Abstract
Scientifically interesting as well as practically important phenotypes often belong to the realm of complex traits. To the extent that these traits are hereditary, they are usually ‘highly polygenic’. The study of such traits presents a challenge for researchers, as the complex genetic architecture of such traits makes it nearly impossible to utilise many of the usual methods of reverse genetics, which often focus on specific genes. In recent years, thousands of genome-wide association studies (GWAS) were undertaken to explore the relationships between complex traits and a large number of genetic factors, most of which are characterised by tiny effects. In this review, we aim to familiarise ‘wet biologists’ with approaches for the interpretation of GWAS results, to clarify some issues that may seem counterintuitive and to assess the possibility of using GWAS results in experiments on various complex traits.
1. Introduction
Complex traits, by definition, depend on a large number of genetic and environmental factors and the interaction between these factors. Complex traits are both the most common and the most interesting phenotypes to study. The key problem in the study of complex traits is the difficulty of researching them using reverse genetics methods, the purpose of which is to search for unknown functions of a molecular sequence by introducing mutations and tracking emerging phenotypes. Naturally, when the expected effect of each of the numerous genes on the phenotype is very weak, such studies are extremely difficult to conduct, as they involve multiple directional changes in the genetic apparatus of the cell that are not possible with current directed genome editing technologies. In addition, for complex traits, it is often unclear what specific changes should be made in cells to obtain the desired phenotypes.
This question can be answered by classical forward genetics, which aims to establish the genetic variability associated with the variability in the studied trait by, for example, searching for mutations responsible for a specific phenotype in screens conducted in model organisms using artificial mutagenesis. The field of forward genetics has been enriched with new tools in the last ten years owing to the development of new technologies: genome sequencing and hybridisation arrays. Single-nucleotide polymorphism (SNP) hybridisation arrays make it possible to perform experiments such as genome-wide association studies (GWAS) with simultaneous interrogation of the allelic status of hundreds and later hundreds of thousands of polymorphisms. Recently, thousands of GWAS have been carried out, the results of which can serve as a foundation for subsequent biological experiments. However, it is difficult not to notice the weak interest of experimental biologists in the results of GWAS. We believe that this is largely due to a cultural barrier between geneticists and experimental biologists. From the point of view of devoted experimentalists, GWAS are often perceived as overpriced experiments aimed at exploring the genetics of overly specific, reductionist or bizarre traits such as exhaustion in shift workers [1], restless leg syndrome [2], household income [3], colour of meat of Atlantic salmon [4], self-reported habitual walking pace [5], only to find a bunch of tiny effects that individually have almost no influence on a trait. Why these studies are completely acceptable—and they are—demands an explanation. The purpose of this review is to acquaint ‘wet biologists’ with what is happening on the other side of the barrier, why the data obtained in GWAS can be taken seriously and, most importantly, how it could be used in their experimental work.
2. GWAS Is a Major Tool for the Genetics of Complex Traits
The emergence of GWAS as a full-fledged experimental design has already been described in excellent reviews [6,7]. In short, at the end of the 20th century, the key method for identifying the genetic factors responsible for a studied trait was the analysis of the linkage of the regions of the genome co-segregated in families with this trait (‘linkage analysis’). This analysis was capable of identifying risk factors with strong effects, such as the association of the ε4 allele of the APOE gene with Alzheimer’s disease (AD) [8,9] and genetic risk factors for breast cancer in the BRCA1 [10] and BRCA2 genes [11]. However, linkage analysis was unsuccessful for most of the complex traits studied. A new approach that would allow the identification of genetic factors with small effects on polygenic complex traits was needed. Genome-wide testing of genotypes should rely on associations of genotypes with traits, and not on linkage analysis, since the former has significantly higher statistical power (Figure 1a) [12]. The ability to carry out genome-wide association analysis was one of the motivations for the Human Genome Project [13] and subsequent projects to study widespread inheritance in human populations, most notably the HapMap Consortium [14].
It is expected that variability that is causal for the complex trait under study will be detected experimentally based on the trait-dependent frequency of the marker alleles with which the causal genotype is linked. Since there are several hundred thousand independent linkage groups for polymorphisms in the human genome, in GWAS, genotypes for all of these polymorphisms must be obtained and tested for association with a trait. The direct result of GWAS is a list of statistically significant associations mapped to specific genome regions (‘GWAS linkage regions’ or ‘GWAS hits’). As we will see below, the complete GWAS results (i.e., what effects [their size, direction and statistical significance] were obtained for each of the studied polymorphisms) are equal if not more important, regardless of whether they reach the genome-wide significance level.
Performing GWAS implies that common genetic variation for an organism of interest is known beforehand. The typical linkage disequilibrium (LD) structure should also provide a sufficient (yet not extensive) number of tagging polymorphisms to capture genetic variability. In most cases, this requires efficient and cheap technology for mass genotyping. Though it was possible to perform large-scale genotyping with conventional methods [15], GWAS were kick-started in the late 2000s, when mass genotyping with hybridisation arrays became available [16,17]. Custom arrays were created for genotyping of many organisms, such as Arabidopsis thaliana [18], mouse [19] and Atlantic salmon [20]. Currently, Illumina provides specialised genotyping arrays for human, porcine, bovine, equine, maize, mouse, potato, ovine and Pacific white shrimp genetic research. The genetic information required for GWAS can also be obtained directly from DNA sequencing, which is a convenient method for organisms with smaller genomes, such as yeast [21] or Caenorhabditis elegans [22]. As a rule, the use of the conventional whole-genome sequencing for GWAS is overkill for organisms with large genomes, but it could be justified in some cases. First, it is useful when a researcher’s primary goal is to collect data on rare mutations, as was done in massive studies on the genetics of blood metabolite levels [23], early onset atrial fibrillation [24] and Lewy body dementia [25]. Second, sequencing is required if GWAS is coupled with research on an unknown genetic variation, as in the case of the genetics of sand pear fruit quality [26]. Finally, for organisms with extensively researched haplotype population structure, such as humans, it is possible to perform genotyping with low-pass full-genome sequencing with coverage <1 followed by imputation with reference haplotypes. The cost of genotyping with this method could be substantially less than with even the cheapest available genotyping arrays and will probably replace them as the principal genotyping method in the future [27,28].
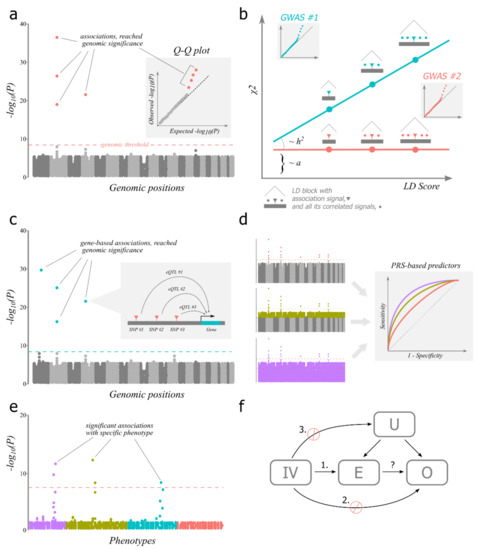
Figure 1.
Main concepts discussed in the text. (a) GWAS. The scheme depicts the Manhattan plot, a main visualisation of GWAS results. Manhattan plot shows distribution of observed p-levels of individual association tests across genomic positions (represented as dots). Manhattan plot allows to quickly assess how many associations pass the genomic significance threshold (dashed line). Inset depicts the corresponding quantile-quantile (Q-Q) plot which shows the distribution of observed versus expected p-levels. (b) LDSC. The scheme shows differences in two GWAS experiments both with population bias and only one has true genetic effects (turquoise). The Q-Q plots for both experiments are depicted as insets to illustrate that they look the same. LD score is a sum of correlations between tested SNPs for a given SNP. Chi-squared is a measure of effect for a given SNP, modelled as a random variable. The slope of the regression is proportional to heritability (h2) and the intercept (a) is proportional to bias. (c) TWAS. The scheme depicts the Manhattan plot with gene-based associations. The inset shows how an individual association is produced with external eQTL data which are used to predict gene expression by genotypes in GWAS data. (d) PRS. The scheme demonstrates a typical situation in GWAS when polygenic scores calculated with relaxed genomic thresholds perform better than with strict threshold. Three ways of selecting associations from the same GWAS for PRS calculation are presented (left). ROC curves (right) represent predictive models based on PRS, calculated using significant (index) associations only (salmon), with relaxed threshold (olive) and all SNPs (‘omnigenic’, lilac). (e) PheWAS. Association tests of a broad range of phenotypes grouped by similarity for a specific genotype are depicted. The typical situation where similar phenotypes have similar degree of association is depicted. (f) Mendelian randomisation. The scheme shows the experiment of studying the possible causal relationship (an arrow, marked with the question mark) between exposure (E) and outcome (O) with instrumental variable (IV) and possible unknown confounders (U). Numbered are conditions for IV to be valid, stop signs symbolise forbidden relations.
The first conventional human GWAS appeared in 2005, when the genetics of age-related macular degeneration was investigated in a modest sample of 50 patients and 96 controls [29]. Thousands of GWAS have been performed since then, with the sample size of the typical GWAS growing over time to provide more statistical power for detection of smaller effects (Figure 2). The need for sufficient samples for GWAS led to an unprecedented degree of collaboration in complex traits genetics. Many consortia of researchers were created to achieve the goal, including the Genetic Investigation of ANthropometric Traits (GIANT) Consortium [30], the Psychiatric Genomics Consortium (PGC) [31], the DIAbetes Genetics Replication And Meta-analysis (DIAGRAM) [32], the International Parkinson’s Disease Genomics Consortium (IPDGC) [33] and many others. The efforts of hundreds of laboratories working together in these consortia allowed them to carry out research on an enormous number of samples. At the time of this writing, 31 of the studies in the GWAS Catalog had a sample size exceeding one million participants, though most of the large studies are meta-analyses. Some examples of the largest studies to demonstrate the diversity of human phenotypes studied with GWAS include those focusing on: (1) Physiological traits such as blood pressure [34], cholesterol level [35] and concentration of liver enzymes in blood serum [36]; (2) medical conditions such as breast cancer [37], chronic renal failure [38], osteoporosis [39], Parkinson’s disease [40], diabetes [41], cataract [42] and dental caries [43]; (3) anthropometric traits such as height [44], longevity [45], handedness [46], body fat distribution [47]; (4) lifestyle traits such as alcohol consumption [48], smoking [49] and chronotype [50]; (5) psychological traits such as self-reported depression [51], risk tolerance [52], intelligence [53], well-being [54], ‘Big Five’ personality traits [55] and even (6) socioeconomic traits such as educational attainment [56], family income [3] and being fired from work [57]. It is difficult to find a common human phenotype that has not yet been studied with GWAS.
It is a rare situation when an analytical framework is developing primarily for a human model, especially for anthropometric traits and psychiatric and common autoimmune diseases. However, non-human GWAS are also developing rapidly. Non-human models could provide unique advantages for complex traits genetics research. For organisms that can be maintained as isogenic lines (for example, those capable of self-fertilisation), it is especially convenient to conduct GWAS for new traits with preliminarily genotyped collections of specimens [58,59]. For example, such collections have been generated for A. thaliana [18], sorghum [60], Drosophila melanogaster [61], rice [62] and many others. When extreme genetic transgression can be achieved, the collections of natural variation are not even required, as it is possible to easily create artificial genetic and phenotypic variation using genetically diverse strains, as can be done with yeasts [63], C. elegans [22] and even mice [64]. For some species, it could be beneficial to create a synthetic population with interspecies hybrid crossing, as was done in a study of hypoxia in catfish [65].

Figure 2.
Published studies with available summary statistics that are included in the GWAS Catalog. (a) Number of studies added to the GWAS Catalog by year. (b) Sample sizes in the GWAS Catalog over time. Some studies mentioned in the text are highlighted. The data were accessed through the GWAS Catalog site on 21 September 2021.
In human genetics, raw genotyping data for specific individuals are almost never available to unauthorised researchers. However, to reuse GWAS results, it is often enough to have access to published GWAS summary statistics (i.e., information about allelic effects and statistical significance over all tested polymorphisms relative to a studied trait). However, many studies do not report summary statistics, although the situation is improving [66]. The severity of the problem depends on the field; for example, the share of public GWAS summary statistics in oncology is currently the lowest among all of human biomedical research [67]. Currently, the UK Biobank stands out from the large individual research centres, as it provides access to GWAS results for 4541 (at the time of the writing) different biomedical, psychological and socioeconomic phenotypes with freely available and harmonised summary statistics files. In general, the database of large-scale genetic research (NHGRI-EBI GWAS Catalog) [68] has, at the time of writing, 5329 records of published studies for over ten thousand traits. For other organisms, the universal resources of GWAS data are not yet available, with the noticeable exception of A. thaliana genetics: the AraGWAS Catalog currently contains data for 462 phenotypes from approximately 20 individual studies [69].
3. Assumptions of GWAS
3.1. Heritability of Complex Traits
GWA studies are often referred to as ‘hypothesis-free’ research, as opposed to the classic ‘candidate’ association analysis, in which the researcher initially assumes a link between a specific locus and a disease. The hypothesis-free approach to genetic research has a number of advantages; in particular, it allows the discovery of new unexpected genes associated with a trait, thereby acting as ‘hypothesis-generating machine’, and is not subject to the effect of overrepresentation of positive results in the literature (publication bias) characteristic of candidate studies [70]. Nevertheless, there is a hypothesis at the heart of GWAS: that a complex trait has a specific hereditary nature. A typical genetic experiment is devoted to the study of diversity associated with heredity. It is often possible to know in advance what proportion of the variability of a trait is determined by genetic factors. This number, the heritability of the trait, can be determined from, for example, twin or adoption studies. Estimates of the heritability of a trait are useful for planning a GWA study—the lower the heritability, the more samples needed for a successful experiment [71]. Note that very often our intuition about the role of genetics in a trait is wrong, especially when it comes to behavioural and socioeconomic traits [72].
It is important to note some properties of heritability. First, heritability is not a measure of how much a trait is determined by genes; rather, it indicates how much the variability of a trait is explained by genetic variation in the studied population. Variability may be low and heritability high, as in the case of schizophrenia, which, despite being a ‘common disease’, still affects only a small fraction (around 1%) of the general population. Conversely, for traits correlated with fitness, for example, such as fecundity and longevity, variation is big, but heritability is low [73]. Second, heritability is not constant, and it depends on the specific population in which it is measured. For example, a study of the heritability of tobacco dependence in the Dutch population at the beginning of the 21st century showed that it is as high as 0.75 [74]. However, such a study would have made no sense until the Dutch became acquainted with tobacco in the 17th century. On the other hand, it can be assumed that estimates of the heritability of adult literacy in the late 19th century were higher than they are today, since the variability in adult literacy in most countries has been reduced to zero by modern secondary education systems. This does not apply to literacy in preschool children, among whom there is indeed such variability and measuring the heritability of this trait makes sense. In a joint twin study on populations in Australia, Norway, Sweden and the United States, the heritability estimate for literacy in preschoolers was approximately 0.7 [75].
Modern GWAS with negative results in large samples are relatively rare. This led to the practice of performing GWAS without having preliminary data on the heritability of a trait. For example, genetic susceptibility to COVID-19 was quickly explored as early as in 2020 without prior understanding of whether heredity plays a role in the spread of the new virus causing this disease [76,77,78]. While this practice seems questionable, it should be noted that the GWAS results themselves could be employed to estimate the heritability of the trait (h2(SNP), heritability in the narrow sense or the theoretical limit of heritability, which could be explained by additive common variation; see the Section 3.3 below).
3.2. Population Structure
In a genetic experiment, the difference in the distribution of allele frequencies depending on the studied phenotype may be associated not with the phenotype, but with the unequal population structure relative to a trait, leading to false positive associations. The problem is more serious in small-scale genetic experiments, however. Massive genetic data often already contain controls for population structure, which can be obtained using principal component analysis or similar methods. A widely used practice in population genetics is to use genetic markers to map DNA samples from known genetic information. The accuracy of this method is even sufficient for use in forensic medicine. For example, the data on genetic markers of African elephants were enough to establish the specific site of poaching activity using DNA isolated from confiscated ivory [79]. The accuracy of genome-wide genetic data is even higher. Principal component analysis of genotypes even allows, for example, the reproduction of the main outlines of the geographical map of Europe from the genotypes of European ethnic groups [80]. When studying human populations, owing to the available reference panels of genotypes, such as International HapMap Consortium [14], 1000 Genomes Consortium [81] or The Haplotype Reference Consortium [82], it is easy to determine which sample belongs to which ethnic group. Thus, in a GWAS experiment, the data itself contain a control for the population structure.
A rigorous method for controlling population structure was developed based on the difference between how linkage disequilibrium relates to the true effects and to the effects caused by uneven population structure. The effects associated with uneven distribution of alleles due to differences in population structure should not, in contrast to true associations, depend on the length of the region with SNPs in high linkage disequilibrium (‘LD blocks’). It can be assumed, at least in the first approximation, that the position of causal polymorphisms for a trait under study is not related to how genotypes were recombined in the past; that is, the position of causal polymorphisms is random relative to the genetic recombination map. This means that the larger the LD block, the greater the chance of finding a higher level of significance for that region. If the effects identified are associated with segregation of haplotypes due to differences in population structure, the association signal should not depend on the length of the linkage region in which it is located (Figure 1b). The method based on this idea is called LD score regression, or LDSC [83].
LD score regression turned out to be one of the most influential ideas in the analysis of GWAS results. The use of methods based on LD score regression, as we will see below, allows researchers not only to control the population structure, but also to determine heritability according to GWAS data, the contribution to the heritability of a trait by functional classes of polymorphisms and the relationship of different traits to each other, as well as meet other objectives.
3.3. Common Additive Variation
The important assumption of GWAS is that the genetics of the trait under study is largely associated with common polymorphisms. Conceptually, when the fitness of an organism is associated with a complex trait, genetic factors with small effects have a great chance of avoiding the effect of negative selection, thus shifting the genetic architecture of the trait towards greater polygenicity with more common genotypes [84,85]. This was tested experimentally in the yeast model, and indeed, rare mutations with large effects were found to be more likely to be recent variants [86].
