NR3C1 Glucocorticoid Receptor Gene Polymorphisms Are Associated with Membranous and IgA Nephropathies
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
2.2. Patients and Sample Collection
2.3. Selection of NR3C1 Polymorphisms
2.4. Genotyping of the NR3C1 Gene rs6198, rs41423247 and rs17209237 Polymorphisms
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Floege, J.; Amann, K. Primary glomerulonephritides. Lancet 2016, 387, 2036–2048. [Google Scholar] [CrossRef]
- Kiryluk, K.; Li, Y.; Sanna-Cherchi, S.; Rohanizadegan, M.; Suzuki, H.; Eitner, F.; Snyder, H.J.; Choi, M.; Hou, P.; Scolari, F.; et al. Geographic Differences in Genetic Susceptibility to IgA Nephropathy: GWAS Replication Study and Geospatial Risk Analysis. PLoS Genet. 2012, 8, e1002765. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Chuang, P.Y.; Liu, Z.-H.; He, J.C. The primary glomerulonephritides: A systems biology approach. Nat. Rev. Nephrol. 2013, 9, 500–512. [Google Scholar] [CrossRef] [PubMed]
- Kiryluk, K.; Li, Y.; Scolari, F.; Sanna-Cherchi, S.; Choi, M.; Verbitsky, M.; Fasel, D.; Lata, S.; Prakash, S.; Shapiro, S.; et al. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat. Genet. 2014, 46, 1187–1196. [Google Scholar] [CrossRef]
- Mucha, K.; Bakun, M.; Jaźwiec, R.; Dadlez, M.; Florczak, M.; Bajor, M.; Gala, K.; Pączek, L. Complement components, proteolysis?related, and cell communication?related proteins detected in urine proteomics are associated with IgA nephropathy. Pol. Arch. Intern. Med. 2014, 124, 380–386. [Google Scholar] [CrossRef]
- Kaleta, B.; Krata, N.; Zagożdżon, R.; Mucha, K. Osteopontin Gene Polymorphism and Urinary OPN Excretion in Patients with Immunoglobulin A Nephropathy. Cells 2019, 8, 524. [Google Scholar] [CrossRef]
- Xie, J.; Liu, L.; Mladkova, N.; Li, Y.; Ren, H.; Wang, W.; Cui, Z.; Lin, L.; Hu, X.; Yu, X.; et al. The genetic architecture of membranous nephropathy and its potential to improve non-invasive diagnosis. Nat. Commun. 2020, 11, 1600. [Google Scholar] [CrossRef]
- Quax, R.A.; Manenschijn, L.; Koper, J.W.; Hazes, J.M.; Lamberts, S.W.J.; Van Rossum, E.F.C.; Feelders, R.A. Glucocorticoid sensitivity in health and disease. Nat. Rev. Endocrinol. 2013, 9, 670–686. [Google Scholar] [CrossRef]
- Evans, W.E.; McLeod, H.L. Pharmacogenomics—Drug Disposition, Drug Targets, and Side Effects. N. Engl. J. Med. 2003, 348, 538–549. [Google Scholar] [CrossRef]
