J. The Primary Mechanism of the IL-10-Regulated Antiinflammatory Response Is to Selectively Inhibit Transcription. Proc. Natl. Acad. Sci. USA 2005, 102, 8686–8691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchins, A.P.; Diez, D.; Miranda-Saavedra, D. The IL-10/STAT3-Mediated Anti-Inflammatory Response: Recent Developments and Future Challenges. Brief. Funct. Genom. 2013, 12, 489–498. [Google Scholar] [CrossRef] [Green Version]
- Sanjabi, S.; Oh, S.A.; Li, M.O. Regulation of the Immune Response by TGF-β: From Conception to Autoimmunity and Infection. Cold Spring Harb. Perspect. Biol. 2017, 9, a022236. [Google Scholar] [CrossRef] [Green Version]
- Lindner, H.A.; Velásquez, S.Y.; Thiel, M.; Kirschning, T. Lung Protection vs. Infection Resolution: Interleukin 10 Suspected of Double-Dealing in COVID-19. Front. Immunol. 2021, 12, 602130. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Zhang, H.; Dauphars, D.J.; He, Y.-W. A Potential Role of Interleukin 10 in COVID-19 Pathogenesis. Trends Immunol. 2021, 42, 3–5. [Google Scholar] [CrossRef]
- McElvaney, O.J.; Hobbs, B.D.; Qiao, D.; McElvaney, O.F.; Moll, M.; McEvoy, N.L.; Clarke, J.; O’Connor, E.; Walsh, S.; Cho, M.H.; et al. A Linear Prognostic Score Based on the Ratio of Interleukin-6 to Interleukin-10 Predicts Outcomes in COVID-19. EBioMedicine 2020, 61, 103026. [Google Scholar] [CrossRef]
- Sharov, K.S. HIV/SARS-CoV-2 Co-Infection: T Cell Profile, Cytokine Dynamics and Role of Exhausted Lymphocytes. Int. J. Infect. Dis. 2021, 102, 163–169. [Google Scholar] [CrossRef]
- Chen, H.; Liu, W.; Wang, Y.; Liu, D.; Zhao, L.; Yu, J. SARS-CoV-2 Activates Lung Epithelial Cell Proinflammatory Signaling and Leads to Immune Dysregulation in COVID-19 Patients. EBioMedicine 2021, 70, 103500. [Google Scholar] [CrossRef]
- Xu, G.; Qi, F.; Li, H.; Yang, Q.; Wang, H.; Wang, X.; Liu, X.; Zhao, J.; Liao, X.; Liu, Y.; et al. The Differential Immune Responses to COVID-19 in Peripheral and Lung Revealed by Single-Cell RNA Sequencing. Cell Discov. 2020, 6, 73. [Google Scholar] [CrossRef]
- Couper, K.N.; Blount, D.G.; Riley, E.M. IL-10: The Master Regulator of Immunity to Infection. J. Immunol. 2008, 180, 5771–5777. [Google Scholar] [CrossRef]
- Brown, L.B.; Spinelli, M.A.; Gandhi, M. The Interplay between HIV and COVID-19: Summary of the Data and Responses to Date. Curr. Opin. HIV AIDS 2021, 16, 63–73. [Google Scholar] [CrossRef]


| H+ (85) | H+/S+ (4) | H−/S+ (7) | |
|---|---|---|---|
| Median age (range) in years | 22.3 (1–35) | 22.5 (19–30) | 22.8 (10.8–40) |
| Male | 35 | 1 | 4 |
| Comorbidities | 10 (11%) | 0 | 0 |
| Coinfections | 5 (4.25%) | 0 | 0 |
| CD4 > 350 mm3 | 81 (95.3%) | 4 (100%) | - |
| HIV RNA < 20 CP | 79 (93%) | 4 (100%) | - |
| Integrase inhibitors | 40 (47%) | 0 | - |
| Protease inhibitors | 31 (36.5%) | 4 (100%) | - |
| Nucleoside reverse transcriptase Inhibitors | 1 (1.17%) | 0 | - |
| Non-nucleoside reverse transcriptase inhibitors | 13 (11%) | 0 | - |
| Positive RT-PCR on sputum | 1 (1.17%) | 1 | 7 |
| IgG Anti-SARS-CoV-2 | 3 (3.53%) | 3 | - |
| Asymptomatic | 85 (100%) | 4 (100%) | 0 |
| Paucisyntomatic/moderate | 0 | 0 | 7 (100%) |
| Severe/critical | 0 | 0 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vanetti, C.; Trabattoni, D.; Stracuzzi, M.; Amendola, A.; Fappani, C.; Rubinacci, V.; Fenizia, C.; Gianolio, L.; Biasin, M.; Dighera, A.; et al. Immunological Characterization of HIV and SARS-CoV-2 Coinfected Young Individuals. Cells 2021, 10, 3187. https://doi.org/10.3390/cells10113187
Vanetti C, Trabattoni D, Stracuzzi M, Amendola A, Fappani C, Rubinacci V, Fenizia C, Gianolio L, Biasin M, Dighera A, et al. Immunological Characterization of HIV and SARS-CoV-2 Coinfected Young Individuals. Cells. 2021; 10(11):3187. https://doi.org/10.3390/cells10113187
Chicago/Turabian StyleVanetti, Claudia, Daria Trabattoni, Marta Stracuzzi, Antonella Amendola, Clara Fappani, Valeria Rubinacci, Claudio Fenizia, Laura Gianolio, Mara Biasin, Anna Dighera, and et al. 2021. "Immunological Characterization of HIV and SARS-CoV-2 Coinfected Young Individuals" Cells 10, no. 11: 3187. https://doi.org/10.3390/cells10113187
APA StyleVanetti, C., Trabattoni, D., Stracuzzi, M., Amendola, A., Fappani, C., Rubinacci, V., Fenizia, C., Gianolio, L., Biasin, M., Dighera, A., Saulle, I., Tanzi, E., Zuccotti, G., Clerici, M., & Giacomet, V. (2021). Immunological Characterization of HIV and SARS-CoV-2 Coinfected Young Individuals. Cells, 10(11), 3187. https://doi.org/10.3390/cells10113187