This does not mean that rare mutations with large effects do not affect complex traits; on the contrary, for many complex traits, such rare mutations with large effects have been found. The distinction between rare and common mutations is arbitrary and motivated largely by the methods used to explore these two types of variation. For example, it was found that common and rare mutations contribute to the variability of a trait in an additive manner for autism spectrum disorder [87] and obesity [88]. However, for a number of such traits (for example, coronary artery disease, type II diabetes and breast cancer), using GWAS has led to a situation in which comparable risk groups can be identified in a much larger number of people than can be identified using well-known mutations with large effects [89]. Moreover, GWAS for many traits with ‘simple’ genetics such as rare monogenic diseases were able to reveal additional genetic factors that can influence the severity of the disease. Such factors were identified in, for example, sickle cell anaemia [90], cystic fibrosis [91], acne [92] and Huntington’s chorea [93]. In fact, it is likely that all genetic traits are complex to some extent and can be studied with GWAS.
GWAS suggests that the underlying inheritance of the trait studied by GWAS should be mainly additive. This often seems counterintuitive to an experimental biologist, as it is natural to expect that the multilevel regulatory pathways characteristic of biological systems should be reflected in the genetics of a trait as epistatic interactions. Thus, it seems strange that it is difficult to find an example of clear epistasis in GWAS results. This could be explained by the fact that, in GWAS data, a weak statistical signal from non-additive interactions will be difficult to detect with a simple search due to the curse of dimensionality. Even for simple two-level interactions for N genotypes, there are (N2/2 − N) combinations times four types of epistatic interactions. The search for an optimal methodological approach aside from simple enumeration, including using machine learning and/or external functional data, has been the subject of a large number of studies (see reviews [94,95]).
However, finding multilevel epistasis in GWAS data can be challenging for reasons beyond statistical limitations. Even all of the people who have ever lived on earth are hardly enough to label all of the combinations of even a two-level epistasis for all LD-independent human polymorphisms. Intuitively, when a complex trait affects fitness, the evolutionary process will also be limited in the selection of non-additive effects. The defining importance of additive variability for evolution is emphasised in classical works on evolutionary biology: Fisher’s fundamental theorem postulates that the rate of evolutionary change is proportional to the additive genetic variability [96,97]. Conversely, epistatic effects are more likely to be found if variation of a complex trait violates assumptions of Fisher’s fundamental theorem. Indeed, in populations existing under artificial selection, like domesticated animals, epistatic effects can still be quite high [98,99]. The other case in which epistatic effects are observed is when a trait does not undergo selective pressure at all. It could be argued that human neurodegenerative diseases could be considered such traits, since they manifest well beyond reproductive age. Curiously, one of the best-known examples of epistasis in human GWAS data is the genetic interaction of the KHDRBS2 and CRYL1 genes in GWAS of AD [100]. In any case, it is unlikely that any complex trait will have genetics completely unrelated to fitness due to high polygenicity and pervasive pleiotropy of genetic factors of complex traits (see Section 4.4 below).
How functional interactions are reflected on a genetic level was tested directly in a 2021 study by Sinnott-Armstrong et al. In this study, GWAS was performed using data from the UK Biobank on metabolic traits (uric acid, IGF-1 and testosterone concentrations) that were deliberately selected to assess how consistent the GWAS data were with well-studied biological traits. As expected, most of the GWAS signals were found in the genes of the known biochemical pathways for the respective substances. However, epistatic interactions even between genetic factors associated with genes in the same biochemical pathway were either not detected or made a negligible contribution to the heritability [101]. It is possible to determine the importance of epistasis for the variability of complex traits by studying the genetics of organisms with small genomes. Yeasts are a convenient model for this type of experiment, since they allow the creation of a synthetic population for genetic testing with crossing of unrelated strains and phenotyping of easily measured traits like growth in the presence of various substances. This artificial system, simulating GWAS conducted under ideal conditions, revealed that even when the experiment has the necessary power to detect epistasis, additive genetic effects still make a decisive contribution to the heritability of a trait [63,86,102]. In this model experiment, the estimate of the contribution of non-additive heritability is probably even higher than expected in natural populations, since only the descendants from the first crosses were studied. Theoretical models predict that in natural populations, the contribution of epistasis to the variability of complex traits is even lower [103].
The underrepresentation of non-additive effects in GWAS data is an important feature of complex traits genetics. Without it, GWAS results would be substantially more difficult to interpret, compare and use in practice.
4. Arguments for GWAS
4.1. Reproducibility
GWA studies are characterised by high reproducibility in independent experiments. They tend to be carried out by large consortia that can bring together most of the scientists interested in studying a given trait. Therefore, the largest study in the field is often the only GWA study of comparable size, with most of the previously identified effects being reproduced at a new level [104]. An example of an impressive replication of the results of comparable GWAS can be found in recent genetic studies of vulnerability to COVID-19, as major independent studies found broadly the same loci associated with an increased risk of coronavirus infection [76,77,78]. The UK Biobank provides alternative GWAS results for a wide range of human traits, which allows the determination of how well the GWAS data from the UK Biobank reproduces previously conducted independent GWAS. Such an analysis was conducted on pairs of independent studies for nine traits and, in general, for polymorphisms that reached the genomic threshold in GWAS, the reproducibility was 85%. For stricter thresholds limiting the selected polymorphisms, reproducibility was even higher, with increases in both statistical significance and effect size [105].
It is normal for the same results to be obtained in studies on different human superpopulations, as evidenced by, for example, comparing GWAS on samples of people of European descent with GWAS on samples of people of East Asian or sub-Saharan African descent. The effects found for the same polymorphisms in such experiments are usually co-directional [106]. Moreover, associations with phenotypes are often located at the same loci for populations in which there are no same polymorphisms. Examples include height [107], blood lipid levels [108], type II diabetes [32,109,110], myopia [111] and schizophrenia [112]. In addition, in a new population, local genetic variability may be more advantageous than in an already tested one, as the new population may offer more common polymorphisms and a different linkage region. For example, the polymorphisms responsible for the association of the NOD2 gene with Crohn’s disease in Europeans are absent in East Asians, and these important associations could not be found in GWAS in this population [113]. Such effects allowed the creation of a new experimental design of the GWAS, trans-ethnic GWAS, which has several advantages over the traditional approach: in particular, it allows more accurate mapping of causal variants [114,115].
For some traits, even interspecies similarity of the GWAS results can be observed. For example, there is strong overlap between the top GWAS results for human growth and the size of some mammalian domestic animals. Associations in the genes LCORL and HMGA2 are in the list of GWAS signals for human height [116], as well as the size of dogs [99], cattle [117] and horses [118]. The use of GWAS to study the genetic causes of increased susceptibility to type II diabetes in Burmese cats has identified the same ANK1 risk gene [119] as that identified in the corresponding study in humans [41]. GWAS for granulomatous colitis in a small sample of boxers and bulldogs identified a single signal in a region [99] for which a homologous region was identified in the human GWAS for inflammatory bowel disease [120].
It is also important to note the reproducibility of genetic effects at the level of the mutation spectrum. The genetic effects found in GWAS are typically small and in general manifest themselves on a regulatory level, while mutations with large effects act on the gene structure directly and are relatively rare. It seems natural to expect that both types of genetic factors are present at the same loci, and for some well-powered studies of both rare and common variations, this indeed turned out to be the case. Examples of traits for which this was observed include height [121], inflammatory bowel disease [122], type II diabetes [123] and schizophrenia [124]. In general, it was demonstrated that rare mutations with significant associations (calculated on a gene level) are enriched by GWAS signals in a large study of exome sequencing in UK Biobank samples [125].
Such effects explain the very well-known cases of comorbidity between monogenic and common diseases. GWAS signals tend to be enriched in the genomic loci linked with Mendelian diseases [126]. Genetic signals for phenotypically matched monogenic and complex traits (for example, growth defect syndromes and height, monogenic mood disorders and schizophrenia and Mendelian cardiovascular diseases and cholesterol level) were found to be enriched in the same loci at a significant rate [127]. It is sometimes possible to use this information to assist in the interpretation of GWAS results. For example, a genome-wide significant SNP associated with coronary artery disease resides in the promoter of the LIPA gene, mutations which are involved in the Mendelian diseases: Wolman disease and cholesteryl ester storage disease. A more complex example is the case of SNPs associated with body mass index (BMI), which through the use of Hi-C data (genome DNA interactions) could be linked to the relatively distant (>100 kbp) CYP19A1 gene, which is phenotypically matched with BMI and involved in aromatase excess syndrome [127].
4.2. Interpretability
The strongest signals in GWAS often correspond to ‘natural’ biological interpretations. Examples include serum calcium concentration, corresponding to the calcium-sensitive receptor gene CASR [128]; dog size and the gene for insulin-like growth factor, IGF1 [99]; exhaustion in shift workers and the gene encoding melatonin receptor [1]; alcohol consumption and the gene for alcohol dehydrogenase, ADH1B [48]; number of cigarettes smoked per day and the gene encoding one of the nicotinic acetylcholine receptor subunits, CHRNA5 [49]; colour of meat of Atlantic salmon and the gene for beta-carotene oxygenase, bco1 [4]; and the anosmia symptom in COVID-19 and the genes encoding the odorant metabolising enzymes UGT2A1 and UGT2A2 [129]. However, most of the GWAS signals have unclear underlying biology, which motivates ‘post-GWAS’ experimental studies.
As with other ‘big’ biological data, there are formal ways to generate functional interpretations of GWAS results. It can be done simply by assigning SNPs to their functional categories individually, which could be enough to yield interesting results. For example, the analysis of early GWAS revealed that GWAS SNP hits are enriched in SNPs that are linked to regulation of gene expression—expression quantitative trait loci (eQTL) [130,131]. The enormous work of fitting cell-specific chromatin markers of ENCODE data to almost 1000 of the biggest GWA studies from the GWAS Catalog provided an expected picture of the distribution of traits to their corresponding affected tissues [132]. A more stringent approach is to consider the LD structure of GWAS data. There are a plethora of such methods that can be applied to GWAS summary statistics. Among the most popular are INRICH [133], DEPICT [134] and MAGMA [135]. The alternative method is to investigate the variability of a trait that could be explained by the variability of SNPs, or the portion of narrow-sense heritability, h2, that was captured in GWAS, h2(GWAS). It is possible to compute the contributions to h2(GWAS) of different groups of SNPs that are linked to a specific functional category. This idea was used to show a remarkable enrichment of genetic effects with DNase I hypersensitive sites, markers of genomic regions involved in transcription regulation [136]. Modern methods for partitioned heritability analysis are based on LD score regression (stratified LDSC, S-LDSC). Since the slope of LDSC regression is proportional to h2(GWAS), LDSC is able to correct for relatedness and population structure; in addition, the use of this method does not require access to raw genotypes, as summary statistics are sufficient [137].
These methods provide an interface between genetic epidemiological data and molecular phenotypes, such as cell-specific gene expression, active chromatin markers, DNA methylation and chromosome interactions, and allow one to assess the enrichment of genetic associations with various functional ontologies. These methods can be used to demonstrate enrichment of specific gene ontologies, like neurogenesis and locomotor behaviour in restless legs syndrome [2], cytokine signalling pathways for COVID-19 [138], cell growth and synapse organisation for volume of lateral nuclei [139] and cell adhesion and transsynaptic signalling for Tourette syndrome [140]. Hormozdiari et al., applied S-LDSC on sets of functional SNPs (eQTLs, etc.,) and found that they are indeed enriched for heritability in an expected, cell type-specific manner [141]. Surprisingly, similar results could easily be obtained with just conventional expression data and other functional data [142]. S-LDSC methods were used to demonstrate enrichment of genetic associations in brain cells, primarily pyramidal and medium spiny neurons of the cortex, in schizophrenia [143,144]; in several brain regions in GWAS of education attainment [56], risk tolerance [52] and household income [3]; in skin cells, particularly melanocytes, for melanoma [145]; in kidneys for serum urea concentration [101]; and in lungs for COVID-19 [78]. More elaborate methods have been used to obtain unexpected results. For example, the H-MAGMA method, a modification of MAGMA for working with chromatin interactions (Hi-C) data [146], was used on autism spectrum disorder GWAS data to identify a number of genes expressed in prenatal human brain [147].
Reference eQTL information allows the integration of GWAS data with specific genes to directly test for gene-based associations. Such an experiment is called a transcriptome-wide association study, or TWAS (Figure 1c) [148,149]. The same approach could be realised with information about the link between genetics and any other functional data (metabolite and protein levels, CpG methylation, splicing, etc.,), but expression reference data are usually much more available. TWA studies could provide insight into which specific genes in a specific cell context are involved in the biology of a trait, provide knowledge of directionality of an effect of gene expression on a trait and, as in typical differential expression studies, be analysed at the gene-set level to reveal underlying biological processes. TWAS could be performed using GWAS summary statistics with tools such as Fusion [149] and S-PrediXcan [150]. However, TWAS have several flaws compared to GWAS, related mostly to imperfections in eQTL reference panels (reviewed in [151]) and the inability to adequately capture eQTL effects in trans (see Section 5.2 below). With eQTL data available for an increasing variety of cell types and tissues via, for example, the GTEx Consortium [152], many traits have already been studied with TWAS, yielding many candidate genes for follow-up studies [153].
4.3. Utility
In principle, GWAS results could provide direct utility via identification of a successful candidate target for a manipulation of a trait. For example, GWAS of drought tolerance in maize yielded a signal in the ZmVPP1 gene, overexpression of which grants the plant drought tolerance [154]. In human genetics, GWAS results could be used to identify promising biomedical targets for common diseases, as many existing drug targets were retrospectively identified as association signals in GWAS (reviewed in [6,155]). Probably the cleanest example of new therapy emerging primarily from GWAS results is the prospective use of deucravacitinib for auto-immune diseases [156,157]. This substance acts on TYK2, the gene for which harboured the association signal in the early GWA studies for lupus [158], type I diabetes [159], psoriasis [160] and Crohn’s disease [161].
Such examples of direct GWAS utility are still relatively rare, but GWAS results are becoming more useful in predicting complex traits by means of genetic data via polygenic risk scores (PRS). The story of PRS is linked to the notorious ‘missing heritability’ problem; taken together, significant signals from early GWAS explained a negligible portion of heritability of a trait (h2(GWAS) << h2) [162]. Later it became clear that heritability, explained by common variation, was not ‘missing’, but ‘hidden’ due to the fact that the polygenicity of the studied traits turned out to be much higher than previously expected [163]. This was first discovered in one of the first GWAS for schizophrenia. Schizophrenia is a common disease and the heritability of schizophrenia, as determined in twin studies, is quite high, so it is reasonable to expect that GWAS on schizophrenia would explain much of this heritability. One can imagine how disappointing the results of the first GWAS for schizophrenia were [164,165,166], as these enormous efforts led to the discovery of only a few significant linkage regions that in total explain a minuscule part of the heritability of the disease.
In one of these GWAS, a previously proposed methodology [167] was used to calculate a generalised number that characterises the genetic risk of a disease as the sum of allelic effects found in GWAS. The calculation included all LD-independent polymorphisms for which a certain threshold level of significance was reached, including thresholds that were less stringent than the genomic threshold of significance. The number calculated using this method was called the polygenic risk score (sometimes called the genome-wide polygenic score and abbreviated as GPS). This PRS turned out to have a peculiar feature: it approximates the risk of the disease much better if a relaxed threshold level of significance is used to calculate it. That is, most of the polymorphisms that determine the risk of disease do not cross the genomic threshold for GWAS (Figure 1d). It is worth noting that all of these conclusions were made on the basis of the PRS, which then explained only about 3% of the variability. Nevertheless, this was still an order of magnitude more than the share of heritability explained by only the significant GWAS hits [164]. Notably, modern techniques for PRS calculation like LDpred2 [168] use advanced procedures for defining independent SNPs and for association threshold optimisation, but the performance is not drastically better than the original approach.
This can be interpreted as follows: (1) Most genetic mutations are linked in some way to functional mutations that affect the development of the disease; (2) for most of these mutations, the effects are too small to be reliably established in a GWAS performed on a sample of a realistic size; and (3) the contribution of the totality of such small genetic effects to h2 outweighs the contribution of mutations with strong effects. This is likely due to the fact that biological systems are able to respond even to mutations that are weakly associated with causal biological mechanisms. This view has been articulated in the ‘omnigenic hypothesis’ of common diseases [169,170]. In the case of schizophrenia, a disease of the brain, this assumption seems realistic, since approximately a third of human genes are associated with the functioning or development of the nervous system (for example, the CL:0002319 ‘neural cell’ ontology group now includes more than 5000 human genes, and GO:0007399 ‘nervous system development’ includes more than 6000 genes), and in each typical linkage group, there is either such a gene or a genomic region (e.g., enhancer) that controls the expression of the gene.
Although polygenic scores were originally used in psychiatric genetics, this methodology has become the standard for interpreting the results of any GWAS. The fact that polygenic scores explain significantly more variability in the trait than the sum of all statistically significant GWAS results has been repeatedly confirmed for other types of complex traits. This holds true even for the most powerful GWAS to date. For example, 3290 significant GWAS hits for human height with a sample size over 700,000 people together explained 24% of the variability in height, while PRS accounted for 34.7% [44]. A previous GWAS on height that included roughly 250,000 individuals estimated 10% of the variability in height, explained by 697 significant associations, while PRS accounted for 29% of the variation [116]. Curiously, such genetic complexity is not consistent across species. As an extreme example, 83% of the phenotypic variability in horse size is attributed to just four common genetic factors—no doubt a consequence of vigorous artificial selection [118].
While more powerful studies could resolve some of uncertainty about previous weak GWAS associations, it is doubtful that any substantial increase in sample size would yield a substantial increase in the variability explained by significant associations. Likewise, more sophisticated analysis might not help either. There have been multiple attempts to build more complex predictive models based on whole-genome genetic data, rather than relying on the sum of weighted effects with simple p-level thresholding as SNP (feature) selection. It is remarkable that such approaches appear not to have significantly improved PRS prediction to date [171,172,173,174]. As discussed above, this could be due to a lack of substantial non-additive (epistatic) effects for a typical complex trait.