- Huizenga, N.A.T.M.; Koper, J.W.; De Lange, P.; Pols, H.A.P.; Stolk, R.P.; Burger, H.; Grobbee, D.E.; Brinkmann, A.O.; De Jong, F.H.; Lamberts, S.W.J. A Polymorphism in the Glucocorticoid Receptor Gene May Be Associated with an Increased Sensitivity to Glucocorticoidsin Vivo1. J. Clin. Endocrinol. Metab. 1998, 83, 144–151. [Google Scholar] [CrossRef][Green Version]
- van Rossum, E.F.C.; Koper, J.W.; Beld, A.W.V.D.; Uitterlinden, A.G.; Arp, P.; Ester, W.A.; Janssen, J.A.; Brinkmann, A.O.; De Jong, F.H.; Grobbee, D.E.; et al. Identification of the Bcl I polymorphism in the glucocorticoid receptor gene: Association with sensitivity to glucocorticoids in vivo and body mass index. Clin. Endocrinol. 2003, 59, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Stevens, A.; Ray, D.W.; Zeggini, E.; John, S.; Richards, H.L.; Griffiths, C.E.M.; Donn, R. Glucocorticoid Sensitivity Is Determined by a Specific Glucocorticoid Receptor Haplotype. J. Clin. Endocrinol. Metab. 2004, 89, 892–897. [Google Scholar] [CrossRef]
- Manenschijn, L.; Van Den Akker, E.L.; Lamberts, S.W.J.; Van Rossum, E.F.C. Clinical Features Associated with Glucocorticoid Receptor Polymorphisms. Ann. N.Y. Acad. Sci. 2009, 1179, 179–198. [Google Scholar] [CrossRef]
- van Rossum, E.F.; Koper, J.W.; Huizenga, N.A.; Uitterlinden, A.G.; Janssen, J.A.; Brinkmann, A.O.; Grobbee, D.E.; de Jong, F.H.; van Duyn, C.M.; Pols, H.A.; et al. A Polymorphism in the Glucocorticoid Receptor Gene, Which Decreases Sensitivity to Glucocorticoids In Vivo, Is Associated With Low Insulin and Cholesterol Levels. Diabetes 2002, 51, 3128–3134. [Google Scholar] [CrossRef] [PubMed]
- Akker, E.V.D.; Russcher, H.; van Rossum, E.; Brinkmann, A.; De Jong, F.; Hokken-Koelega, A.; Pols, H.A.P.; Koper, J.; Lamberts, S. Glucocorticoid Receptor Polymorphism Affects Transrepression But Not Transactivation. J. Clin. Endocrinol. Metab. 2006, 91, 2800–2803. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.-X.; Dong, J.; Zhang, J.; Liu, F.; Wang, W.; Zhang, L.; He, Y. Polymorphisms in NR3C1 gene associated with risk of metabolic syndrome in a Chinese population. Endocrine 2014, 47, 740–748. [Google Scholar] [CrossRef]