However, in many cases, PRS are already the best-known predictors for a complex trait. An example is the use of PRS to identify individuals at increased risk for various common inherited diseases: type II diabetes, inflammatory bowel disease, breast cancer, atrial fibrillation and coronary artery disease [89]. For coronary artery disease, the results were particularly impressive, as the PRS predictor was found to be more accurate than the cumulative genetic markers already used in current clinical practice [89,175]. In a study conducted on a sample of 47,000 people, of which 11,000 were diagnosed with coronary artery disease, it was demonstrated that people who fall into the upper quintile for polygenic risk have twice the risk of coronary artery disease compared to the general population [176]. Examples of studies on the use of PRS in predictive models of disease risk include those evaluating the effectiveness of PRS in predicting breast cancer [177,178], prostate cancer [179], glaucoma [180] and longevity [181]. A study by Zhang et al., focused on what predictive performance could realistically be expected for PRS for 14 types of cancer. They estimated that the range of performance lies between AUCs of 0.63 and 0.89, with the lowest estimate for ovarian cancer and the highest for testicular cancer [182]. In a massive exome study of obesity, it was shown that the prevalence of obesity on a background of risk-increasing mutations in the MC4R gene and protective mutations in GPR75 are greatly modified by PRS of BMI, with an absolute difference in prevalence of approximately 60% between individuals belonging to extreme PRS quintiles [88]. The inclusion of PRS in predictive models for the transition to AD in individuals with mild cognitive decline improves the model compared to models based on APOE alone, with the AUC increasing from 0.68 to 0.84 [183,184]. For Parkinson’s disease, the best models currently predict the disease at an AUC only up to 0.65, which indicates that this is a poor predictor [40,185]; however, stratification by PRS allows the identification of the time of disease manifestation [186].
An impressive result demonstrating the predictive power of PRS was obtained in a GWA study on education (‘educational attainment’ and ‘educational achievement’) [56]. It would seem that a complex social trait like education would be difficult to predict based on genetic data. Nonetheless, this study turned out to be one of the most successful GWAS, largely due to the massive sample of more than one million people. Later, it turned out that this PRS explained approximately 15% of the variability in the duration of education, correlated (r = 0.4) with students’ final grades [187] and predicted educational success almost as well as the best predictor, family’s socioeconomic status [188,189]. One study even showed that genetic differences are entirely responsible for the difference in learning outcomes in UK schools both with and without pre-selection of students [190]. We should note, however, that the utility of these results is mainly in informing effective education policies as opposed to using education PRS on an individual level (see [191]). Prediction of education PRS is partially related to heritable family environment, as evidenced by the drop in the predictive power of PRS for education in adopted individuals [192]. This illustrates that just as for heritability estimates, polygenic scores do depend on the original population in which they were calculated.
The use of PRS can significantly improve the prediction of the outcome of a pleiotropic rare mutation. For example, carriers of a 22q11.2 deletion (DiGeorge syndrome) are developmentally delayed, but in addition to this, approximately 20–25% are at risk of developing schizophrenia [193,194]. The PRS calculated for schizophrenia significantly modifies the risk for carriers of the mutation: if the polygenic risk for schizophrenia is taken into account, then the risk of developing schizophrenia for carriers of 22q11.2 deletions is 33% for the highest decile and only 9% for the lowest decile. Similarly, for lack of intellectual development (measured as IQ < 70 with an a priori risk of approximately 40%), if PRS for IQ is used, then the risk of decreased IQ is 63% for carriers from the lowest decile and 24% for carriers from the highest decile [195].
When using a PRS, one should consider differences between the target population and the population in which the GWAS summary statistics used to compute the PRS was determined. For example, the average prediction accuracy for 17 quantitative traits, calculated for the European GWAS, falls approximately four-fold for the sub-Saharan African population [196]. Note that while PRS have low generalisability in ‘non-native’ populations [197], the use of PRS derived from trans-ethnic GWAS seems to provide an advantage over single-origin GWAS, even in a ‘native’ context [198].
Unfortunately, for many complex traits with low heritability and/or low phenotypic variation, it is impractical to achieve a sample size necessary to reliably establish genomic loci responsible for the majority of the heritable phenotypic variation. It seems that for many of the important complex traits, PRS remains the best measure to approximate genetic predisposition. Fortunately, PRS, though a simplified model of genetic risk, still often proves to be a quite effective complex trait predictor, often better than those already existing or at least able to produce a substantial increase in performance in a joint application.
4.4. Interoperability
Important evidence supporting GWAS is that genetic factors of similar complex traits tend to be similar themselves. This would of course not be the case if GWAS effects were just random noise. We have already noted that genetic factors for some monogenic traits are often found in the same genomic regions identified in GWAS for similar complex traits. While more powerful methods for estimation of co-heritability use individual genotype information [199], it is possible to employ the summary statistics information from GWAS (reviewed in [200] with, for example, LDSC [201,202,203]).
With more data available for different traits, the study of pleiotropy became more common and can be exploited as a starting point for functional studies. Analysis of pleiotropic effects could identify new susceptibility loci responsible for known comorbidities in human diseases. Examples include studies of major depression and loneliness [204], AD and diastolic blood pressure [205] and obesity and walking pace [5]. In GWAS on the volume of the human thalamus and its nuclei, LDSC was used for the discovery of significant positive correlations between the genetics of the volume of posterior nuclei and bipolar disorder, the volume of intralaminar nuclei and multiple sclerosis and the volume of the whole thalamus and Parkinson’s disease [139]. In a 2021 screen by Xicoy et al., for co-heritability between Parkinson’s disease and levels of 370 lipid species and lipid-related molecules in blood, eight specific lipid levels were found to share a significant portion of genetic architecture with Parkinson’s disease, which could have direct implications in explaining the aetiology of the disease and in practical diagnostics [206].
With the abundance of human traits available for analysis to date, it has even become possible to perform ‘reverse GWAS’ analysis, in which a plethora of traits are tested for association with a specific SNP. This analysis is called phenome-wide association, or PheWAS (Figure 1e) [207]. PheWAS of the associations of diverse complex traits in data from the UK Biobank showed that pleiotropic effects are extremely common [208]. In a practical sense, PheWAS could be used to test the pleiotropy of SNPs for a candidate medical target [156,209]. In addition, PheWAS, paired with MAGMA, could be generalised on the level of genes and gene sets [203]. This, in particular, allows researchers to test PheWAS trait enrichment in a xenobiological context with orthologous genes. For example, the GWAS signals for the phenotypes of the quality of ovine wool are, unsurprisingly, found to be associated with the human traits of hair colour and baldness [210].
In many situations, researchers are interested not only in similarity of traits, but also in causal relationships between them. There is an approach, instrumental variable (IV) analysis, that exploits the genetics of complex traits to address this type of question. In social sciences and epidemiology, when studying the causal interaction of exposure and outcome, it is often difficult to isolate them from the influence of other external factors that have not been taken into account in the model. To solve this problem, IV analysis utilises so-called instrumental variables that should be (1) associated with the exposure, (2) associated with the outcome only by means of the exposure and (3) not be associated with any confounders, including unknown ones (Figure 1f). If this is the case, it is possible to study the relationship between outcome and exposure, conditioned on the IV. The beauty of this approach is that it allows one to imply causality regardless of the existence of any unknown confounders or reverse causality. An example is an experiment that studied the causal relationship between maternal smoking and birthweight using various state cigarette taxes in the USA as an IV, in which it could be seen that cigarette taxes clearly meet all three conditions and could be used as an IV for this problem [211]. In practice, it is difficult to find IVs that meet all three conditions. In this regard, genetic factors are amazing IVs because they are perfectly random due to panmixia and depend on hardly any external factors. This application of IV analysis is called Mendelian randomisation [212]. For example, the rs671 SNP in the alcohol dehydrogenase gene ALDH2 strongly affects the ability of organisms to metabolise alcohol. This SNP is common in East Asian populations, and for men, it greatly affects their average alcohol consumption. Thus, the genotypes of rs671 could be used as an IV to study the effect of alcohol consumption on, for example, blood pressure and cardiovascular diseases risk, which turned out to be quite high [213,214]. Before GWAS, the application of this experimental design was highly situational because common individual polymorphisms with strong effects like those observed for rs671 are very rare. Since GWAS has become bigger and PRS tended to explain more of the variability in a trait, PRS became more suitable for use as IV, for example, in studies of the relationships between gout, BMI, urate and triglyceride levels [208]; brain structure and depression [215]; telomere length and type II diabetes [216]; AD and various other traits [217]; and blood pressure and various cardiovascular diseases [218]. While there are important statistical problems related to exposure measurement errors [219] and assumption of absence of measurement error in SNP-exposure association [220], the major concern with genetic IV is pleiotropy, which could violate both conditions 2 and 3. However, there are a number of statistical methods aimed to alleviate this problem with GWAS data, like MR-Egger regression [221], CAUSE [222], MR-APSS [223], cML-MA [224] and others.
5. Bench Use of GWAS
5.1. Narrow-Focus Follow-Up Studies
Typically, GWAS results contain a small number of easily interpretable results, with a majority of loci lacking ‘natural’ functional interpretations. Linkage regions of GWAS can be very expansive, contain many genes or contain genes with unknown functions. Often, a GWAS linkage region does not contain genes at all if, for example, the causal variability of a trait is associated with an enhancer for a gene located far from this region. Such situations motivate further research aimed at interpreting specific GWAS signals. For example, the biological implications of a well-known genetic signal in a very large (approximately 6 million base pairs) linkage group in the MHC cluster for schizophrenia, one of the strongest signals associated with GWAS, remained a mystery for a long time. The mystery was solved, at least partially, in the 2016 work by Sekar et al., in which this association signal was linked with the variability in the gene of complement component 4 (C4) responsible for the degree of synaptic pruning during maturation of the brain [225]. In addition, the rs1421085 polymorphism in one of the FTO gene introns resides within the linkage regions for several GWAS (BMI, obesity, type II diabetes, chronotype), and CRISPR/Cas9 editing of the SNP confirmed its role in the regulation of the expression of the neighbouring IRX3 gene and in abnormal development of adipose tissue [226,227].
Defining the biological interpretation of a specific GWAS signal often involves fine-mapping of a supposed causal SNP. Experimental data could be utilised for this purpose. In a 2021 study by Guan et al., eQTL, mQTL and ATAC-seq data were used to fine-map the region of rs164748, which was identified in GWAS for estimated glomerular filtration rate (eGFR). In this study, genome editing in mice was used to demonstrate that there are at least two genes, DPEP1 and CHMP1A, involved in the regulation of ferroptosis and that they surprisingly had the opposite effects on a trait [228]. The study illustrated that a GWAS LD-region could contain multiple causal genetic factors. Fine-mapping could be performed with genetic instruments that have been discussed above, such as trans-ethnic GWAS or pleiotropy analysis, which could be further enriched with experimental data. For example, analysis of genetic associations for rheumatoid arthritis in a trans-ethnic context with chromatin accessibility data allowed researchers to narrow down credible causal SNP sets [229]. An example of a study based on joint analysis of GWAS data is the beautifully designed study on bone fragility, which is known to be a symptom of type II diabetes. Analysis of the GWAS results for both traits, together with epigenetic data, made it possible to determine the common candidate rs56371916 polymorphism in the intron of the ADCY5 gene. ADCY5 expression in CRISPR/Cas9-edited adipocytes and osteoblasts is highly dependent on the rs56371916 alleles, which appear to be responsible for bone fragility in type II diabetes [230].
5.2. Interpretation of Functional Annotations Using GWAS Results
Experimental data are often processed in bioinformatics analysis using various ontology enrichment techniques. This could of course be done with GWAS data if a researcher wants to check whether, for example, ChIP-seq active chromatin peaks are colocalised with significant SNPs for a specific trait. The modern way to do this, as discussed earlier, is to use methods specialised for functional interpretation of GWAS data, like MAGMA or S-LDSC. While all of these methods were created for the interpretation of GWAS results, they could in some circumstances be used in reverse scenarios aimed to answer questions such as which complex trait genetics corresponds to a given functional annotation or which functional annotation better describes a given complex genetic trait.
This type of analysis is natural to apply to single-cell experiments, since it assists in the interpretation of revealed cell classes. For example, in a 2021 study by Sheng et al., on single-cell characterisation over different cell types in kidneys, MAGMA was applied to gene expression data and S-LDSC was applied to the scATAC-seq data [231]. Both of these analyses uncovered enrichment by GWAS results for eGFR in proximal tubules of nephrons, which mirrors the complementary result obtained with enrichment GWAS for eGFR using LDSC-SEG on expression data in proximal tubules of nephrons [232]. Using the S-LDSC approach on brain scATAC-seq data revealed that microglia are the only cell type enriched in AD GWAS results, which is in line with previous research on the disease [233]. In a 2021 study by Kupari et al., MAGMA analysis was utilised to juxtapose neuronal cell types to specific subjective locations of chronic pain [234]. In the work of Baselmans et al., stratified LDSC was used to locate brain regions associated with the genetics of generalised well-being using brain region-specific expression and DNA methylation data [54]. Such expected enrichment does not always occur; for example, in GWAS on BMI, it was shown that most of the identified genetic factors affect the level of gene expression in neuronal tissues, which is an argument for obesity being at least partially a neurological/psychiatric trait [235].
An interesting example of GWAS-informed interpretation of expression data is described in a 2017 paper by Calderon et al., which describes a method (named ‘RolyPoly’) for cell-specific enrichment in GWAS results using scRNA-seq data on a gene level. Specifically, RolyPoly gene scores for AD GWAS in the most relevant cell type (again, microglia) are correlated with test statistics of genes found in an independent differential expression experiment on AD. The result is quite impressive, as it theoretically allows us to obtain the same information from gene expression data from healthy people with AD GWAS results and from much more elaborate experiments involving collection of specific laser-microdissected brains of deceased AD patients to study differential expression [236].
In experimental biology, the causality between a gene and a phenotype could be established in direct experiments with altered and rescued gene expression via, for example, genomic editing. However, the genetic factors of complex traits are too numerous, and their effects are too small for any existing genome editing technology to be practical to use for phenotypes manifested at the level of the whole organism. There have been attempts to utilise the Mendelian randomisation approach to find causal genes linked to a trait of interest using GWAS summary statistics and eQTL data in relevant cell types. In 2019, Porcu et al., described an approach, called ‘Transcriptome-wide summary statistics-based Mendelian Randomisation approach’, TWMR, that allows the identification of potential causal links between gene expression and a trait. The utility of the method is backed by its ability to predict significant GWAS hits retrospectively and to correctly point to known causal links, for example, between SORT1 gene expression and low-density lipoprotein using eQTL data in liver tissues [237]. It should be noted that Mendelian randomisation with GWAS/eQTL data is not straightforward because nothing prevents SNP from bypassing gene expression and influencing a trait of interest through some unknown confounder effects. For causality estimation, it is possible to employ multiple LD-independent loci within the vicinity of a gene, which could be used to distinguish mediation and pleiotropic effects, since in a true causality situation, eQTL and GWAS effects should be correlated in multiple loci. This approach is realised in the MRLocus method [238].
5.3. Use of PRS in Experimental Biology
Biological perspective gives us an intuition that information that is more relevant to the biology of a trait should be a better predictor; hence, use of functional data could improve or even outperform PRS-based predictions. For example, TWAS results provide a direct link between gene expression and a trait, which is tempting to use for diagnostic purposes. In a 2021 study by Pain et al., gene expression risk scores (GeRS), which are based on imputed gene expression from TWAS data, were constructed for a panel of human diseases: rheumatoid arthritis, inflammatory bowel disease, coronary artery disease, type II diabetes, height, BMI, intelligence and depression. In all cases, while GeRS explained a significant portion of heritability, they all performed worse as a trait predictor than the corresponding PRS [239]. This could be explained as follows: the gain in predictive power from aggregation of weak genetic signals into enhanced signals of functional data is outweighed by the loss of predictive power due to pleiotropy, additional non-genetic factors and pure stochasticity.
Pleiotropy seems to drastically affect the performance of methods based on individual SNPs and individual functional effects. On the other hand, polygenic scores, which integrate all genetic effects into a single number, could surprisingly be effective in informing which genes are associated with a specific trait. In 2021, Võsa et al., studied the role of distant expression–genotype interactions (trans-eQTL) in the genetics of complex traits. Notably, trans-eQTL effects are neglected in conventional eQTL-based analyses like TWAS because potential trans-eQTLs are much more abundant than traditional cis-eQTLs and attempts to identify them all individually would inevitably fail due to the dimensionality curse. The authors found that if testing for eQTL effects was limited by SNPs known to be related to a different trait, trans-eQTL effects became apparent and widespread. They further used PRS to construct expression quantitative trait scores (eQTS), in which PRS was used as a mediator for gene expression in a blood dataset for which gene expression and genotype data were simultaneously available. This allows detection of situations in which multiple weak and trans-eQTL effects converge on a single gene. The eQTS method seems to be more appropriate for the highly polygenic nature of complex traits; for example, it proved to be able to correctly detect genes for lipid metabolism using PRS for high-density lipoprotein concentration [240].
The other way to use PRS in experimental biology is to search for rare mutations relevant to a trait. The reasons why GWAS on a common disease generally does not provide suitable candidate targets for medical research include low effects, unclear genomic location due to LD and, potentially, a too-broad mechanism of action due to pleiotropy. In contrast, rare mutations are more straightforward to interpret, more prone to have large and focused effects and can be easily manipulated with conventional genetic methods. The problem is they are, by definition, rare and in general much more difficult to find and even more difficult to identify as a modificator to a trait. Usually, these mutations are found in a family genetic analysis, massive screening of trios for de novo mutations or even more massive exome-wide screenings. At the same time, candidate regions from GWAS proved to be useful as starting points for the search for meaningful rare mutations in targeted sequencing experiments focused on, for example, inflammatory bowel disease [241,242], age-related macular degeneration [243], type II diabetes [244] and rheumatoid arthritis [245,246]. The use of PRS presents a logical development in the search for rare mutations. For example, patients with schizophrenia who have clinically significant CNVs have been shown to have lower PRS [247,248]; in addition, patients with schizophrenia have lower PRS if they carry loss of function and deleterious de novo mutations [249]. In the work of Zhou et al., this hypothesis was simulated and tested on UK Biobank data to reveal that lower PRS quantiles are expected to be enriched in rare variants with large effects [250]. The use of PRS allows one to check for enrichments in rare mutations against not phenotype (which could be rare or difficult to obtain), but the PRS of the phenotype, since a large proportion of causal rare mutations is expected for individuals with low PRS. Of course, the same logic could be applied not only to rare mutations, but also to other factors like somatic mutations and epistatic, epigenetic and environmental factors.
In general, it seems possible to use PRS in experimental biology for sample stratification. Sometimes it is difficult to generate a good cell model for a trait. For example, in psychiatric genetics, it seems that the most relevant cell model for research is a foetal brain of a future psychiatric patient, which is, of course, close to impossible to obtain. Instead, stem cells from any donor could be procured with the goal of creating a relevant cell model, leveraging PRS to study a trait of interest. For example, this logic motivated a 2020 report by Dobrindt et al., on iPSC for several extreme PRS for schizophrenia [251]. In principle, the PRS stratification can be used to generate cell models of any high-level complex trait such as family income and walking pace, which could be impossible to study at the cellular level otherwise.