- van Oosten, M.J.; Dolhain, R.J.; Koper, J.W.; van Rossum, E.F.; Emonts, M.; Han, K.H.; Wouters, J.M.; Hazes, J.M.; Lamberts, S.W.; Feelders, R.A. Polymorphisms in the glucocorticoid receptor gene that modulate glucocorticoid sensitivity are associated with rheumatoid arthritis. Arthritis Res. Ther. 2010, 12, 1–8. [Google Scholar] [CrossRef]
- Quax, R.A.; De Man, Y.A.; Koper, J.W.; van Rossum, E.F.; Willemsen, S.P.; Lamberts, S.W.; Hazes, J.M.; Dolhain, R.J.; Feelders, R.A. Glucocorticoid receptor gene polymorphisms and disease activity during pregnancy and the postpartum period in rheumatoid arthritis. Arthritis Res. Ther. 2012, 14, R183. [Google Scholar] [CrossRef]
- Fang, S.-Y.; Li, C.-L.; Liu, X.-S.; Chen, F.; Hua, H. Correlation between polymorphisms of the NR3C1 gene and glucocorticoid effectiveness in patients with pemphigus vulgaris. Sci. Rep. 2017, 7, 11890. [Google Scholar] [CrossRef]
- Huang, M.; Inukai, T.; Kagami, K.; Abe, M.; Shinohara, T.; Watanabe, A.; Somazu, S.; Oshiro, H.; Goi, K.; Goto, H.; et al. Splicing variant profiles and single nucleotide polymorphisms of the glucocorticoid receptor gene in relation to glucocorticoid sensitivity of B-cell precursor acute lymphoblastic leukaemia. Hematol. Oncol. 2018, 36, 245–251. [Google Scholar] [CrossRef]
- Keskin, O.; Uluca, Ü.; Birben, E.; Coşkun, Y.; Ozkars, M.Y.; Keskin, M.; Kucukosmanoglu, E.; Kalayci, O. Genetic associations of the response to inhaled corticosteroids in children during an asthma exacerbation. Pediatr. Allergy Immunol. 2016, 27, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Keskin, O.; Farzan, N.; Birben, E.; Akel, H.; Karaaslan, C.; Der Zee, A.H.M.-V.; Wechsler, M.E.; Vijverberg, S.; Kalayci, O. Genetic associations of the response to inhaled corticosteroids in asthma: A systematic review. Clin. Transl. Allergy 2019, 9, 2. [Google Scholar] [CrossRef]
- Ye, J.; Yu, Z.; Ding, J.; Chen, Y.; Huang, J.; Yao, Y.; Xiao, H.; Yang, J.; Shen, Y.; Meng, Q. Genetic variations of the NR3C1 gene in children with sporadic nephrotic syndrome. Biochem. Biophys. Res. Commun. 2006, 348, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Zalewski, G.; Wasilewska, A.; Zoch-Zwierz, W.; Chyczewski, L. Response to prednisone in relation to NR3C1 intron B polymorphisms in childhood nephrotic syndrome. Pediatr. Nephrol. 2008, 23, 1073–1078. [Google Scholar] [CrossRef]
- Teeninga, N.; Holthe, J.E.K.-V.; Akker, E.L.V.D.; Kersten, M.C.; Boersma, E.; Krabbe, H.G.; Knoers, N.V.; van der Heijden, A.J.; Koper, J.W.; Nauta, J. Genetic and in vivo determinants of glucocorticoid sensitivity in relation to clinical outcome of childhood nephrotic syndrome. Kidney Int. 2014, 85, 1444–1453. [Google Scholar] [CrossRef] [PubMed]
- Suvanto, M.; Jahnukainen, T.; Kestilä, M.; Jalanko, H. Single Nucleotide Polymorphisms in Pediatric Idiopathic Nephrotic Syndrome. Int. J. Nephrol. 2016, 2016, 1417456. [Google Scholar] [CrossRef][Green Version]
- Liu, J.; Wan, Z.; Song, Q.; Li, Z.; He, Y.; Tang, Y.; Xie, W.; Xie, Y.; Zhang, J. NR3C1 gene polymorphisms are associated with steroid resistance in patients with primary nephrotic syndrome. Pharmacogenomics 2018, 19, 45–60. [Google Scholar] [CrossRef] [PubMed]
- Perkowska-Ptasinska, A.; Bartczak, A.; Wagrowska-Danilewicz, M.; Halon, A.; Okon, K.; Wozniak, A.; Danilewicz, M.; Karkoszka, H.; Marszałek, A.; Kowalewska, J.; et al. Clinicopathologic correlations of renal pathology in the adult population of Poland. Nephrol. Dial. Transplant. 2017, 32, ii209–ii218. [Google Scholar] [CrossRef]
- Stanescu, H.C.; Arcos-Burgos, M.; Medlar, A.; Bockenhauer, D.; Kottgen, A.; Dragomirescu, L.; Voinescu, C.; Patel, N.; Pearce, K.; Hubank, M.; et al. Risk HLA-DQA1 and PLA2R1 Alleles in Idiopathic Membranous Nephropathy. N. Engl. J. Med. 2011, 364, 616–626. [Google Scholar] [CrossRef]
- Cox, S.N.; Pesce, F.; Moustafa, J.S.E.-S.; Sallustio, F.; Serino, G.; Kkoufou, C.; Giampetruzzi, A.; Ancona, N.; Falchi, M.; Schena, F.P.; et al. Multiple rare genetic variants co-segregating with familial IgA nephropathy all act within a single immune-related network. J. Intern. Med. 2016, 281, 189–205. [Google Scholar] [CrossRef]
- Ukkola, O.; Perusse, L.; Chagnon, Y.; Després, J.-P.; Bouchard, C. Interactions among the glucocorticoid receptor, lipoprotein lipase and adrenergic receptor genes and abdominal fat in the Québec Family Study. Int. J. Obes. 2001, 25, 1332–1339. [Google Scholar] [CrossRef][Green Version]
- Geelen, C.C.; van Greevenbroek, M.M.; van Rossum, E.F.; Schaper, N.C.; Nijpels, G.; ’t Hart, L.M.; Schalkwijk, C.G.; Ferreira, I.; van der Kallen, C.J.; Sauerwein, H.; et al. BclI Glucocorticoid Receptor Polymorphism Is Associated With Greater Body Fatness: The Hoorn and CODAM Studies. J. Clin. Endocrinol. Metab. 2013, 98, E595–E599. [Google Scholar] [CrossRef] [PubMed][Green Version]