6. Conclusions
Studies of the genetics of complex traits have now reached maturity, and powerful new instruments have been developed to measure and interpret their results. These results are usually available for any interested researcher in the form of GWAS summary statistics, which allows researchers to relate complex trait genetics to a broad range of biological experiments and unlock new experimental designs in complex traits research. For instance, LDSC and other methods discussed could be used for functional data interpretation, Mendelian randomisation-based methods could help to infer causality and polygenic scores allow the direct consideration of polygenic inheritance. However, we urge avoiding overestimation of non-additive genetic effects or underestimation of pleiotropy. We hope that after reading this review, it will be clearer for experimental biologists how the results of modern genetic research can help them in their work.
Author Contributions
Conceptualisation, writing, original draft: N.V.K.; writing, reviewing and editing: M.V.A., A.K.G., V.E.G.; visualisation: N.V.K.; supervision: V.E.G. All authors have read and agreed to the published version of the manuscript.
Funding
The study was supported by the Russian Science Foundation, grant No. 21-15-00124, https://rscf.ru/project/21-15-00124/ (accessed on 21 September 2021).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; the collection, analysis or interpretation of data; the writing of the manuscript or the decision to publish the results.
Abbreviations
| GWAS | genome-wide association study or GWA study |
| SNP | single-nucleotide polymorphism |
| PRS | polygenic risk score |
| LD | linkage disequilibrium |
| LDSC | LD score regression |
| S-LDSC | stratified LD score regression |
| IV | instrumental variable |
| ROC | receiver operating characteristic |
| AUC | area under the receiver operating characteristic curve |
| eQTL | expression quantitative trait loci |
| TWAS | transcriptome-wide association study |
| eGFR | estimated glomerular filtration rate |
| AD | Alzheimer’s disease |
References
- Sulkava, S.; Ollila, H.; Alasaari, J.; Puttonen, S.; Härmä, M.; Viitasalo, K.; Lahtinen, A.; Lindström, J.; Toivola, A.; Sulkava, R.; et al. Common Genetic Variation Near Melatonin Receptor 1A Gene Linked to Job-Related Exhaustion in Shift Workers. Sleep 2017, 40, zsw011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schormair, B.; Zhao, C.; Bell, S.; Tilch, E.; Salminen, A.V.; Pütz, B.; Dauvilliers, Y.; Stefani, A.; Högl, B.; Poewe, W.; et al. Identification of Novel Risk Loci for Restless Legs Syndrome in Genome-Wide Association Studies in Individuals of European Ancestry: A Meta-Analysis. Lancet Neurol. 2017, 16, 898–907. [Google Scholar] [CrossRef] [Green Version]
- Hill, W.D.; Davies, N.M.; Ritchie, S.J.; Skene, N.G.; Bryois, J.; Bell, S.; Di Angelantonio, E.; Roberts, D.J.; Xueyi, S.; Davies, G.; et al. Genome-Wide Analysis Identifies Molecular Systems and 149 Genetic Loci Associated with Income. Nat. Commun. 2019, 10, 5741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Helgeland, H.; Sodeland, M.; Zoric, N.; Torgersen, J.S.; Grammes, F.; von Lintig, J.; Moen, T.; Kjøglum, S.; Lien, S.; Våge, D.I. Genomic and Functional Gene Studies Suggest a Key Role of Beta-Carotene Oxygenase 1 like (Bco1l) Gene in Salmon Flesh Color. Sci. Rep. 2019, 9, 20061. [Google Scholar] [CrossRef] [Green Version]
- Timmins, I.R.; Zaccardi, F.; Nelson, C.P.; Franks, P.W.; Yates, T.; Dudbridge, F. Genome-Wide Association Study of Self-Reported Walking Pace Suggests Beneficial Effects of Brisk Walking on Health and Survival. Commun. Biol. 2020, 3, 634. [Google Scholar] [CrossRef]
- Visscher, P.M.; Wray, N.R.; Zhang, Q.; Sklar, P.; McCarthy, M.; Brown, A.M.; Yang, J. 10 Years of GWAS Discovery: Biology, Function, and Translation. Am. J. Hum. Genet. 2017, 101, 5–22. [Google Scholar] [CrossRef] [Green Version]
- Claussnitzer, M.; Cho, J.H.; Collins, R.; Cox, N.J.; Dermitzakis, E.T.; Hurles, M.E.; Kathiresan, S.; Kenny, E.E.; Lindgren, C.M.; MacArthur, D.G.; et al. A Brief History of Human Disease Genetics. Nature 2020, 577, 179–189. [Google Scholar] [CrossRef] [Green Version]
- Corder, E.H.; Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.E.; Gaskell, P.C.; Small, G.W.; Roses, A.D.; Haines, J.L.; Pericak-Vance, M.A. Gene Dose of Apolipoprotein E Type 4 Allele and the Risk of Alzheimer's Disease in Late Onset Families. Science 1993, 261, 921–923. [Google Scholar] [CrossRef]
- Saunders, A.M.; Strittmatter, W.J.; Schmechel, D.; George-Hyslop, P.H.S.; Pericakvance, M.A.; Joo, S.H.; Rosi, B.L.; Gusella, J.F.; Crapper-MacLachlan, D.R.; Alberts, M.J.; et al. Association of Apolipoprotein E Allele ϵ4 with Late-Onset Familial and Sporadic Alzheimer’s Disease. Neurology 1993, 43, 1467–1472. [Google Scholar] [CrossRef] [Green Version]
- Hall, J.M.; Lee, M.K.; Newman, B.; Morrow, J.E.; Anderson, L.A.; Huey, B.; King, M.-C. Linkage of Early-Onset Familial Breast Cancer to Chromosome 17q21. Science 1990, 250, 1684–1689. [Google Scholar] [CrossRef] [Green Version]
- Wooster, R.; Neuhausen, S.L.; Mangion, J.; Quirk, Y.; Ford, D.; Collins, N.; Nguyen, K.; Seal, S.; Tran, T.; Averill, D.; et al. Localization of a Breast Cancer Susceptibility Gene, BRCA2, to Chromosome 13q12-13. Science 1994, 265, 2088–2090. [Google Scholar] [CrossRef]
- Risch, N.; Merikangas, K. The Future of Genetic Studies of Complex Human Diseases. Science 1996, 273, 1516–1517. [Google Scholar] [CrossRef] [Green Version]
- Lander, E.S.; Schork, N.J. Genetic Dissection of Complex Traits. Science 1994, 265, 2037–2048. [Google Scholar] [CrossRef] [Green Version]
- Gibbs, R.A.; Belmont, J.W.; Hardenbol, P.; Willis, T.D.; Yu, F.; Yang, H.; Ch’ang, L.-Y.; Huang, W.; Liu, B.; Shen, Y.; et al. The International HapMap Project. Nature 2003, 426, 789–796. [Google Scholar] [CrossRef] [Green Version]
- Ozaki, K.; Ohnishi, Y.; Iida, A.; Sekine, A.; Yamada, R.; Tsunoda, T.; Sato, H.; Sato, H.; Hori, M.; Nakamura, Y.; et al. Functional SNPs in the Lymphotoxin-A Gene that are Associated with Susceptibility to Myocardial Infarction. Nat. Genet. 2002, 32, 650–654. [Google Scholar] [CrossRef]
- Kennedy, G.C.; Matsuzaki, H.; Dong, S.; Liu, W.-M.; Huang, J.; Liu, G.; Su, X.; Cao, M.; Chen, W.; Zhang, J.; et al. Large-Scale Genotyping of Complex DNA. Nat. Biotechnol. 2003, 21, 1233–1237. [Google Scholar] [CrossRef]
- Manolio, T.A.; Collins, F.S. The HapMap and Genome-Wide Association Studies in Diagnosis and Therapy. Annu. Rev. Med. 2009, 60, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Plagnol, V.; Hu, T.T.; Toomajian, C.; Clark, R.M.; Ossowski, S.; Ecker, J.; Weigel, D.; Nordborg, M. Recombination and Linkage Disequilibrium in Arabidopsis thaliana. Nat. Genet. 2007, 39, 1151–1155. [Google Scholar] [CrossRef]
- Yang, H.; Ding, Y.; Hutchins, L.N.; Szatkiewicz, J.; Bell, A.T.; Paigen, B.J.; Graber, J.; De Villena, F.P.-M.; Churchill, G.A. A Customized and Versatile High-Density Genotyping Array for the Mouse. Nat. Methods 2009, 6, 663–666. [Google Scholar] [CrossRef]
- Barson, N.J.; Aykanat, T.; Hindar, K.; Baranski, M.; Bolstad, G.H.; Fiske, P.; Jacq, C.; Jensen, A.J.; Johnston, S.; Karlsson, S.; et al. Sex-Dependent Dominance at a Single Locus Maintains Variation in Age at Maturity in Salmon. Nature 2015, 528, 405–408. [Google Scholar] [CrossRef]
- Peter, J.; De Chiara, M.; Friedrich, A.; Yue, J.-X.; Pflieger, D.; Bergström, A.; Sigwalt, A.; Barre, B.; Freel, K.; Llored, A.; et al. Genome Evolution across 1011 Saccharomyces cerevisiae isolates. Nature 2018, 556, 339–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, A.; Sterken, M.G.; De Bos, J.U.; Van Creij, J.; Kamble, R.; Snoek, B.; Kammenga, J.E.; Houtkooper, R.H. Natural Genetic Variation in C. Elegansidentified Genomic Loci Controlling Metabolite Levels. Genome Res. 2018, 28, 1296–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Long, T.; Hicks, M.; Yu, H.-C.; Biggs, W.H.; Kirkness, E.F.; Menni, C.; Zierer, J.; Small, K.S.; Mangino, M.; Messier, H.; et al. Whole-Genome Sequencing Identifies Common-to-Rare Variants Associated with Human Blood Metabolites. Nat. Genet. 2017, 49, 568–578. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.H.; Weng, L.-C.; Roselli, C.; Lin, H.; Haggerty, C.; Shoemaker, M.B.; Barnard, J.; Arking, D.E.; Chasman, D.I.; Albert, C.; et al. Association Between Titin Loss-of-Function Variants and Early-Onset Atrial Fibrillation. JAMA 2018, 320, 2354–2364. [Google Scholar] [CrossRef]
- Chia, R.; Center, T.A.G.; Sabir, M.S.; Bandres-Ciga, S.; Saez-Atienzar, S.; Reynolds, R.H.; Gustavsson, E.; Walton, R.L.; Ahmed, S.; Viollet, C.; et al. Genome Sequencing Analysis Identifies New Loci Associated with Lewy Body Dementia and Provides Insights into its Genetic Architecture. Nat. Genet. 2021, 53, 294–303. [Google Scholar] [CrossRef]
- Zhang, M.-Y.; Xue, C.; Hu, H.; Li, J.; Xue, Y.; Wang, R.; Fan, J.; Zou, C.; Tao, S.; Qin, M.; et al. Genome-Wide Association Studies Provide Insights into the Genetic Determination of Fruit Traits of Pear. Nat. Commun. 2021, 12, 1144. [Google Scholar] [CrossRef]
- Li, J.H.; Mazur, C.A.; Berisa, T.; Pickrell, J.K. Low-Pass Sequencing Increases the Power of GWAS and Decreases Measurement Error of Polygenic Risk Scores Compared to Genotyping Arrays. Genome Res. 2021, 31, 529–537. [Google Scholar] [CrossRef]
- Martin, A.R.; Atkinson, E.G.; Chapman, S.B.; Stevenson, A.; Stroud, R.E.; Abebe, T.; Akena, D.; Alemayehu, M.; Ashaba, F.K.; Atwoli, L.; et al. Low-Coverage Sequencing Cost-Effectively Detects Known and Novel Variation in Underrepresented Populations. Am. J. Hum. Genet. 2021, 108, 656–668. [Google Scholar] [CrossRef]
- Klein, R.J.; Zeiss, C.; Chew, E.Y.; Tsai, J.-Y.; Sackler, R.S.; Haynes, C.; Henning, A.K.; SanGiovanni, J.P.; Mane, S.M.; Mayne, S.T.; et al. Complement Factor H Polymorphism in Age-Related Macular Degeneration. Science 2005, 308, 385–389. [Google Scholar] [CrossRef]
- Allen, H.L.; Estrada, K.; Lettre, G.; Berndt, S.I.; Weedon, M.N.; Rivadeneira, F.; Willer, C.J.; Jackson, A.U.; Vedantam, S.; Raychaudhuri, S.; et al. Hundreds of Variants Clustered in Genomic Loci and Biological Pathways Affect Human Height. Nature 2010, 467, 832–838. [Google Scholar] [CrossRef] [Green Version]
- Corvin, A.; Sullivan, P.F. What Next in Schizophrenia Genetics for the Psychiatric Genomics Consortium? Schizophr. Bull. 2016, 42, 538–541. [Google Scholar] [CrossRef] [Green Version]
- Mahajan, A.; Go, M.J.; Zhang, W.; Below, E.J.; Gaulton, K.J.; Ferreira, T.; Horikoshi, M.; Johnson, A.D.; Ng, M.C.Y.; Prokopenko, I.; et al. Genome-Wide Trans-Ancestry Meta-Analysis Provides Insight into the Genetic Architecture of Type 2 Diabetes Susceptibility. Nat. Genet. 2014, 46, 234–244. [Google Scholar] [CrossRef]
- The International Parkinson Disease Genomics Consortium (IPDGC). Ten Years of the International Parkinson Disease Genomics Consortium: Progress and Next Steps. J. Park. Dis. 2020, 10, 19–30. [Google Scholar] [CrossRef] [Green Version]
- Evangelou, E.; Program, T.M.V.; Warren, H.R.; Mosen-Ansorena, D.; Mifsud, B.; Pazoki, R.; Gao, H.; Ntritsos, G.; Dimou, N.; Cabrera, C.P.; et al. Genetic Analysis of over 1 Million People Identifies 535 New Loci Associated with Blood Pressure Traits. Nat. Genet. 2018, 50, 1412–1425. [Google Scholar] [CrossRef] [Green Version]
- Klarin, D.; Damrauer, S.; Cho, K.; Sun, Y.V.; Teslovich, T.M.; Honerlaw, J.; Gagnon, D.R.; Duvall, S.L.; Li, J.; Peloso, G.M.; et al. Genetics of Blood Lipids among ~300,000 Multi-Ethnic Participants of the Million Veteran Program. Nat. Genet. 2018, 50, 1514–1523. [Google Scholar] [CrossRef]
- Chen, V.L.; Du, X.; Chen, Y.; Kuppa, A.; Handelman, S.K.; Vohnoutka, R.B.; Peyser, P.A.; Palmer, N.D.; Bielak, L.F.; Halligan, B.; et al. Genome-Wide Association Study of Serum Liver Enzymes Implicates Diverse Metabolic and Liver Pathology. Nat. Commun. 2021, 12, 816. [Google Scholar] [CrossRef]
- Ferreira, M.A.; Collaborators, E.; Gamazon, E.R.; Al-Ejeh, F.; Aittomäki, K.; Andrulis, I.L.; Anton-Culver, H.; Arason, A.; Arndt, V.; Aronson, K.J.; et al. Genome-Wide Association and Transcriptome Studies Identify Target Genes and Risk Loci for Breast Cancer. Nat. Commun. 2019, 10, 1741. [Google Scholar] [CrossRef] [Green Version]
- Wuttke, M.; Li, Y.; Li, M.; Sieber, K.B.; Feitosa, M.F.; Gorski, M.; Tin, A.; Wang, L.; Chu, A.Y.; Hoppmann, A.; et al. A Catalog of Genetic Loci associated with Kidney Function from Analyses of a Million Individuals. Nat. Genet. 2019, 51, 957–972. [Google Scholar] [CrossRef] [Green Version]
- Morris, J.A.; Kemp, J.P.; Youlten, S.E.; Laurent, L.; Logan, J.G.; Chai, R.C.; Vulpescu, N.A.; Forgetta, V.; Kleinman, A.; Mohanty, S.T.; et al. An Atlas of Genetic Influences on Osteoporosis in Humans and Mice. Nat. Genet. 2019, 51, 258–266. [Google Scholar] [CrossRef]
- Foo, J.N.; Chew, E.G.Y.; Chung, S.J.; Peng, R.; Blauwendraat, C.; Nalls, M.A.; Mok, K.Y.; Satake, W.; Toda, T.; Chao, Y.; et al. Identification of Risk Loci for Parkinson Disease in Asians and Comparison of Risk Between Asians and Europeans. JAMA Neurol. 2020, 77, 746–754. [Google Scholar] [CrossRef]
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 New Risk Loci for Type 2 Diabetes and Related Vascular Outcomes among 1.4 Million Participants in a Multi-Ancestry Meta-Analysis. Nat. Genet. 2020, 52, 680–691. [Google Scholar] [CrossRef]
- Choquet, H.; Melles, R.B.; Anand, D.; Yin, J.; Cuellar-Partida, G.; Wang, W.; Hoffmann, T.J.; Nair, K.S.; Hysi, P.G.; Lachke, S.A.; et al. A Large Multiethnic GWAS Meta-Analysis of Cataract Identifies New Risk Loci and Sex-Specific Effects. Nat. Commun. 2021, 12, 3595. [Google Scholar] [CrossRef]
- Shungin, D.; Haworth, S.; Divaris, K.; Agler, C.; Kamatani, Y.; Lee, M.K.; Grinde, K.; Hindy, G.; Alaraudanjoki, V.; Pesonen, P.; et al. Genome-Wide Analysis of Dental Caries and Periodontitis Combining Clinical and Self-Reported Data. Nat. Commun. 2019, 10, 2773. [Google Scholar] [CrossRef] [Green Version]
- Yengo, L.; Sidorenko, J.; Kemper, E.K.; Zheng, Z.; Wood, A.R.; Weedon, M.; Frayling, T.; Hirschhorn, J.; Yang, J.; Visscher, P.M.; et al. Meta-Analysis of Genome-Wide Association Studies for Height and Body Mass Index In ∼700,000 Individuals of European Ancestry. Hum. Mol. Genet. 2018, 27, 3641–3649. [Google Scholar] [CrossRef]