- van Rossum, L.; Lamberts, S. Polymorphisms in the Glucocorticoid Receptor Gene and Their Associations with Metabolic Parameters and Body Composition. Recent Prog. Horm. Res. 2004, 59, 333–357. [Google Scholar] [CrossRef] [PubMed]
- Rosmond, R.; Holm, G. A 5-Year Follow-Up Study of 3 Polymorphisms in the Human Glucocorticoid Receptor Gene in Relation to Obesity, Hypertension, and Diabetes. J. CardioMetabolic Syndr. 2008, 3, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Syed, A.A.; Halpin, C.G.; Irving, J.A.E.; Unwin, N.; White, M.; Bhopal, R.S.; Redfern, C.P.F.; Weaver, J.U. A common intron 2 polymorphism of the glucocorticoid receptor gene is associated with insulin resistance in men. Clin. Endocrinol. 2008, 68, 879–884. [Google Scholar] [CrossRef]
- Moradi, M.; Gharesouran, J.; Ghafouri-Fard, S.; Noroozi, R.; Talebian, S.; Taheri, M.; Rezazadeh, M. Role of NR3C1 and GAS5 genes polymorphisms in multiple sclerosis. Int. J. Neurosci. 2019, 130, 407–412. [Google Scholar] [CrossRef] [PubMed]
- Quax, R.A.M.; Koper, J.W.; Huisman, A.M.; Weel, A.; Hazes, J.M.W.; Lamberts, S.W.J.; Feelders, R.A. Polymorphisms in the glucocorticoid receptor gene and in the glucocorticoid-induced transcript 1 gene are associated with disease activity and response to glucocorticoid bridging therapy in rheumatoid arthritis. Rheumatol. Int. 2015, 35, 1325–1333. [Google Scholar] [CrossRef]
- Boyle, B.; Koranyi, K.; Patocs, A.; Liko, I.; Szappanos, A.; Bertalan, R.; Racz, K.; Balazs, C. Polymorphisms of the glucocorticoid receptor gene in Graves ophthalmopathy. Br. J. Ophthalmol. 2007, 92, 131–134. [Google Scholar] [CrossRef]
- Herrera, C.; Marcos, M.; Carbonell, C.; Miron-Canelo, J.A.; Espinosa, G.; Cervera, R.; Chamorro, A.-J. Association between allelic variants of the human glucocorticoid receptor gene and autoimmune diseases: A systematic review and meta-analysis. Autoimmun. Rev. 2018, 17, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Chatzikyriakidou, A.; Georgiou, I.; Voulgari, P.V.; Georgiadis, A.N.; Argyriou, E.S.; Drosos, A.A. Glucocorticoid receptor variants may predispose to rheumatoid arthritis susceptibility. Scand. J. Rheumatol. 2009, 38, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Donn, R.; Payne, D.; Ray, D. Glucocorticoid receptor gene polymorphisms and susceptibility to rheumatoid arthritis. Clin. Endocrinol. 2007, 67, 342–345. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Meng, Y.; Li, H.-F.; Hong, Y.; Sun, L.; Zhu, X.; Yue, Y.-X.; Gao, X.; Wang, S.; Li, Y.; et al. GRgene polymorphism is associated with inter-subject variability in response to glucocorticoids in patients with myasthenia gravis. Eur. J. Neurol. 2016, 23, 1372–1379. [Google Scholar] [CrossRef] [PubMed]