- Wright, K.M.; Rand, K.A.; Kermany, A.; Noto, K.; Curtis, D.; Garrigan, D.; Slinkov, D.; Dorfman, I.; Granka, J.M.; Byrnes, J.; et al. A Prospective Analysis of Genetic Variants Associated with Human Lifespan. G3 Genes Genomes Genet. 2019, 9, 2863–2878. [Google Scholar] [CrossRef] [Green Version]
- Cuellar-Partida, G.; Tung, J.Y.; Eriksson, N.; Albrecht, E.; Aliev, F.; Andreassen, O.A.; Barroso, I.; Beckmann, J.S.; Boks, M.P.; Boomsma, D.I.; et al. Genome-Wide Association Study Identifies 48 Common Genetic Variants Associated with Handedness. Nat. Hum. Behav. 2021, 5, 59–70. [Google Scholar] [CrossRef]
- Pulit, S.L.; Stoneman, C.; Morris, A.P.; Wood, A.R.; Glastonbury, C.A.; Tyrrell, J.; Yengo, L.; Ferreira, T.; Marouli, E.; Ji, Y.; et al. Meta-Analysis of Genome-Wide Association Studies for Body Fat Distribution in 694 649 Individuals of European Ancestry. Hum. Mol. Genet. 2019, 28, 166–174. [Google Scholar] [CrossRef] [Green Version]
- Kranzler, H.R.; Zhou, H.; Kember, R.L.; Smith, R.V.; Justice, A.C.; Damrauer, S.; Tsao, P.S.; Klarin, D.; Baras, A.; Reid, J.; et al. Genome-Wide Association Study of Alcohol Consumption and Use Disorder in 274,424 Individuals from Multiple Populations. Nat. Commun. 2019, 10, 1499. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Jiang, Y.; Wedow, R.; Li, Y.; Brazel, D.M.; Chen, F.; Datta, G.; Davila-Velderrain, J.; McGuire, D.; Tian, C.; et al. Association Studies of up to 1.2 Million Individuals Yield New Insights into the Genetic Etiology of Tobacco and Alcohol Use. Nat. Genet. 2019, 51, 237–244. [Google Scholar] [CrossRef]
- Jones, S.E.; Lane, J.M.; Wood, A.R.; Van Hees, V.T.; Tyrrell, J.; Beaumont, R.N.; Jeffries, A.R.; Dashti, H.S.; Hillsdon, M.; Ruth, K.S.; et al. Genome-Wide Association Analyses of Chronotype in 697,828 Individuals Provides Insights into Circadian Rhythms. Nat. Commun. 2019, 10, 343. [Google Scholar] [CrossRef] [Green Version]
- Howard, D.M.; Adams, M.J.; Clarke, T.-K.; Hafferty, J.D.; Gibson, J.; Shirali, M.; Coleman, J.R.I.; Hagenaars, S.P.; Ward, J.; Wigmore, E.M.; et al. Genome-Wide Meta-Analysis of Depression Identifies 102 Independent Variants and Highlights the Importance of the Prefrontal Brain Regions. Nat. Neurosci. 2019, 22, 343–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linnér, R.K.; Biroli, P.; Kong, E.; Meddens, S.F.W.; Wedow, R.; Fontana, M.A.; Lebreton, M.; Tino, S.P.; Abdellaoui, A.; Hammerschlag, A.R.; et al. Genome-Wide Association Analyses of Risk Tolerance and Risky Behaviors in over 1 Million Individuals Identify Hundreds of Loci and Shared Genetic Influences. Nat. Genet. 2019, 51, 245–257. [Google Scholar] [CrossRef] [PubMed]
- Savage, J.E.; Jansen, P.R.; Stringer, S.; Watanabe, K.; Bryois, J.; de Leeuw, C.; Nagel, M.; Awasthi, S.; Barr, P.B.; Coleman, J.R.I.; et al. Genome-Wide Association Meta-Analysis in 269,867 Individuals Identifies New Genetic and Functional Links to Intelligence. Nat. Genet. 2018, 50, 912–919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baselmans, B.M.L.; Jansen, R.; Ip, H.F.; Van Dongen, J.; Abdellaoui, A.; van de Weijer, M.; Bao, Y.; Smart, M.; Kumari, M.; Willemsen, G.; et al. Multivariate Genome-Wide Analyses of the Well-Being Spectrum. Nat. Genet. 2019, 51, 445–451. [Google Scholar] [CrossRef]
- Lo, M.-T.; Hinds, A.D.; Tung, J.Y.; Franz, C.; Fan, C.-C.; Wang, Y.; Smeland, O.B.; Schork, A.; Holland, D.; Kauppi, K.; et al. Genome-Wide Analyses for Personality Traits Identify Six Genomic Loci and Show Correlations with Psychiatric Disorders. Nat. Genet. 2017, 49, 152–156. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.J.; Wedow, R.; Okbay, A.; Kong, E.; Maghzian, O.; Zacher, M.; Nguyen-Viet, T.A.; Bowers, P.; Sidorenko, J.; Linnér, R.K.; et al. Gene Discovery and Polygenic Prediction from a Genome-Wide Association Study of Educational Attainment in 1.1 Million Individuals. Nat. Genet. 2018, 50, 1112–1121. [Google Scholar] [CrossRef] [Green Version]
- Karlsson Linnér, R.; Mallard, T.T.; Barr, P.B.; Sanchez-Roige, S.; Madole, J.W.; Driver, M.N.; Poore, H.E.; de Vlaming, R.; Grotzinger, A.D.; Tielbeek, J.J.; et al. Multivariate Analysis of 1.5 Million People Identifies Genetic Associations with Traits Related to Self-Regulation and Addiction. Nat. Neurosci. 2021, 24, 1367–1376. [Google Scholar] [CrossRef]
- Atwell, S.; Huang, Y.S.; Vilhjalmsson, B.; Willems, G.; Horton, M.; Li, Y.; Meng, D.; Platt, A.; Tarone, A.; Hu, T.T.; et al. Genome-wide Association Study of 107 Phenotypes in Arabidopsis Thaliana Inbred Lines. Nature 2010, 465, 627–631. [Google Scholar] [CrossRef]
- Alonso-Blanco, C.; Andrade, J.; Becker, C.; Bemm, F.; Bergelson, J.; Borgwardt, K.M.; Cao, J.; Chae, E.; Dezwaan, T.M.; Ding, W.; et al. 1,135 Genomes Reveal the Global Pattern of Polymorphism in Arabidopsis thaliana. Cell 2016, 166, 481–491. [Google Scholar] [CrossRef] [Green Version]
- Morris, G.P.; Ramu, P.; Deshpande, S.P.; Hash, C.T.; Shah, T.; Upadhyaya, H.D.; Riera-Lizarazu, O.; Brown, P.J.; Acharya, C.B.; Mitchell, S.E.; et al. Population Genomic and Genome-Wide Association Studies of Agroclimatic Traits in Sorghum. Proc. Natl. Acad. Sci. USA 2013, 110, 453–458. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.; Massouras, A.; Inoue, Y.; Peiffer, J.; Ràmia, M.; Tarone, A.; Turlapati, L.; Zichner, T.; Zhu, D.; Lyman, R.F.; et al. Natural Variation in Genome Architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 2014, 24, 1193–1208. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Mauleon, R.; Hu, Z.; Chebotarov, D.; Tai, S.; Wu, Z.; Li, M.; Zheng, T.; Fuentes, R.R.; Zhang, F.; et al. Genomic Variation in 3010 Diverse Accessions of Asian Cultivated Rice. Nature 2018, 557, 43–49. [Google Scholar] [CrossRef]
- Bloom, J.S.; Kotenko, I.; Sadhu, M.; Treusch, S.; Albert, F.; Kruglyak, L. Genetic Interactions Contribute Less than Additive Effects to Quantitative Trait Variation in Yeast. Nat. Commun. 2015, 6, 8712. [Google Scholar] [CrossRef] [Green Version]
- Burke, D.T.; Kozloff, K.M.; Chen, S.; West, J.L.; Wilkowski, J.M.; Goldstein, S.A.; Miller, R.A.; Galecki, A.T. Dissection of Complex Adult Traits in a Mouse Synthetic Population. Genome Res. 2012, 22, 1549–1557. [Google Scholar] [CrossRef] [Green Version]
- Zhong, X.; Wang, X.; Zhou, T.; Jin, Y.; Tan, S.; Jiang, C.; Geng, X.; Li, N.; Shi, H.; Zeng, Q.; et al. Genome-Wide Association Study Reveals Multiple Novel QTL Associated with Low Oxygen Tolerance in Hybrid Catfish. Mar. Biotechnol. 2017, 19, 379–390. [Google Scholar] [CrossRef]
- Thelwall, M.; Munafo, M.; Mas-Bleda, A.; Stuart, E.; Makita, M.; Weigert, V.; Keene, C.; Khan, N.; Drax, K.; Kousha, K. Is Useful Research Data Usually shared? An Investigation of Genome-Wide Association Study Summary Statistics. PLoS ONE 2020, 15, e0229578. [Google Scholar] [CrossRef] [Green Version]
- Buniello, A. Why We Need More Freely Available Cancer GWAS Summary Statistics. Available online: https://blog.opentargets.org/open-sharing-of-cancer-summary-statistics/ (accessed on 16 July 2021).
- Buniello, A.; MacArthur, J.A.L.; Cerezo, M.; Harris, L.W.; Hayhurst, J.; Malangone, C.; McMahon, A.; Morales, J.; Mountjoy, E.; Sollis, E.; et al. The NHGRI-EBI GWAS Catalog of Published Genome-Wide Association Studies, Targeted Arrays and Summary Statistics 2019. Nucleic Acids Res. 2019, 47, D1005–D1012. [Google Scholar] [CrossRef] [Green Version]
- Togninalli, M.; Seren, A.; Freudenthal, A.J.; Monroe, J.G.; Meng, D.; Nordborg, M.; Weigel, D.; Borgwardt, K.M.; Korte, A.; Grimm, D.G. AraPheno and the AraGWAS Catalog 2020: A Major Database Update Including RNA-Seq and Knockout Mutation Data for Arabidopsis thaliana. Nucleic Acids Res. 2020, 48, D1063–D1068. [Google Scholar] [CrossRef]
- Collins, A.L.; Kim, Y.; Sklar, P.; O'Donovan, M.; Sullivan, P.F. International Schizophrenia Consortium Hypothesis-Driven Candidate Genes for Schizophrenia Compared to Genome-Wide Association Results. Psychol. Med. 2012, 42, 607–616. [Google Scholar] [CrossRef]
- Sullivan, P.F.; Geschwind, D.H. Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell 2019, 177, 162–183. [Google Scholar] [CrossRef] [Green Version]
- Willoughby, E.A.; Love, A.; McGue, M.; Iacono, W.G.; Quigley, J.; Lee, J.J. Free Will, Determinism, and Intuitive Judgments About the Heritability of Behavior. Behav. Genet. 2019, 49, 136–153. [Google Scholar] [CrossRef] [Green Version]
- Kruuk, L.E.B.; Clutton-Brock, T.H.; Slate, J.; Pemberton, J.M.; Brotherstone, S.; Guinness, F.E. Heritability of Fitness in a Wild Mammal Population. Proc. Natl. Acad. Sci. USA 2000, 97, 698–703. [Google Scholar] [CrossRef] [Green Version]
- Vink, J.M.; Willemsen, G.; Boomsma, D.I. Heritability of Smoking Initiation and Nicotine Dependence. Behav. Genet. 2005, 35, 397–406. [Google Scholar] [CrossRef]
- Byrne, B.; Coventry, W.L.; Olson, R.K.; Samuelsson, S.; Corley, R.; Willcutt, E.G.; Wadsworth, S.; DeFries, J.C. Genetic and Environmental Influences on Aspects of Literacy and Language in Early Childhood: Continuity and Change from Preschool to Grade 2. J. Neurolinguist. 2009, 22, 219–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellinghaus, D.; Degenhardt, F.; Bujanda, L.; Buti, M.; Albillos, A.; Invernizzi, P.; Fernandez, J.; Prati, D.; Baselli, G.; Asselta, R.; et al. Genomewide Association Study of Severe Covid-19 with Respiratory Failure. N. Engl. J. Med. 2020, 383, 1522–1534. [Google Scholar] [CrossRef]
- Pairo-Castineira, E.; Clohisey, S.; Klaric, L.; Bretherick, A.D.; Rawlik, K.; Pasko, D.; Walker, S.; Parkinson, N.; Fourman, M.H.; Russell, C.D.; et al. Genetic Mechanisms of Critical Illness in COVID-19. Nature 2020, 591, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Ganna, A.; COVID-19 Host Genetics Initiative. Mapping the human genetic architecture of COVID-19. Nature 2021, 1626. [Google Scholar] [CrossRef]
- Wasser, S.K.; Clark, W.J.; Drori, O.; Kisamo, E.S.; Mailand, C.; Mutayoba, B.; Stephens, M. Combating the Illegal Trade in African Elephant Ivory with DNA Forensics. Conserv. Biol. 2008, 22, 1065–1071. [Google Scholar] [CrossRef]
- Novembre, J.; Johnson, T.; Bryc, K.; Kutalik, Z.; Boyko, A.R.; Auton, A.; Indap, A.; King, K.S.; Bergmann, S.; Nelson, M.R.; et al. Genes Mirror Geography within Europe. Nature 2008, 456, 98–101. [Google Scholar] [CrossRef] [Green Version]
- The 1000 Genomes Project Consortium; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, S.; Das, S.; Kretzschmar, W.; Delaneau, O.; Wood, A.R.; Teumer, A.; Kang, H.M.; Fuchsberger, C.; Danecek, P.; Sharp, K.; et al. A Reference Panel of 64,976 Haplotypes for Genotype Imputation. Nat. Genet. 2016, 48, 1279–1283. [Google Scholar] [CrossRef] [Green Version]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price1, A.L.; Neale, B.M. LD Score Regression Distinguishes Confounding from Polygenicity in Genome-Wide Association Studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Gazal, S.; Loh, P.-R.; Finucane, H.K.; Ganna, A.; Schoech, A.; Sunyaev, S.; Price, A.L. Functional Architecture of Low-Frequency Variants Highlights Strength of Negative Selection across Coding and Non-Coding Annotations. Nat. Genet. 2018, 50, 1600–1607. [Google Scholar] [CrossRef]
- O'Connor, L.J.; Schoech, A.P.; Hormozdiari, F.; Gazal, S.; Patterson, N.; Price, A.L. Extreme Polygenicity of Complex Traits Is Explained by Negative Selection. Am. J. Hum. Genet. 2019, 105, 456–476. [Google Scholar] [CrossRef] [Green Version]
- Bloom, J.S.; Boocock, J.; Treusch, S.; Sadhu, M.J.; Day, L.; Oates-Barker, H.; Kruglyak, L. Rare Variants Contribute Disproportionately to Quantitative Trait Variation in Yeast. eLife 2019, 8, e49212. [Google Scholar] [CrossRef]
- Weiner, D.J.; Wigdor, E.M.; Ripke, S.; Walters, R.K.; Kosmicki, J.A.; Grove, J.; Samocha, K.E.; Goldstein, J.I.; Okbay, A.; Bybjerg-Grauholm, J.; et al. Polygenic Transmission Disequilibrium Confirms that Common and Rare Variation Act Additively to Create Risk for Autism Spectrum Disorders. Nat. Genet. 2017, 49, 978–985. [Google Scholar] [CrossRef] [Green Version]
- Akbari, P.; Gilani, A.; Sosina, O.; Kosmicki, J.A.; Khrimian, L.; Fang, Y.-Y.; Persaud, T.; Garcia, V.; Sun, D.; Li, A.; et al. Sequencing of 640,000 Exomes identifies GPR75 Variants Associated with Protection from Obesity. Science 2021, 373, eabf8683. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-Wide Polygenic Scores for Common Diseases Identify Individuals with Risk Equivalent to Monogenic Mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef]
- Sebastiani, P.; Solovieff, N.; Hartley, S.W.; Milton, J.N.; Riva, A.; Dworkis, D.A.; Melista, E.; Klings, E.; Garrett, M.E.; Telen, M.J.; et al. Genetic Modifiers of the Severity of Sickle Cell Anemia Identified through a Genome-Wide Association Study. Am. J. Hematol. 2010, 85, 29–35. [Google Scholar] [CrossRef] [Green Version]
- Wright, F.A.; Strug, L.J.; Doshi, V.K.; Commander, C.; Blackman, S.; Sun, L.; Berthiaume, Y.; Cutler, D.M.; Cojocaru, A.; Collaco, J.M.; et al. Genome-Wide Association and Linkage Identify Modifier Loci of Lung Disease Severity in Cystic Fibrosis at 11p13 and 20q13.2. Nat. Genet. 2011, 43, 539–546. [Google Scholar] [CrossRef]
- Navarini, A.A.; Simpson, M.A.; Weale, M.; Knight, J.; Carlavan, I.; Reiniche, P.; Burden, D.A.; Layton, A.; Bataille, V.; Allen, M.; et al. Genome-wide association study identifies three novel susceptibility loci for severe Acne vulgaris. Nat. Commun. 2014, 5, 4020. [Google Scholar] [CrossRef] [Green Version]
- Moss, D.J.H.; Pardiñas, A.F.; Langbehn, D.; Lo, K.; Leavitt, B.R.; Roos, R.; Durr, A.; Mead, S.; Holmans, P.; Jones, L.; et al. Identification of Genetic Variants Associated with Huntington’s Disease Progression: A Genome-Wide Association Study. Lancet Neurol. 2017, 16, 701–711. [Google Scholar] [CrossRef]
- Wei, W.; Hemani, G.; Haley, C. Detecting Epistasis in Human Complex Traits. Nat. Rev. Genet. 2014, 15, 722–733. [Google Scholar] [CrossRef]
- Chatelain, C.; Durand, G.; Thuillier, V.; Auge, F. Performance of Epistasis Detection Methods in Semi-Simulated GWAS. BMC Bioinform. 2018, 19, 231. [Google Scholar] [CrossRef]
- Fisher Sir, R.A.; Fisher, R.A. The Genetical Theory of Natural Selection: A Complete Variorum Edition; OUP: Oxford, UK, 1999; ISBN 9780198504405. [Google Scholar]