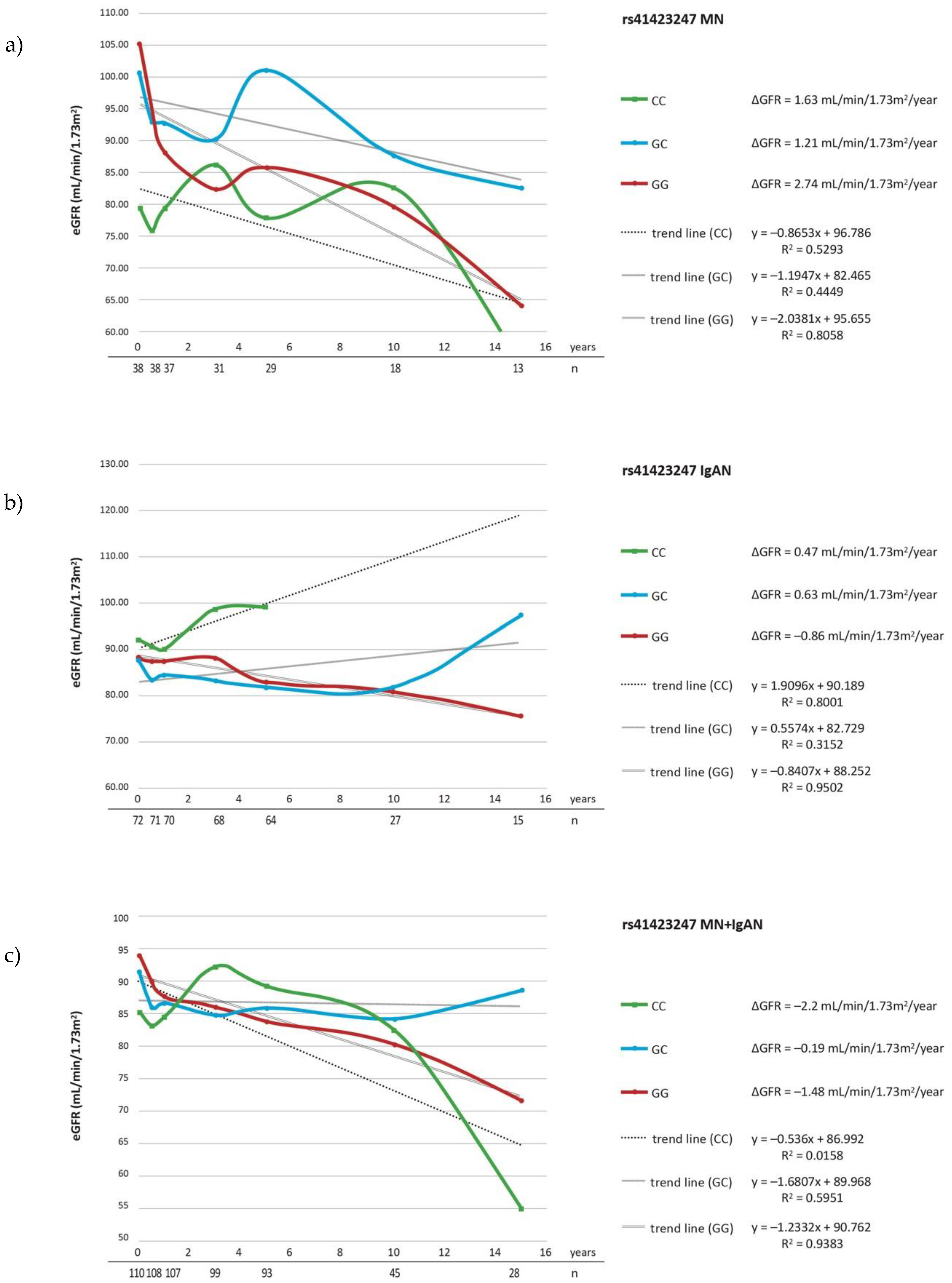
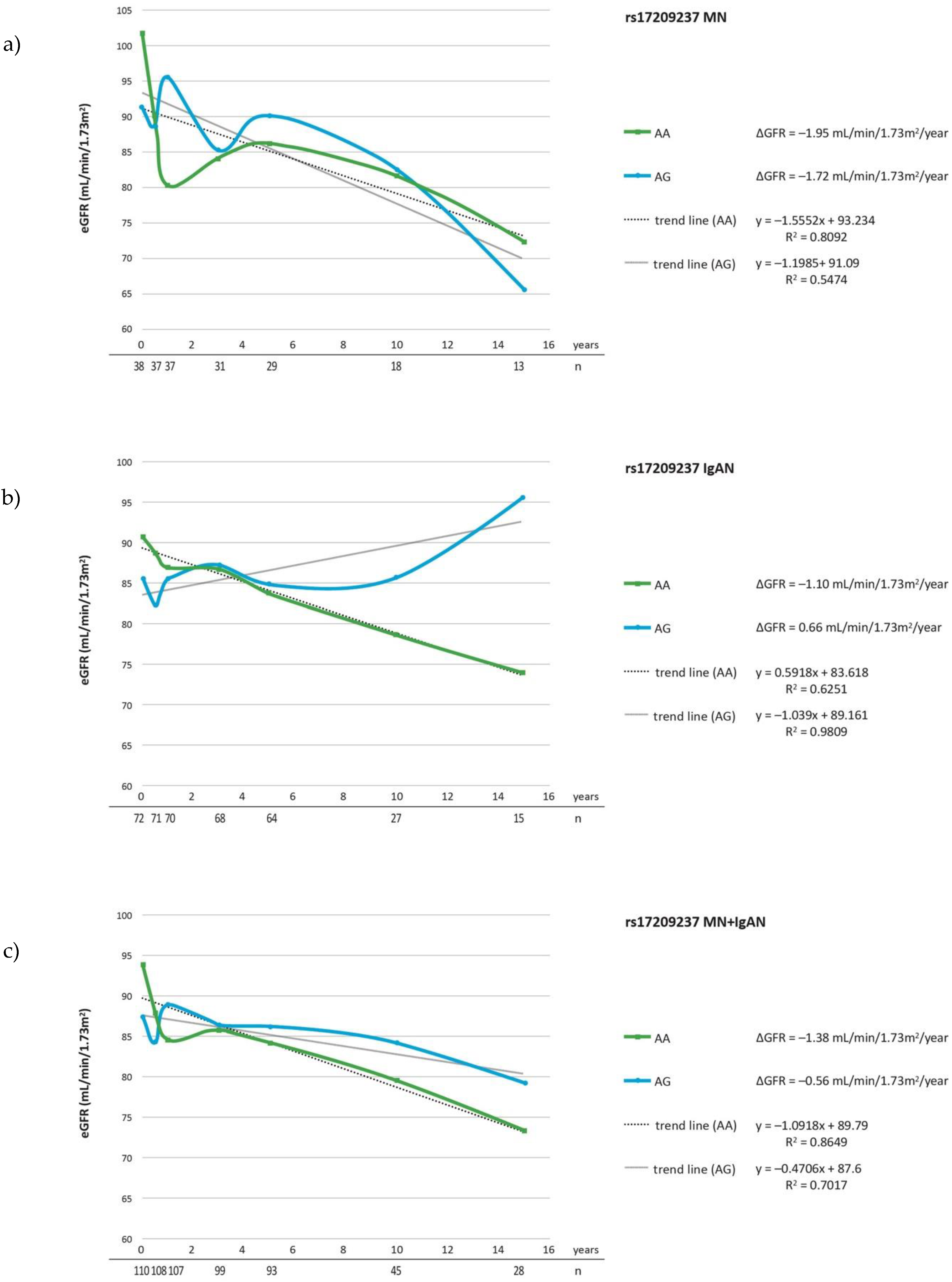
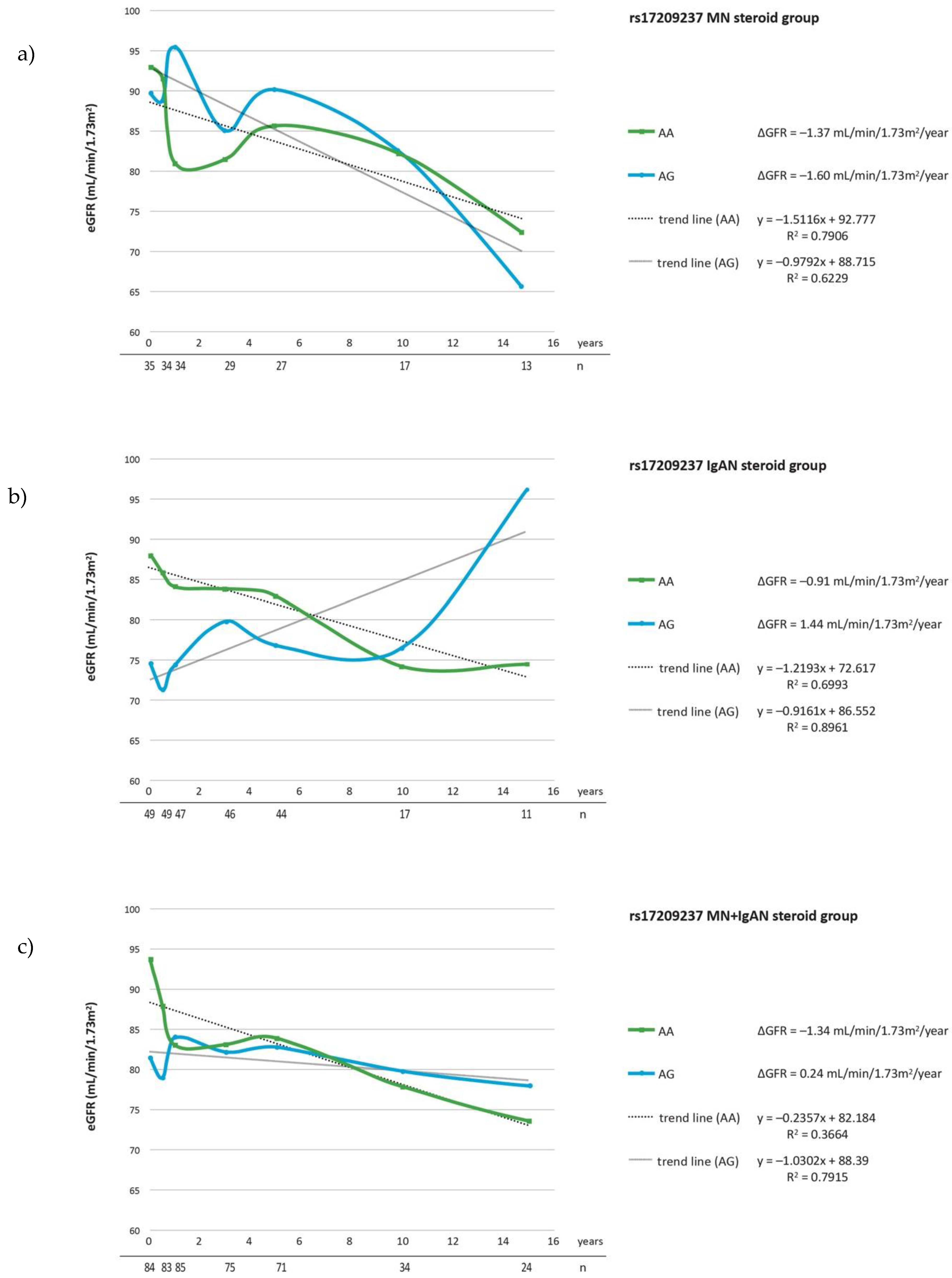
| Parameter | IgAN n = 72 | MN n = 38 | Controls n = 175 |
|---|---|---|---|
| Age, year | 33.96 ± 12.03 | 42.89 ± 14.37 | 48.7 ± 17.9 |
| Gender (male/female, n) | 38/34 | 24/14 | 86/89 |
| Creatinine (mg/dL) | 1.09 ± 0.48 | 0.92 ± 0.45 | 0.93 ± 0.20 |
| GFR (mL/min/1.73 m2) | 88.38 ± 30.98 | 96.89 ± 29.31 | 85.84 ± 19.30 |
| 24-h urine protein excretion, mg/d | 1.73 ± 2.07 | 4.44 ± 3.37 | - |
| ACE-I (n) | 44 | 26 | - |
| ARB (n) | 19 | 8 | - |
| CsA (n) | 2 | 7 | - |
| TAC (n) | 0 | 1 | - |
| CYF (n) | 0 | 4 | - |
| AZA (n) | 4 | 1 | - |
| MMF (n) | 1 | 0 | - |
| Stages n = 30 | |
|---|---|
| I | 8 |
| I/II | 4 |
| II | 13 |
| II/III | 2 |
| III | 3 |
| IV | 0 |
| Oxford Classification n = 48 | Hass Classification n = 56 | ||||
|---|---|---|---|---|---|
| I | II | III | IV | V | |
| n = 1 | n = 5 | n = 25 | n = 18 | n = 7 | |
| M | |||||
| 0 | 1 | 5 | 20 | 1 | 3 |
| 1 | 0 | 0 | 0 | 14 | 4 |
| E | |||||
| 0 | 1 | 5 | 11 | 12 | 4 |
| 1 | 0 | 0 | 9 | 3 | 3 |
| S | |||||
| 0 | 1 | 1 | 8 | 1 | 0 |
| 1 | 0 | 4 | 12 | 14 | 7 |
| T | |||||
| 0 | 1 | 4 | 18 | 11 | 1 |
| 1 | 0 | 1 | 2 | 4 | 3 |
| 2 | 0 | 0 | 0 | 0 | 3 |
| C | |||||
| 0 | 1 | 5 | 13 | 9 | 5 |
| 1 | 0 | 0 | 7 | 6 | 2 |
| 2 | 0 | 0 | 0 | 0 | 0 |
| Genotypes | ||||||
|---|---|---|---|---|---|---|
| rs6198 | ||||||
| CC | CT | TT | ||||
| IgAN + MN | 0 | 0% | 36 | 33% | 74 | 67% |
| MN | 0 | 0% | 12 | 32% | 26 | 68% |
| IgAN | 0 | 0% | 24 | 33% | 48 | 67% |
| Control | 2 | 1% | 48 | 27% | 125 | 71% |
| rs41423247 | ||||||
| CC | GC | GG | ||||
| IgAN + MN | 19 | 17% | 44 | 40% | 47 | 43% |
| MN | 10 | 26% | 12 | 32% | 16 | 42% |
| IgAN | 9 | 13% | 32 | 44% | 31 | 43% |
| Control | 23 | 13% | 78 | 45% | 74 | 42% |
| OR = 0.349, p = 0.026 | ||||||
| rs17209237 | ||||||
| GG | AG | AA | ||||
| IgAN + MN | 1 | 1% | 42 | 42% | 56 | 57% |
| MN | 2 | 5% | 16 | 42% | 20 | 38% |
| IgAN | 0 | 0% | 30 | 42% | 42 | 58% |