- Crow, J.F. On Epistasis: Why it is Unimportant in Polygenic Directional Selection. Philos. Trans. R. Soc. B Biol. Sci. 2010, 365, 1241–1244. [Google Scholar] [CrossRef] [Green Version]
- Pettersson, M.; Besnier, F.; Siegel, P.B.; Carlborg, O. Replication and Explorations of High-Order Epistasis Using a Large Advanced Intercross Line Pedigree. PLoS Genet. 2011, 7, e1002180. [Google Scholar] [CrossRef] [Green Version]
- Hayward, J.J.; Castelhano, M.; Oliveira, K.C.; Corey, E.; Balkman, C.; Baxter, T.L.; Casal, M.L.; Center, S.A.; Fang, M.; Garrison, S.J.; et al. Complex Disease and Phenotype Mapping in the Domestic Dog. Nat. Commun. 2016, 7, 10460. [Google Scholar] [CrossRef]
- Gusareva, E.S.; Carrasquillo, M.M.; Bellenguez, C.; Cuyvers, E.; Colon, S.; Graff-Radford, N.R.; Petersen, R.C.; Dickson, D.W.; John, J.M.M.; Bessonov, K.; et al. Genome-Wide Association Interaction Analysis for Alzheimer's Disease. Neurobiol. Aging 2014, 35, 2436–2443. [Google Scholar] [CrossRef] [Green Version]
- Sinnott-Armstrong, N.; Naqvi, S.; Rivas, M.; Pritchard, J.K. GWAS of Three Molecular Traits Highlights Core Genes and Pathways Alongside a Highly Polygenic Background. eLife 2021, 10, e58615. [Google Scholar] [CrossRef]
- Fournier, T.; Saada, O.A.; Hou, J.; Peter, J.; Caudal, E.; Schacherer, J. Extensive Impact of Low-Frequency Variants on the Phenotypic Landscape at Population-Scale. eLife 2019, 8, e49258. [Google Scholar] [CrossRef]
- Mäki-Tanila, A.; Hill, W.G. Influence of Gene Interaction on Complex Trait Variation with Multilocus Models. Genetics 2014, 198, 355–367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marigorta, U.M.; Rodríguez, J.A.; Gibson, G.; Navarro, A. Replicability and Prediction: Lessons and Challenges from GWAS. Trends Genet. 2018, 34, 504–517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Sullivan, J.W.; Ioannidis, J.P.A. Reproducibility in the UK Biobank of Genome-Wide Significant Signals Discovered in Earlier Genome-Wide Association Studies. Sci. Rep. 2021, 11, 18625. [Google Scholar] [CrossRef] [PubMed]
- Marigorta, U.M.; Navarro, A. High Trans-Ethnic Replicability of GWAS Results Implies Common Causal Variants. PLoS Genet. 2013, 9, e1003566. [Google Scholar] [CrossRef] [Green Version]
- N'Diaye, A.; Chen, G.K.; Palmer, C.D.; Ge, B.; Tayo, B.; Mathias, R.A.; Ding, J.; Nalls, M.A.; Adeyemo, A.; Adoue, V.; et al. Identification, Replication, and Fine-Mapping of Loci Associated with Adult Height in Individuals of African Ancestry. PLoS Genet. 2011, 7, e1002298. [Google Scholar] [CrossRef]
- Coram, M.; Duan, Q.; Hoffmann, T.J.; Thornton, T.; Knowles, J.W.; Johnson, N.A.; Ochs-Balcom, H.M.; Donlon, T.A.; Martin, L.W.; Eaton, C.; et al. Genome-wide Characterization of Shared and Distinct Genetic Components that Influence Blood Lipid Levels in Ethnically Diverse Human Populations. Am. J. Hum. Genet. 2013, 92, 904–916. [Google Scholar] [CrossRef] [Green Version]
- Adeyemo, A.; Tekola-Ayele, F.; Doumatey, A.P.; Bentley, A.R.; Chen, G.; Huang, H.; Zhou, J.; Shriner, D.; Fasanmade, O.; Okafor, G.; et al. Evaluation of Genome Wide Association Study Associated Type 2 Diabetes Susceptibility Loci in Sub Saharan Africans. Front. Genet. 2015, 6, 335. [Google Scholar] [CrossRef] [Green Version]
- Spracklen, C.N.; Horikoshi, M.; Kim, Y.J.; Lin, K.; Bragg, F.; Moon, S.; Suzuki, K.; Tam, C.H.T.; Tabara, Y.; Kwak, S.-H.; et al. Identification of Type 2 Diabetes Loci in 433,540 East Asian Individuals. Nat. Cell Biol. 2020, 582, 240–245. [Google Scholar] [CrossRef]
- Tedja, M.S.; Wojciechowski, R.; Hysi, P.G.; Eriksson, N.; Furlotte, N.A.; Verhoeven, V.J.M.; Iglesias, A.I.; Meester-Smoor, M.A.; Tompson, S.W.; Fan, Q.; et al. Genome-Wide Association Meta-Analysis Highlights Light-Induced Signaling as a Driver for Refractive Error. Nat. Genet. 2018, 50, 834–848. [Google Scholar] [CrossRef]
- Lam, M.; Chen, C.-Y.; Li, Z.; Martin, A.R.; Bryois, J.; Ma, X.; Gaspar, H.; Ikeda, M.; Benyamin, B.; Brown, B.C.; et al. Comparative genetic architectures of schizophrenia in East Asian and European populations. Nat. Genet. 2019, 51, 1670–1678. [Google Scholar] [CrossRef]
- Liu, J.Z.; Van Sommeren, S.; Huang, H.; Ng, S.C.; Alberts, R.; Takahashi, A.; Ripke, S.; Lee, J.; Jostins, L.; Shah, T.; et al. Association Analyses Identify 38 Susceptibility Loci for Inflammatory Bowel Disease and Highlight Shared Genetic Risk across Populations. Nat. Genet. 2015, 47, 979–986. [Google Scholar] [CrossRef]
- Li, Y.R.; Keating, B.J. Trans-Ethnic Genome-Wide Association Studies: Advantages and Challenges of Mapping in Diverse Populations. Genome Med. 2014, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Schaid, D.J.; Chen, W.; Larson, N.B. From Genome-Wide Associations to Candidate Causal Variants by Statistical Fine-Mapping. Nat. Rev. Genet. 2018, 19, 491–504. [Google Scholar] [CrossRef]
- Wood, A.R.; The Electronic Medical Records and Genomics (eMERGE) Consortium; Esko, T.; Yang, J.; Vedantam, S.; Pers, T.H.; Gustafsson, S.; Chu, A.Y.; Estrada, K.; Luan, J.; et al. Defining the Role of Common Variation in the Genomic and Biological Architecture of Adult Human Height. Nat. Genet. 2014, 46, 1173–1186. [Google Scholar] [CrossRef] [Green Version]
- Bouwman, A.C.; Daetwyler, H.D.; Chamberlain, A.J.; Ponce, C.H.; Sargolzaei, M.; Schenkel, F.S.; Sahana, G.; Govignon-Gion, A.; Boitard, S.; Dolezal, M.; et al. Meta-Analysis of Genome-Wide Association Studies for Cattle Stature Identifies Common Genes that Regulate Body Size in Mammals. Nat. Genet. 2018, 50, 362–367. [Google Scholar] [CrossRef]
- Makvandi-Nejad, S.; Hoffman, G.E.; Allen, J.; Chu, E.; Gu, E.; Chandler, A.M.; Loredo, A.I.; Bellone, R.R.; Mezey, J.G.; Brooks, S.; et al. Four Loci Explain 83% of Size Variation in the Horse. PLoS ONE 2012, 7, e39929. [Google Scholar] [CrossRef] [Green Version]
- Samaha, G.; Wade, C.M.; Beatty, J.; Lyons, L.A.; Fleeman, L.M.; Haase, B. Mapping the Genetic Basis of Diabetes Mellitus in the Australian Burmese Cat (Felis catus). Sci. Rep. 2020, 10, 19194. [Google Scholar] [CrossRef]
- Jostins, L.; Ripke, S.; Weersma, R.K.; Duerr, R.H.; McGovern, D.P.; Hui, K.Y.; Lee, J.C.; Schumm, L.P.; Sharma, Y.; Anderson, C.A.; et al. Host–Microbe Interactions have Shaped the Genetic Architecture of Inflammatory Bowel Disease. Nature 2012, 491, 119–124. [Google Scholar] [CrossRef] [Green Version]
- Marouli, E.; Graff, M.; Medina-Gomez, C.; Lo, K.S.; Wood, A.R.; Kjaer, T.R.; Fine, R.S.; Lu, Y.; Schurmann, C.; Highland, H.M.; et al. Rare and Low-Frequency Coding Variants alter Human Adult Height. Nature 2017, 542, 186–190. [Google Scholar] [CrossRef] [Green Version]
- Luo, Y.; de Lange, K.M.; Jostins, L.; Moutsianas, L.; Randall, J.; Kennedy, A.N.; Lamb, A.C.; McCarthy, S.; Ahmad, T.; Edwards, C.; et al. Exploring the Genetic Architecture of Inflammatory Bowel Disease by Whole-Genome Sequencing identifies Association at ADCY7. Nat. Genet. 2017, 49, 186–192. [Google Scholar] [CrossRef] [Green Version]
- Flannick, J.; Mercader, J.M.; Fuchsberger, C.; Udler, M.S.; Mahajan, A.; Wessel, J.; Teslovich, T.M.; Caulkins, L.; Koesterer, R.; Barajas-Olmos, F.; et al. Exome Sequencing of 20,791 Cases of Type 2 Diabetes and 24,440 Controls. Nature 2019, 570, 71–76. [Google Scholar] [CrossRef] [Green Version]
- Singh, T.; Poterba, T.; Curtis, D.; Akil, H.; Al Eissa, M.; Barchas, J.D.; Bass, N.; Bigdeli, T.B.; Breen, G.; Bromet, E.J.; et al. Exome Sequencing Identifies Rare Coding Variants in 10 Genes Which Confer Substantial Risk for Schizophrenia. medRxiv 2020. [Google Scholar] [CrossRef]
- Wang, Q.; Dhindsa, R.S.; Carss, K.; Harper, A.R.; Nag, A.; Tachmazidou, I.; Vitsios, D.; Deevi, S.V.V.; Mackay, A.; Muthas, D.; et al. Rare Variant Contribution to Human Disease in 281,104 UK Biobank Exomes. Nature 2021, 597, 527–532. [Google Scholar] [CrossRef]
- Blair, D.R.; Lyttle, C.S.; Mortensen, J.M.; Bearden, C.F.; Jensen, A.B.; Khiabanian, H.; Melamed, R.; Rabadan, R.; Bernstam, E.V.; Brunak, S.; et al. A Nondegenerate Code of Deleterious Variants in Mendelian Loci Contributes to Complex Disease Risk. Cell 2013, 155, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Freund, M.K.; Burch, K.S.; Shi, H.; Mancuso, N.; Kichaev, G.; Garske, K.M.; Pan, D.Z.; Miao, Z.; Mohlke, K.L.; Laakso, M.; et al. Phenotype-Specific Enrichment of Mendelian Disorder Genes near GWAS Regions across 62 Complex Traits. Am. J. Hum. Genet. 2018, 103, 535–552. [Google Scholar] [CrossRef] [Green Version]
- O’Seaghdha, C.M.; Wu, H.; Yang, Q.; Kapur, K.; Guessous, I.; Zuber, A.M.; Köttgen, A.; Stoudmann, C.; Teumer, A.; Kutalik, Z.; et al. Meta-Analysis of Genome-Wide Association Studies Identifies Six New Loci for Serum Calcium Concentrations. PLoS Genet. 2013, 9, e1003796. [Google Scholar] [CrossRef] [Green Version]
- Shelton, J.F.; Shastri, A.J.; Aslibekyan, S.; Auton, A. The 23andMe COVID-19 Team the UGT2A1/UGT2A2 Locus is Associated with COVID-19-Related Anosmia. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nicolae, D.; Gamazon, E.; Zhang, W.; Duan, S.; Dolan, M.E.; Cox, N.J. Trait-Associated SNPs Are More Likely to be eQTLs: Annotation to Enhance Discovery from GWAS. PLoS Genet. 2010, 6, e1000888. [Google Scholar] [CrossRef]
- Nica, A.C.; Montgomery, S.; Dimas, A.S.; Stranger, B.; Beazley, C.; Barroso, I.; Dermitzakis, E.T. Candidate Causal Regulatory Effects by Integration of Expression QTLs with Complex Trait Genetic Associations. PLoS Genet. 2010, 6, e1000895. [Google Scholar] [CrossRef] [Green Version]
- Boix, C.A.; James, B.T.; Park, Y.P.; Meuleman, W.; Kellis, M. Regulatory Genomic Circuitry of Human Disease Loci by Integrative Epigenomics. Nature 2021, 590, 300–307. [Google Scholar] [CrossRef]
- Lee, P.H.; O’Dushlaine, C.; Thomas, B.; Purcell, S.M. INRICH: Interval-based Enrichment Analysis for Genome-Wide Association Studies. Bioinformatics 2012, 28, 1797–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pers, T.H.; Karjalainen, J.M.; Chan, Y.; Westra, H.-J.; Wood, A.R.; Yang, J.; Lui, J.C.; Vedantam, S.; Gustafsson, S.; Esko, T.; et al. Biological Interpretation of Genome-Wide Association Studies Using Predicted Gene Functions. Nat. Commun. 2015, 6, 5890. [Google Scholar] [CrossRef] [PubMed]
- De Leeuw, C.A.; Mooij, J.M.; Heskes, T.; Posthuma, D. MAGMA: Generalized Gene-Set Analysis of GWAS Data. PLoS Comput. Biol. 2015, 11, e1004219. [Google Scholar] [CrossRef] [PubMed]
- Gusev, A.; Lee, S.H.; Trynka, G.; Finucane, H.; Vilhjalmsson, B.; Xu, H.; Zang, C.; Ripke, S.; Bulik-Sullivan, B.; Stahl, E.; et al. Partitioning Heritability of Regulatory and Cell-Type-Specific Variants across 11 Common Diseases. Am. J. Hum. Genet. 2014, 95, 535–552. [Google Scholar] [CrossRef] [Green Version]
- Finucane, H.K.; Gusev, A.; Trynka, G.; Reshef, Y.; Loh, P.-R.; Anttila, V.; Xu, H.; Zang, C.; Farh, K.; Ripke, S.; et al. Partitioning Heritability by Functional Annotation Using Genome-Wide Association Summary Statistics. Nat. Genet. 2015, 47, 1228–1235. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Huang, Y.; Zhao, S.; Yao, Y.; Zhang, Y.; Qu, J.; Wu, N.; Su, J. Integrative Genomics Analysis reveals a 21q22.11 Locus Contributing Risk to COVID-19. Hum. Mol. Genet. 2021, 30, 1247–1258. [Google Scholar] [CrossRef]
- Elvsåshagen, T.; Shadrin, A.; Frei, O.; van der Meer, D.; Bahrami, S.; Kumar, V.J.; Smeland, O.; Westlye, L.T.; Andreassen, O.A.; Kaufmann, T. The Genetic Architecture of the Human Thalamus and its Overlap with Ten Common Brain Disorders. Nat. Commun. 2021, 12, 2909. [Google Scholar] [CrossRef]
- Tsetsos, F.; Yu, D.; Sul, J.H.; Huang, A.Y.; Illmann, C.; Osiecki, L.; Darrow, S.M.; Hirschtritt, M.E.; Greenberg, E.; Muller-Vahlet, K.R.; et al. Synaptic Processes and Immune-Related Pathways Implicated in Tourette Syndrome. Transl. Psychiatry 2021, 11, 56. [Google Scholar] [CrossRef]
- Hormozdiari, F.; Gazal, S.; Van De Geijn, B.; Finucane, H.K.; Ju, C.J.-T.; Loh, P.-R.; Schoech, A.; Reshef, Y.; Liu, X.; O’Connor, L.; et al. Leveraging Molecular Quantitative Trait Loci to Understand the Genetic Architecture of Diseases and Complex Traits. Nat. Genet. 2018, 50, 1041–1047. [Google Scholar] [CrossRef]
- Finucane, H.K.; Reshef, Y.A.; Anttila, V.; Slowikowski, K.; Gusev, A.; Byrnes, A.; Gazal, S.; Loh, P.-R.; Lareau, C.; Shoresh, N.; et al. Heritability Enrichment of Specifically Expressed Genes Identifies Disease-Relevant Tissues and Cell Types. Nat. Genet. 2018, 50, 621–629. [Google Scholar] [CrossRef]
- Fromer, M.; Roussos, P.; Sieberts, S.K.; Johnson, J.; Kavanagh, D.H.; Perumal, T.M.; Ruderfer, D.M.; Oh, E.C.; Topol, A.; Shah, H.R.; et al. Gene Expression Elucidates Functional Impact of Polygenic Risk for Schizophrenia. Nat. Neurosci. 2016, 19, 1442–1453. [Google Scholar] [CrossRef] [Green Version]
- Schizophrenia Working Group of the Psychiatric Genomics Consortium; Ripke, S.; Walters, J.T.R.; O’Donovan, M.C. Schizophrenia Working Group of the Psychiatric Genomics Consortium; Ripke, S.; Walters, J.T.R.; O’Donovan, M.C. Mapping Genomic Loci Prioritises Genes and Implicates Synaptic Biology in Schizophrenia. medRxiv 2020. [Google Scholar] [CrossRef]
- Landi, M.T.; Bishop, D.T.; MacGregor, S.; Machiela, M.J.; Stratigos, A.J.; Ghiorzo, P.; Brossard, M.; Calista, D.; Choi, J.; Fargnoli, M.C.; et al. Genome-Wide Association Meta-Analyses Combining Multiple Risk Phenotypes Provide Insights into the Genetic Architecture of Cutaneous Melanoma Susceptibility. Nat. Genet. 2020, 52, 494–504. [Google Scholar] [CrossRef]
- Sey, N.Y.A.; Hu, B.; Mah, W.; Fauni, H.; McAfee, J.C.; Rajarajan, P.; Brennand, K.J.; Akbarian, S.; Won, H. A Computational Tool (H-MAGMA) for Improved Prediction of Brain-Disorder Risk Genes by Incorporating Brain Chromatin Interaction Profiles. Nat. Neurosci. 2020, 23, 583–593. [Google Scholar] [CrossRef]
- Matoba, N.; Liang, D.; Sun, H.; Aygün, N.; McAfee, J.C.; Davis, J.E.; Raffield, L.M.; Qian, H.; Piven, J.; Li, Y.; et al. Common Genetic Risk Variants Identified in the SPARK Cohort Support DDHD2 as a Candidate Risk Gene for Autism. Transl. Psychiatry 2020, 10, 265. [Google Scholar] [CrossRef]