| Control | 2 | 1% | 58 | 33% | 115 | 66% |
| (a) | MN | ||||||||||||
| SNP | Genotype | ΔeGFR≥ −1 per Year (n) | ΔeGFR< −1 per Year (n) | RR | OR | Chi2 | p | Proteinuria < 1 g/day (n) | Proteinuria≥ 1 g/day (n) | RR | OR | Chi2 | p |
| rs6198 | CT | 5 | 7 | n.s. | n.s. | 0.433 | 0.510 | 7 | 5 | n.s. | n.s. | 0.175 | 0.675 |
| TT | 8 | 18 | n.s. | n.s. | 17 | 9 | n.s. | n.s. | |||||
| rs41423247 | CC | 4 | 6 | n.s. | n.s. | 1.9 | 0.388 | 7 | 3 | n.s. | n.s. | 0.324 | 0.850 |
| GC | 6 | 6 | n.s. | n.s. | 7 | 5 | n.s. | n.s. | |||||
| GG | 4 | 12 | n.s. | n.s. | 10 | 6 | n.s. | n.s. | |||||
| rs17209237 | AA | 4 | 16 | 1.6 | 4 | 3.6 | 0.058 | 13 | 7 | 0.8 | 0.69 | 0.286 | 0.593 |
| AG | 8 | 8 | 9 | 7 | |||||||||
| (b) | IgAN | ||||||||||||
| SNP | Genotype | ΔeGFR≥ −1 per Year (n) | ΔeGFR< −1 per Year (n) | RR | OR | Chi2 | p | Proteinuria < 1 g/day (n) | Proteinuria ≥ 1 g/day (n) | RR | OR | Chi2 | p |
| rs6198 | CT | 8 | 16 | n.s. | n.s. | 2.266 | 0.132 | 18 | 5 | n.s. | n.s. | 0.156 | 0.693 |
| TT | 25 | 23 | n.s. | n.s. | 34 | 12 | n.s. | n.s. | |||||
| rs41423247 | CC | 5 | 4 | n.s. | n.s. | 0.546 | 0.761 | 8 | 1 | n.s. | n.s. | 4.251 | 0.119 |
| GC | 15 | 17 | n.s. | n.s. | 19 | 11 | n.s. | n.s. | |||||
| GG | 13 | 18 | n.s. | n.s. | 25 | 5 | n.s. | n.s. | |||||
| rs17209237 | AA | 16 | 26 | 1.429 | 2.125 | 2.431 | 0.119 | 36 | 6 | 0.351 | 0.242 | 6.195 | 0.013 |
| AG | 17 | 13 | 16 | 11 | |||||||||
| (c) | All Patients | ||||||||||||
| SNP | Genotype | ΔeGFR≥ −1 per Year (n) | ΔeGFR< −1 per Year (n) | RR | OR | Chi2 | p | Proteinuria < 1 g/day (n) | Proteinuria ≥ 1 g/day (n) | RR | OR | Chi2 | p |
| rs6198 | CT | 13 | 23 | n.s. | n.s. | 0.716 | 0.397 | 25 | 10 | n.s. | n.s. | 0.004 | 0.095 |
| TT | 33 | 41 | n.s. | n.s. | 51 | 21 | n.s. | n.s. | |||||
| rs41423247 | CC | 8 | 11 | n.s. | n.s. | 1.248 | 0.536 | 15 | 4 | n.s. | n.s. | 2.85 | 0.241 |
| GC | 21 | 33 | n.s. | n.s. | 26 | 16 | n.s. | n.s. | |||||
| GG | 17 | 30 | n.s. | n.s. | 35 | 11 | n.s. | n.s. | |||||
| rs17209237 | AA | 20 | 42 | 1.484 | 2.5 | 5.302 | 0.021 | 49 | 13 | 0.501 | 0.369 | 5.33 | 0.021 |
| AG | 25 | 21 | 25 | 18 | |||||||||
| GG | 1 | 1 | 2 | 0 | |||||||||
| (a) | MN | ||||||||||||
| SNP | Genotype | ΔeGFR≥ −1 per Year (n) | ΔeGFR< −1 per Year (n) | RR | OR | Chi2 | p | Proteinuria< 1 g/day (n) | Proteinuria≥ 1 g/day (n) | RR | OR | Chi2 | p |