- Gamazon, E.R.; GTEx Consortium; Wheeler, H.; Shah, K.P.; Mozaffari, S.; Aquino-Michaels, K.; Carroll, R.J.; Eyler, A.E.; Denny, J.C.; Nicolae, D.; et al. A Gene-Based Association Method for Mapping Traits Using Reference Transcriptome Data. Nat. Genet. 2015, 47, 1091–1098. [Google Scholar] [CrossRef] [Green Version]
- Gusev, A.; Ko, A.; Shi, H.; Bhatia, G.; Chung, W.; Penninx, B.W.J.H.; Jansen, R.; De Geus, E.J.C.; Boomsma, I.D.; Wright, A.F.; et al. Integrative approaches for Large-Scale Transcriptome-Wide Association Studies. Nat. Genet. 2016, 48, 245–252. [Google Scholar] [CrossRef] [Green Version]
- Barbeira, A.N.; GTEx Consortium; Dickinson, S.P.; Bonazzola, R.; Zheng, J.; Wheeler, H.E.; Torres, J.M.; Torstenson, E.S.; Shah, K.P.; Garcia, T.; et al. Exploring the Phenotypic Consequences of Tissue Specific Gene Expression Variation Inferred from GWAS Summary Statistics. Nat. Commun. 2018, 9, 1825. [Google Scholar] [CrossRef]
- Wainberg, M.; Sinnott-Armstrong, N.; Mancuso, N.; Barbeira, A.N.; Knowles, D.A.; Golan, D.; Ermel, R.; Ruusalepp, A.; Quertermous, T.; Hao, K.; et al. Opportunities and Challenges for Transcriptome-Wide Association Studies. Nat. Genet. 2019, 51, 592–599. [Google Scholar] [CrossRef]
- Kim-Hellmuth, S.; Aguet, F.; Oliva, M.; Muñoz-Aguirre, M.; Kasela, S.; Wucher, V.; Castel, S.E.; Hamel, A.R.; Viñuela, A.; Roberts, A.L.; et al. Cell Type–Specific Genetic Regulation of Gene Expression across Human Tissues. Science 2020, 369, eaaz8528. [Google Scholar] [CrossRef]
- Mancuso, N.; Shi, H.; Goddard, P.; Kichaev, G.; Gusev, A.; Pasaniuc, B. Integrating Gene Expression with Summary Association Statistics to Identify Genes Associated with 30 Complex Traits. Am. J. Hum. Genet. 2017, 100, 473–487. [Google Scholar] [CrossRef]
- Wang, X.W.H.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, J.L.X.; Qin, F. Genetic Variation in ZmVPP1 contributes to Drought Tolerance in Maize Seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Plenge, R.M.; Scolnick, E.M.; Altshuler, D. Validating Therapeutic Targets through Human Genetics. Nat. Rev. Drug Discov. 2013, 12, 581–594. [Google Scholar] [CrossRef]
- Diogo, D.; Bastarache, L.; Liao, K.P.; Graham, R.R.; Fulton, R.S.; Greenberg, J.D.; Eyre, S.; Bowes, J.; Cui, J.; Lee, A.; et al. TYK2 Protein-Coding Variants Protect against Rheumatoid Arthritis and Autoimmunity, with No Evidence of Major Pleiotropic Effects on Non-Autoimmune Complex Traits. PLoS ONE 2015, 10, e0122271. [Google Scholar] [CrossRef] [Green Version]
- Burke, J.R.; Cheng, L.; Gillooly, K.M.; Strnad, J.; Zupa-Fernandez, A.; Catlett, I.M.; Zhang, Y.; Heimrich, E.M.; McIntyre, K.W.; Cunningham, M.D.; et al. Autoimmune Pathways in Mice and Humans are Blocked by Pharmacological Stabilization of the TYK2 Pseudokinase Domain. Sci. Transl. Med. 2019, 11, eaaw1736. [Google Scholar] [CrossRef]
- Suarez-Gestal, M.; Calaza, M.; Endreffy, E.; Pullmann, R.; Ordi-Ros, J.; Sebastiani, G.D.; Ruzickova, S.; Santos, M.J.; Papasteriades, C.; Marchini, M.; et al. Replication of Recently Identified Systemic Lupus Erythematosus Genetic Associations: A Case–Control Study. Arthritis Res. Ther. 2009, 11, R69. [Google Scholar] [CrossRef] [Green Version]
- Wallace, C.; Smyth, D.J.; Maisuria-Armer, M.; Walker, N.M.; Todd, A.J.A.; Clayton, D.G. The Imprinted DLK1-MEG3 Gene Region on Chromosome 14q32.2 Alters Susceptibility to Type 1 Diabetes. Nat. Genet. 2010, 42, 68–71. [Google Scholar] [CrossRef] [Green Version]
- Genetic Analysis of Psoriasis Consortium & the Wellcome Trust Case Control Consortium 2 A Genome-Wide Association Study Identifies New Psoriasis Susceptibility Loci and an Interaction between HLA-C and ERAP1. Nat. Genet. 2010, 42, 985–990. [CrossRef] [Green Version]
- Franke, A.; McGovern, D.P.B.; Barrett, J.C.; Wang, K.; Radford-Smith, G.L.; Ahmad, T.; Lees, C.W.; Balschun, T.; Lee, J.; Roberts, R.; et al. Genome-Wide Meta-Analysis increases to 71 the Number of Confirmed Crohn's Disease Susceptibility Loci. Nat. Genet. 2010, 42, 1118–1125. [Google Scholar] [CrossRef] [Green Version]
- Maher, B. Personal Genomes: The Case of the Missing Heritability. Nature 2008, 456, 18–21. [Google Scholar] [CrossRef]
- Gibson, G. Hints of Hidden Heritability in GWAS. Nat. Genet. 2010, 42, 558–560. [Google Scholar] [CrossRef] [PubMed]
- The International Schizophrenia Consortium; Purcell, S.M.; Wray, N.R.; Stone, J.L.; Visscher, P.M.; O’Donovan, M.C.; Sullivan, P.F.; Sklar, P. Common Polygenic Variation Contributes to Risk of Schizophrenia and Bipolar Disorder. Nature 2009, 460, 748–752. [Google Scholar] [CrossRef] [PubMed]
- Genetic Risk and Outcome in Psychosis (GROUP); Stefansson, H.; Ophoff, R.A.; Steinberg, S.; Andreassen, O.A.; Cichon, S.; Rujescu, D.; Werge, T.; Pietiläinen, O.P.H.; Mors, O.; et al. Common Variants Conferring Risk of Schizophrenia. Nature 2009, 460, 744–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, J.; Levinson, D.F.; Duan, J.; Sanders, A.R.; Zheng, Y.; Pe’Er, I.; Dudbridge, F.; Holmans, P.; Whittemore, A.S.; Mowry, B.J.; et al. Common Variants on Chromosome 6p22.1 are Associated with Schizophrenia. Nature 2009, 460, 753–757. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Goddard, M.; Visscher, P. Prediction of Individual Genetic Risk to Disease from Genome-Wide Association Studies. Genome Res. 2007, 17, 1520–1528. [Google Scholar] [CrossRef] [Green Version]
- Privé, F.; Arbel, J.; Vilhjálmsson, B.J. LDpred2: Better, Faster, Stronger. bioRxiv 2020. [Google Scholar] [CrossRef]
- Visscher, P.M.; Hill, W.G.; Wray, N.R. Heritability in the Genomics Era—Concepts and Misconceptions. Nat. Rev. Genet. 2008, 9, 255–266. [Google Scholar] [CrossRef]
- Boyle, E.A.; Li, Y.I.; Pritchard, J.K. An Expanded View of Complex Traits: From Polygenic to Omnigenic. Cell 2017, 169, 1177–1186. [Google Scholar] [CrossRef] [Green Version]
- Wei, Z.; Wang, W.; Bradfield, J.; Li, J.; Cardinale, C.; Frackelton, E.; Kim, C.; Mentch, F.; Van Steen, K.; Visscher, P.M.; et al. Large Sample Size, Wide Variant Spectrum, and Advanced Machine-Learning Technique Boost Risk Prediction for Inflammatory Bowel Disease. Am. J. Hum. Genet. 2013, 92, 1008–1012. [Google Scholar] [CrossRef] [Green Version]
- Romagnoni, A.; International Inflammatory Bowel Disease Genetics Consortium (IIBDGC); Jégou, S.; Van Steen, K.; Wain, L.; Hugot, J.-P. Comparative Performances of Machine Learning Methods for Classifying Crohn Disease Patients Using Genome-Wide Genotyping Data. Sci. Rep. 2019, 9, 1035. [Google Scholar] [CrossRef]
- Mieth, B.; Rozier, A.; Rodriguez, J.A.; Höhne, M.M.C.; Görnitz, N.; Müller, K.-R. DeepCOMBI: Explainable Artificial Intelligence for the Analysis and Discovery in Genome-Wide Association Studies. NAR Genom. Bioinform. 2021, 3, lqab065. [Google Scholar] [CrossRef]
- Wang, H.; Bennett, D.A.; De Jager, P.L.; Zhang, Q.-Y.; Zhang, H.-Y. Genome-Wide Epistasis Analysis for Alzheimer’s Disease and Implications for Genetic Risk Prediction. Alzheimer's Res. Ther. 2021, 13, 55. [Google Scholar] [CrossRef]
- Fahed, A.C.; Wang, M.; Homburger, J.R.; Patel, A.P.; Bick, A.G.; Neben, C.L.; Lai, C.; Brockman, D.; Philippakis, A.; Ellinor, P.T.; et al. Polygenic Background Modifies Penetrance of Monogenic Variants for Tier 1 Genomic Conditions. Nat. Commun. 2020, 11, 3635. [Google Scholar] [CrossRef]
- Aragam, K.G.; Dobbyn, A.; Judy, R.; Chaffin, M.; Chaudhary, K.; Hindy, G.; Cagan, A.; Finneran, P.; Weng, L.-C.; Loos, R.J.; et al. Limitations of Contemporary Guidelines for Managing Patients at High Genetic Risk of Coronary Artery Disease. J. Am. Coll. Cardiol. 2020, 75, 2769–2780. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; McGuffog, L.; Barrowdale, D.; Lee, A.; Soucy, P.; Dennis, J.; Domchek, S.M.; Robson, M.; Spurdle, A.B.; Ramus, S.; et al. Evaluation of Polygenic Risk Scores for Breast and Ovarian Cancer Risk Prediction in BRCA1 and BRCA2 Mutation Carriers. J. Natl. Cancer Inst. 2017, 109, djw302. [Google Scholar] [CrossRef] [Green Version]
- Barnes, D.R.; Rookus, M.A.; McGuffog, L.; Leslie, G.; Mooij, T.M.; Dennis, J.; Mavaddat, N.; Adlard, J.; Ahmed, M.; Aittomäki, K.; et al. Polygenic Risk Scores and Breast and Epithelial Ovarian Cancer Risks for Carriers of BRCA1 and BRCA2 Pathogenic Variants. Genet. Med. 2020, 22, 1653–1666. [Google Scholar] [CrossRef]
- Lecarpentier, J.; Silvestri, V.; Kuchenbaecker, K.B.; Barrowdale, D.; Dennis, J.; McGuffog, L.; Soucy, P.; Leslie, G.; Rizzolo, P.; Navazio, A.S.; et al. Prediction of Breast and Prostate Cancer Risks in Male BRCA1 and BRCA2 Mutation Carriers Using Polygenic Risk Scores. J. Clin. Oncol. 2017, 35, 2240–2250. [Google Scholar] [CrossRef]
- Han, X.; Qassim, A.; An, J.; Marshall, H.; Zhou, T.; Ong, J.-S.; Hassall, M.M.; Hysi, P.G.; Foster, P.J.; Khaw, P.T.; et al. Genome-Wide Association Analysis of 95 549 Individuals Identifies Novel Loci and Genes Influencing Optic Disc Morphology. Hum. Mol. Genet. 2019, 28, 3680–3690. [Google Scholar] [CrossRef]
- Liu, X.; Song, Z.; Li, Y.; Yao, Y.; Fang, M.; Bai, C.; An, P.; Chen, H.; Chen, Z.; Tang, B.; et al. Integrated Genetic Analyses Revealed Novel Human Longevity Loci and Reduced Risks of Multiple Diseases in a Cohort Study of 15,651 Chinese Individuals. Aging Cell 2021, 20, e13323. [Google Scholar] [CrossRef]
- Zhang, Y.D.; Breast Cancer Association Consortium (BCAC); Hurson, A.N.; Zhang, H.; Choudhury, P.P.; Easton, D.F.; Milne, R.L.; Simard, J.; Hall, P.; Michailidou, K.; et al. Assessment of Polygenic Architecture and Risk Prediction Based on Common Variants across Fourteen Cancers. Nat. Commun. 2020, 11, 3353. [Google Scholar] [CrossRef]
- Escott-Price, V.; Myers, A.J.; Huentelman, M.; Hardy, J. Polygenic Risk Score Analysis of Pathologically confirmed Alzheimer Disease. Ann. Neurol. 2017, 82, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.; Escott-Price, V. Genes, Pathways and Risk Prediction in Alzheimer's Disease. Hum. Mol. Genet. 2019, 28, 235–240. [Google Scholar] [CrossRef] [PubMed]
- Nalls, M.A.; Blauwendraat, C.; Vallerga, C.L.; Heilbron, K.; Bandres-Ciga, S.; Chang, D.; Tan, M.; Kia, D.A.; Noyce, A.J.; Xue, A.; et al. Identification of Novel Risk Loci, Causal Insights, and Heritable Risk for Parkinson’s Disease: A Meta-Analysis of Genome-Wide Association Studies. Lancet Neurol. 2019, 18, 1091–1102. [Google Scholar] [CrossRef]
- Han, Y.; Teeple, E.; Shankara, S.; Sadeghi, M.; Zhu, C.; Liu, D.; FinnGen; Wang, C.; Frau, F.; Klinger, K.W.; et al. Genome-Wide Polygenic Risk Score Identifies Individuals at Elevated Parkinson’s Disease Risk. medRxiv 2020. [Google Scholar] [CrossRef]
- Allegrini, A.G.; Selzam, S.; Rimfeld, K.; von Stumm, S.; Pingault, J.B.; Plomin, R. Genomic Prediction of Cognitive Traits in Childhood and Adolescence. Mol. Psychiatry 2019, 24, 819–827. [Google Scholar] [CrossRef]
- Von Stumm, S.; Smith-Woolley, E.; Ayorech, Z.; McMillan, A.; Rimfeld, K.; Dale, P.S.; Plomin, R. Predicting Educational Achievement from Genomic Measures and Socioeconomic Status. Dev. Sci. 2019, 23, e12925. [Google Scholar] [CrossRef] [Green Version]
- Morris, T.T.; Davies, N.M.; Smith, G.D. Can Education be Personalised using Pupils’ Genetic Data? eLife 2020, 9, e49962. [Google Scholar] [CrossRef]
- Smith-Woolley, E.; Pingault, J.-B.; Selzam, S.; Rimfeld, K.; Krapohl, E.; Von Stumm, S.; Asbury, K.; Dale, P.S.; Young, T.; Allen, R.; et al. Differences in Exam Performance Between Pupils Attending Selective and Non-Selective Schools Mirror the Genetic Differences between Them. npj Sci. Learn. 2018, 3, 3. [Google Scholar] [CrossRef]
- Richardson, K.; Jones, M.C. Why Genome-Wide Associations with Cognitive Ability Measures are Probably Spurious. New Ideas Psychol. 2019, 55, 35–41. [Google Scholar] [CrossRef]
- Cheesman, R.; Hunjan, A.; Coleman, J.; Ahmadzadeh, Y.; Plomin, R.; McAdams, T.; Eley, T.C.; Breen, G. Comparison of Adopted and Nonadopted Individuals Reveals Gene–Environment Interplay for Education in the UK Biobank. Psychol. Sci. 2020, 31, 582–591. [Google Scholar] [CrossRef]
- Murphy, K.C.; Jones, L.A.; Owen, M.J. High Rates of Schizophrenia in Adults with Velo-Cardio-Facial Syndrome. Arch. Gen. Psychiatry 1999, 56, 940–945. [Google Scholar] [CrossRef] [Green Version]
- Zinkstok, J.; Van Amelsvoort, T. Neuropsychological Profile and Neuroimaging in Patients with 22Q11.2 Deletion Syndrome: A Review Keywords. Child Neuropsychol. 2005, 11, 21–37. [Google Scholar] [CrossRef]
- Davies, R.W.; Fiksinski, A.M.; Breetvelt, E.J.; Williams, N.M.; Hooper, S.R.; Monfeuga, T.; Bassett, A.S.; Owen, M.J.; Gur, R.E.; Morrow, B.E.; et al. Using Common Genetic Variation to Examine Phenotypic Expression and Risk Prediction in 22q11.2 Deletion Syndrome. Nat. Med. 2020, 26, 1912–1918. [Google Scholar] [CrossRef]
- Martin, A.R.; Kanai, M.; Kamatani, Y.; Okada, Y.; Neale, B.M.; Daly, M.J. Clinical Use of Current Polygenic Risk Scores May Exacerbate Health Disparities. Nat. Genet. 2019, 51, 584–591. [Google Scholar] [CrossRef]
- Majara, L.; Kalungi, A.; Koen, N.; Zar, H.; Stein, D.J.; Kinyanda, E.; Atkinson, E.G.; Martin, A.R. Low Generalizability of Polygenic Scores in African Populations due to Genetic and Environmental Diversity. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bigdeli, T.B.; Consortium on the Genetics of Schizophrenia (COGS) Investigators; Genovese, G.; Georgakopoulos, P.; Meyers, J.L.; Peterson, R.; Iyegbe, C.O.; Medeiros, H.; Valderrama, J.; Achtyes, E.D.; et al. Contributions of Common Genetic Variants to Risk of Schizophrenia among Individuals of African and Latino Ancestry. Mol. Psychiatry 2019, 25, 2455–2467. [Google Scholar] [CrossRef] [Green Version]
- Cross-Disorder Group of the Psychiatric Genomics Consortium Genetic relationship between Five Psychiatric Disorders Estimated from Genome-Wide SNPs. Nat. Genet. 2013, 45, 984–994. [CrossRef] [Green Version]
- Van Rheenen, W.; Peyrot, W.J.; Schork, A.J.; Lee, S.H.; Wray, N.R. Genetic Correlations of Polygenic Disease Traits: From Theory to Practice. Nat. Rev. Genet. 2019, 20, 567–581. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.; Finucane, H.K.; Anttila, V.; Gusev, A.; Day, F.R.; Loh, P.-R.; Duncan, E.L.; Perry, J.R.; Patterson, N.; Robinson, E.; et al. An Atlas of Genetic Correlations across Human Diseases and Traits. Nat. Genet. 2015, 47, 1236–1241. [Google Scholar] [CrossRef] [Green Version]
- Zheng, J.; Erzurumluoglu, A.M.; Elsworth, B.L.; Kemp, J.P.; Howe, L.; Haycock, P.C.; Hemani, G.; Tansey, K.; Laurin, C.; Pourcain, B.S.; et al. LD Hub: A Centralized Database and Web Interface to Perform LD Score Regression that Maximizes the Potential of Summary Level GWAS Data for SNP Heritability and Genetic Correlation Analysis. Bioinformatics 2017, 33, 272–279. [Google Scholar] [CrossRef]
- Watanabe, K.; Stringer, S.; Frei, O.; Mirkov, M.U.; de Leeuw, C.; Polderman, T.J.C.; van der Sluis, S.; Andreassen, O.A.; Neale, B.M.; Posthuma, D. A Global Overview of Pleiotropy and Genetic Architecture in Complex Traits. Nat. Genet. 2019, 51, 1339–1348. [Google Scholar] [CrossRef]
- Gao, J.; Davis, L.K.; Hart, A.B.; Sanchez-Roige, S.; Han, L.; Cacioppo, J.T.; Palmer, A.A. Genome-Wide Association Study of Loneliness Demonstrates a Role for Common Variation. Neuropsychopharmacology 2017, 42, 811–821. [Google Scholar] [CrossRef] [Green Version]
- Bone, W.P.; Program, T.V.M.V.; Siewert, K.M.; Jha, A.; Klarin, D.; Damrauer, S.M.; Chang, K.-M.; Tsao, P.S.; Assimes, T.L.; Ritchie, M.D.; et al. Multi-Trait Association Studies Discover Pleiotropic Loci Between Alzheimer’s Disease and Cardiometabolic Traits. Alzheimer's Res. Ther. 2021, 13, 34. [Google Scholar] [CrossRef]
- Xicoy, H.; Klemann, C.J.; De Witte, W.; Martens, M.B.; Martens, G.J.; Poelmans, G. Shared Genetic Etiology between Parkinson’s Disease and Blood Levels of Specific Lipids. npj Park. Dis. 2021, 7, 23. [Google Scholar] [CrossRef] [PubMed]
- Denny, J.; Bastarache, L.; Ritchie, M.D.; Carroll, R.J.; Zink, R.; Mosley, J.; Field, J.R.; Pulley, J.M.; Ramirez, A.H.; Bowton, E.; et al. Systematic Comparison of Phenome-Wide Association Study of Electronic Medical Record Data and Genome-Wide Association Study Data. Nat. Biotechnol. 2013, 31, 1102–1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richardson, T.G.; Harrison, S.; Hemani, G.; Smith, G.D. An Atlas of Polygenic Risk Score Associations to Highlight Putative Causal Relationships across the Human Phenome. eLife 2019, 8, e43657. [Google Scholar] [CrossRef] [PubMed]
- Robinson, J.R.; Denny, J.C.; Roden, D.M.; Van Driest, S.L. Genome-wide and Phenome-wide Approaches to Understand Variable Drug Actions in Electronic Health Records. Clin. Transl. Sci. 2018, 11, 112–122. [Google Scholar] [CrossRef] [Green Version]
- Zhao, B.; Luo, H.; Huang, X.; Wei, C.; Di, J.; Tian, Y.; Fu, X.; Li, B.; Liu, G.E.; Fang, L.; et al. Integration of a Single-Step Genome-Wide Association Study with a Multi-Tissue Transcriptome Analysis Provides Novel Insights into the Genetic Basis of Wool and Weight Traits in Sheep. Genet. Sel. Evol. 2021, 53, 56. [Google Scholar] [CrossRef]
- Evans, W.N.; Ringel, J.S. Can Higher Cigarette Taxes Improve Birth Outcomes? J. Public Econ. 1999, 72, 135–154. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.D.; Ebrahim, S. ‘Mendelian Randomization’: Can Genetic Epidemiology Contribute to Understanding Environmental Determinants of Disease? Int. J. Epidemiol. 2003, 32, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Smith, G.D.; Harbord, R.M.; Lewis, S.J. Alcohol Intake and Blood Pressure: A Systematic Review Implementing a Mendelian Randomization Approach. PLoS Med. 2008, 5, e52. [Google Scholar] [CrossRef] [Green Version]
- Cho, Y.; Shin, S.-Y.; Won, S.; Relton, C.L.; Smith, G.D.; Shin, M.-J. Alcohol Intake and Cardiovascular Risk Factors: A Mendelian Randomisation Study. Sci. Rep. 2015, 5, 18422. [Google Scholar] [CrossRef] [Green Version]
- Shen, X.; Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium; Howard, D.M.; Adams, M.J.; Hill, W.D.; Clarke, T.-K.; Deary, I.J.; Whalley, H.C.; McIntosh, A.M. A Phenome-Wide Association and Mendelian Randomisation Study of Polygenic Risk for Depression in UK Biobank. Nat. Commun. 2020, 11, 230. [Google Scholar] [CrossRef]
- Cao, L.; Li, Z.Q.; Shi, Y.Y.; Liu, Y. Telomere Length and Type 2 Diabetes: Mendelian Randomization Study and Polygenic Risk Score Analysis. Yi Chuan 2020, 42, 882–888. [Google Scholar] [CrossRef]
- Andrews, S.J.; Fulton-Howard, B.; O'Reilly, P.; Marcora, E.; Goate, A.M.; Farrer, L.A.; Haines, J.L.; Mayeux, R.; Naj, A.C.; Pericak-Vance, M.A.; et al. Causal Associations Between Modifiable Risk Factors and the Alzheimer's Phenome. Ann. Neurol. 2020, 89, 54–65. [Google Scholar] [CrossRef]
- Higgins, H.; Mason, A.M.; Larsson, S.C.; Gill, D.; Langenberg, C.; Burgess, S. Estimating the Population Benefits of Blood Pressure Lowering: A Wide-Angled Mendelian Randomization Study in UK Biobank. J. Am. Hear. Assoc. 2021, 10, e021098. [Google Scholar] [CrossRef]
- Pierce, B.L.; VanderWeele, T.J. The Effect of Non-Differential Measurement Error on Bias, Precision and Power in Mendelian Randomization Studies. Int. J. Epidemiol. 2012, 41, 1383–1393. [Google Scholar] [CrossRef]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Smith, G.D.; Sheehan, N.; Thompson, J. A Framework for the Investigation of Pleiotropy in Two-Sample Summary Data Mendelian Randomization. Stat. Med. 2017, 36, 1783–1802. [Google Scholar] [CrossRef] [Green Version]
- Bowden, J.; Smith, G.D.; Burgess, S. Mendelian Randomization with Invalid Instruments: Effect Estimation and Bias Detection through Egger Regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [Green Version]
- Morrison, J.; Knoblauch, N.; Marcus, J.H.; Stephens, M.; He, X. Mendelian Randomization Accounting for Correlated and Uncorrelated Pleiotropic Effects Using Genome-Wide Summary Statistics. Nat. Genet. 2020, 52, 740–747. [Google Scholar] [CrossRef]
- Hu, X.; Zhao, J.; Lin, Z.; Wang, Y.; Peng, H.; Zhao, H.; Wan, X.; Yang, C. MR-APSS: A Unified Approach to Mendelian Randomization Accounting for Pleiotropy and Sample Structure Using Genome-Wide Summary Statistics. bioRxiv 2021. [Google Scholar] [CrossRef]
- Xue, H.; Shen, X.; Pan, W. Constrained Maximum Likelihood-Based Mendelian Randomization Robust to Both Correlated and Uncorrelated Pleiotropic Effects. Am. J. Hum. Genet. 2021, 108, 1251–1269. [Google Scholar] [CrossRef]
- Sekar, A.; Adolfsson, R.; Bialas, A.R.; De Rivera, H.; Davis, A.; Hammond, T.R.; Kamitaki, N.; Tooley, K.; Presumey, J.; Baum, M.; et al. Schizophrenia Risk from Complex Variation of Complement Component 4. Nature 2016, 530, 177–183. [Google Scholar] [CrossRef] [Green Version]
- Smemo, S.; Tena, J.J.; Kim, K.-H.; Gamazon, E.; Sakabe, N.J.; Gómez-Marín, C.; Aneas, I.; Credidio, F.L.; Sobreira, D.R.; Wasserman, N.F.; et al. Obesity-Associated Variants within FTO form Long-Range Functional Connections with IRX3. Nature 2014, 507, 371–375. [Google Scholar] [CrossRef] [Green Version]
- Claussnitzer, M.; Dankel, S.N.; Kim, K.-H.; Quon, G.; Meuleman, W.; Haugen, C.; Glunk, V.; Sousa, I.S.; Beaudry, J.L.; Puviindran, V.; et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N. Engl. J. Med. 2015, 373, 895–907. [Google Scholar] [CrossRef] [Green Version]
- Guan, Y.; Liang, X.; Ma, Z.; Hu, H.; Liu, H.; Miao, Z.; Linkermann, A.; Hellwege, J.N.; Voight, B.F.; Susztak, K. A Single Genetic Locus Controls Both Expression of DPEP1/CHMP1A and Kidney Disease Development via Ferroptosis. Nat. Commun. 2021, 12, 5078. [Google Scholar] [CrossRef]
- Kichaev, G.; Pasaniuc, B. Leveraging Functional-Annotation Data in Trans-ethnic Fine-Mapping Studies. Am. J. Hum. Genet. 2015, 97, 260–271. [Google Scholar] [CrossRef] [Green Version]
- Sinnott-Armstrong, N.; Sousa, I.S.; Laber, S.; Rendina-Ruedy, E.; Dankel, S.E.N.; Ferreira, T.; Mellgren, G.; Karasik, D.; Rivas, M.; Pritchard, J.; et al. A Regulatory Variant at 3q21.1 Confers an Increased Pleiotropic Risk for Hyperglycemia and Altered Bone Mineral Density. Cell Metab. 2021, 33, 615–628. [Google Scholar] [CrossRef] [PubMed]
- Sheng, X.; Guan, Y.; Ma, Z.; Wu, J.; Liu, H.; Qiu, C.; Vitale, S.; Miao, Z.; Seasock, M.J.; Palmer, M.; et al. Mapping the Genetic Architecture of Human Traits to Cell Types in the Kidney Identifies Mechanisms of Disease and Potential Treatments. Nat. Genet. 2021, 53, 1322–1333. [Google Scholar] [CrossRef] [PubMed]
- Stanzick, K.J.; Li, Y.; Schlosser, P.; Gorski, M.; Wuttke, M.; Thomas, L.F.; Rasheed, H.; Rowan, B.X.; Graham, S.E.; Vanderweff, B.R.; et al. Discovery and Prioritization of Variants and Genes for Kidney Function in >1.2 million Individuals. Nat. Commun. 2021, 12, 4350. [Google Scholar] [CrossRef] [PubMed]
- Corces, M.R.; Shcherbina, A.; Kundu, S.; Gloudemans, M.J.; Frésard, L.; Granja, J.M.; Louie, B.H.; Eulalio, T.; Shams, S.; Bagdatli, S.T.; et al. Single-Cell Epigenomic Analyses Implicate Candidate Causal Variants at Inherited Risk Loci for Alzheimer’s and Parkinson’s Diseases. Nat. Genet. 2020, 52, 1158–1168. [Google Scholar] [CrossRef]
- Kupari, J.; Usoskin, D.; Parisien, M.; Lou, D.; Hu, Y.; Fatt, M.; Lönnerberg, P.; Spångberg, M.; Eriksson, B.; Barkas, N.; et al. Single Cell Transcriptomics of Primate Sensory Neurons Identifies Cell Types Associated with Chronic Pain. Nat. Commun. 2021, 12, 1510. [Google Scholar] [CrossRef]
- Locke, A.E.; Kahali, B.; Berndt, S.I.; Justice, A.E.; Pers, T.H.; Day, F.R.; Powell, C.; Vedantam, S.; Buchkovich, M.L.; Yang, J.; et al. Genetic Studies of Body Mass Index Yield New Insights for Obesity Biology. Nature 2015, 518, 197–206. [Google Scholar] [CrossRef] [Green Version]
- Calderon, D.; Bhaskar, A.; Knowles, D.A.; Golan, D.; Raj, T.; Fu, A.Q.; Pritchard, J.K. Inferring Relevant Cell Types for Complex Traits by Using Single-Cell Gene Expression. Am. J. Hum. Genet. 2017, 101, 686–699. [Google Scholar] [CrossRef] [Green Version]
- Porcu, E.; Rüeger, S.; Lepik, K.; Santoni, F.A.; Reymond, A.; Kutalik, Z.; eQTLGen Consortium. BIOS Consortium Mendelian Randomization Integrating GWAS and eQTL Data Reveals Genetic Determinants of Complex and Clinical Traits. Nat. Commun. 2019, 10, 3300. [Google Scholar] [CrossRef] [Green Version]
- Zhu, A.; Matoba, N.; Wilson, E.P.; Tapia, A.L.; Li, Y.; Ibrahim, J.G.; Stein, J.L.; Love, M.I. MRLocus: Identifying causal genes mediating a trait through Bayesian Estimation of Allelic Heterogeneity. PLoS Genet. 2021, 17, e1009455. [Google Scholar] [CrossRef]
- Pain, O.; Glanville, K.P.; Hagenaars, S.; Selzam, S.; Fürtjes, A.; Coleman, I.J.R.; Rimfeld, K.; Breen, G.; Folkersen, L.; Lewis, C.M. Imputed Gene Expression Risk Scores: A Functionally Informed Component of Polygenic Risk. Hum. Mol. Genet. 2021, 30, 727–738. [Google Scholar] [CrossRef]
- Võsa, U.; Claringbould, A.; Westra, H.-J.; Bonder, M.J.; Deelen, P.; Zeng, B.; Kirsten, H.; Saha, A.; Kreuzhuber, R.; Yazar, S.; et al. Large-Scale Cis- and Trans-Eqtl Analyses Identify Thousands of Genetic Loci and Polygenic Scores that Regulate Blood Gene Expression. Nat. Genet. 2021, 53, 1300–1310. [Google Scholar] [CrossRef]
- Finkel, Y.; Gassull, A.M.; Goossens, D.; Laukens, D.; Lémann, M.; Libioulle, C.; O’Morain, C.; Reenaers, C.; Rutgeerts, P.; Tysk, C.; et al. Resequencing of Positional Candidates Identifies Low Frequency IL23R Coding Variants Protecting against Inflammatory Bowel Disease. Nat. Genet. 2011, 43, 43–47. [Google Scholar] [CrossRef]
- Rivas, M.A.; National Institute of Diabetes and Digestive Kidney Diseases Inflammatory Bowel Disease Genetics Consortium (NIDDK IBDGC); Beaudoin, M.; Gardet, A.; Stevens, C.; Sharma, Y.; Zhang, C.K.; Boucher, G.; Ripke, S.; Ellinghaus, D.; et al. Deep Resequencing of GWAS Loci Identifies Independent Rare Variants Associated with Inflammatory Bowel Disease. Nat. Genet. 2011, 43, 1066–1073. [Google Scholar] [CrossRef] [Green Version]
- Seddon, J.M.; Yu, Y.; Miller, E.C.; Reynolds, R.; Tan, P.L.; Gowrisankar, S.; Goldstein, J.; Triebwasser, M.; Anderson, E.H.; Zerbib, J.; et al. Rare Variants in CFI, C3 and C9 are Associated with High Risk of Advanced Age-Related Macular Degeneration. Nat. Genet. 2013, 45, 1366–1370. [Google Scholar] [CrossRef] [Green Version]
- Flannick, J.; Thorleifsson, G.; Beer, N.L.; Jacobs, S.B.R.; Grarup, N.; Burtt, N.P.; Mahajan, A.; Fuchsberger, C.; Atzmon, G.; Benediktsson, R.; et al. Loss-of-Function Mutations in SLC30A8 Protect against Type 2 Diabetes. Nat. Genet. 2014, 46, 357–363. [Google Scholar] [CrossRef]
- Diogo, D.; Kurreeman, F.; Stahl, E.A.; Liao, K.P.; Gupta, N.; Greenberg, J.D.; Rivas, M.A.; Hickey, B.; Flannick, J.; Thomson, B.; et al. Rare, Low-Frequency, and Common Variants in the Protein-Coding Sequence of Biological Candidate Genes from GWASs Contribute to Risk of Rheumatoid Arthritis. Am. J. Hum. Genet. 2013, 92, 15–27. [Google Scholar] [CrossRef] [Green Version]
- Motegi, T.; Kochi, Y.; Matsuda, K.; Kubo, M.; Yamamoto, K.; Momozawa, Y. Identification of Rare Coding Variants in TYK2 Protective for Rheumatoid Arthritis in the Japanese Population and their Effects on Cytokine Signalling. Ann. Rheum. Dis. 2019, 78, 1062–1069. [Google Scholar] [CrossRef]
- Bergen, S.E.; Ploner, A.; Howrigan, D.; O’Donovan, M.C.; Smoller, J.W.; Sullivan, P.F.; Sebat, J.; Neale, B.; Kendler, K.S. CNV Analysis Group and the Schizophrenia Working Group of the Psychiatric Genomics Consortium Joint Contributions of Rare Copy Number Variants and Common SNPs to Risk for Schizophrenia. Am. J. Psychiatry 2019, 176, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, S.; Ninomiya, K.; Kushima, I.; Saito, T.; Shimasaki, A.; Sakusabe, T.; Momozawa, Y.; Kubo, M.; Kamatani, Y.; Ozaki, N.; et al. Polygenic Risk Scores in Schizophrenia with Clinically Significant Copy Number Variants. Psychiatry Clin. Neurosci. 2020, 74, 35–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, E.; GROUP Investigators; Han, J.; Morgan, J.; Carrera, N.; Escott-Price, V.; Pocklington, A.J.; Duffield, M.; Hall, L.S.; Legge, S.E.; et al. De Novo Mutations Identified by Exome Sequencing Implicate Rare Missense Variants in SLC6A1 in Schizophrenia. Nat. Neurosci. 2020, 23, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Yu, D.; Scharf, J.M.; Mathews, C.A.; McGrath, L.; Cook, E.; Lee, S.H.; Davis, L.K.; Gamazon, E.R. Contextualizing Genetic Risk Score for Disease Screening and Rare Variant Discovery. Nat. Commun. 2021, 12, 4418. [Google Scholar] [CrossRef]
- Dobrindt, K.; Zhang, H.; Das, D.; Abdollahi, S.; Prorok, T.; Ghosh, S.; Weintraub, S.; Genovese, G.; Powell, S.K.; Lund, A.; et al. Publicly Available hiPSC Lines with Extreme Polygenic Risk Scores for Modeling Schizophrenia. Complex Psychiatry 2020, 6, 68–82. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).