| rs17209237 | AA | 3 | 15 | 1.79 | 5.714 | 4.95 | 0.026 | 12 | 6 | 0.714 | 0.571 | 0.609 | 0.435 |
| AG | 8 | 7 | 8 | 7 | |||||||||
| GG | 1 | 1 | 2 | 0 | |||||||||
| (b) | IgAN | ||||||||||||
| SNP | Genotype | ΔeGFR≥ −1 per Year (n) | ΔeGFR< −1 per Year (n) | RR | OR | Chi2 | p | Proteinuria< 1 g/day (n) | Proteinuria≥ 1 g/day (n) | RR | OR | Chi2 | p |
| rs17209237 | AA | 10 | 21 | 1.4 | 2.1 | 1.510 | 0.219 | 21 | 6 | 0.404 | 0.23 | 5.347 | 0.021 |
| AG | 9 | 9 | 9 | 11 | |||||||||
| GG | 0 | 0 | 0 | 0 | |||||||||
| (c) | All patients | ||||||||||||
| SNP | Genotype | ΔeGFR≥ −1 per Year (n) | ΔeGFR< −1 per Year (n) | RR | OR | Chi2 | p | Proteinuria< 1 g/day (n) | Proteinuria≥ 1 g/day (n) | RR | OR | Chi2 | p |
| rs17209237 | AA | 13 | 36 | 1.515 | 2.94 | 5.306 | 0.021 | 39 | 10 | 0.435 | 0.291 | 6.355 | 0.012 |
| AG | 17 | 16 | 17 | 15 | |||||||||
| GG | 1 | 1 | 2 | 0 | |||||||||
| Oxford Classification n = 44 | ||||||
|---|---|---|---|---|---|---|
| SNP | Genotype | M0 | M1 | OR | Chi2 | p |
| rs41423247 | GG | 10 | 12 | 5.40 | 6.29 | 0.012 |
| GC | 18 | 4 | ||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pac, M.; Krata, N.; Moszczuk, B.; Wyczałkowska-Tomasik, A.; Kaleta, B.; Foroncewicz, B.; Rudnicki, W.; Pączek, L.; Mucha, K. NR3C1 Glucocorticoid Receptor Gene Polymorphisms Are Associated with Membranous and IgA Nephropathies. Cells 2021, 10, 3186. https://doi.org/10.3390/cells10113186
Pac M, Krata N, Moszczuk B, Wyczałkowska-Tomasik A, Kaleta B, Foroncewicz B, Rudnicki W, Pączek L, Mucha K. NR3C1 Glucocorticoid Receptor Gene Polymorphisms Are Associated with Membranous and IgA Nephropathies. Cells. 2021; 10(11):3186. https://doi.org/10.3390/cells10113186
Chicago/Turabian StylePac, Michał, Natalia Krata, Barbara Moszczuk, Aleksandra Wyczałkowska-Tomasik, Beata Kaleta, Bartosz Foroncewicz, Witold Rudnicki, Leszek Pączek, and Krzysztof Mucha. 2021. "NR3C1 Glucocorticoid Receptor Gene Polymorphisms Are Associated with Membranous and IgA Nephropathies" Cells 10, no. 11: 3186. https://doi.org/10.3390/cells10113186
APA StylePac, M., Krata, N., Moszczuk, B., Wyczałkowska-Tomasik, A., Kaleta, B., Foroncewicz, B., Rudnicki, W., Pączek, L., & Mucha, K. (2021). NR3C1 Glucocorticoid Receptor Gene Polymorphisms Are Associated with Membranous and IgA Nephropathies. Cells, 10(11), 3186. https://doi.org/10.3390/cells10113186






