Conditional CRISPR-Cas Genome Editing in Drosophila to Generate Intestinal Tumors
Abstract
:1. Introduction
2. Material and Methods
2.1. Fly Stocks
2.2. CRISPR/Cas9 Mutagenesis
2.3. Immunostaining and Image Acquisition
2.4. Quantification of Midgut Mitosis
2.5. Quantification of GFP Positive Cells
2.6. Measurement of Midgut Length and Diameter
2.7. Tumor Incidence
2.8. Statistical Analysis
3. Results
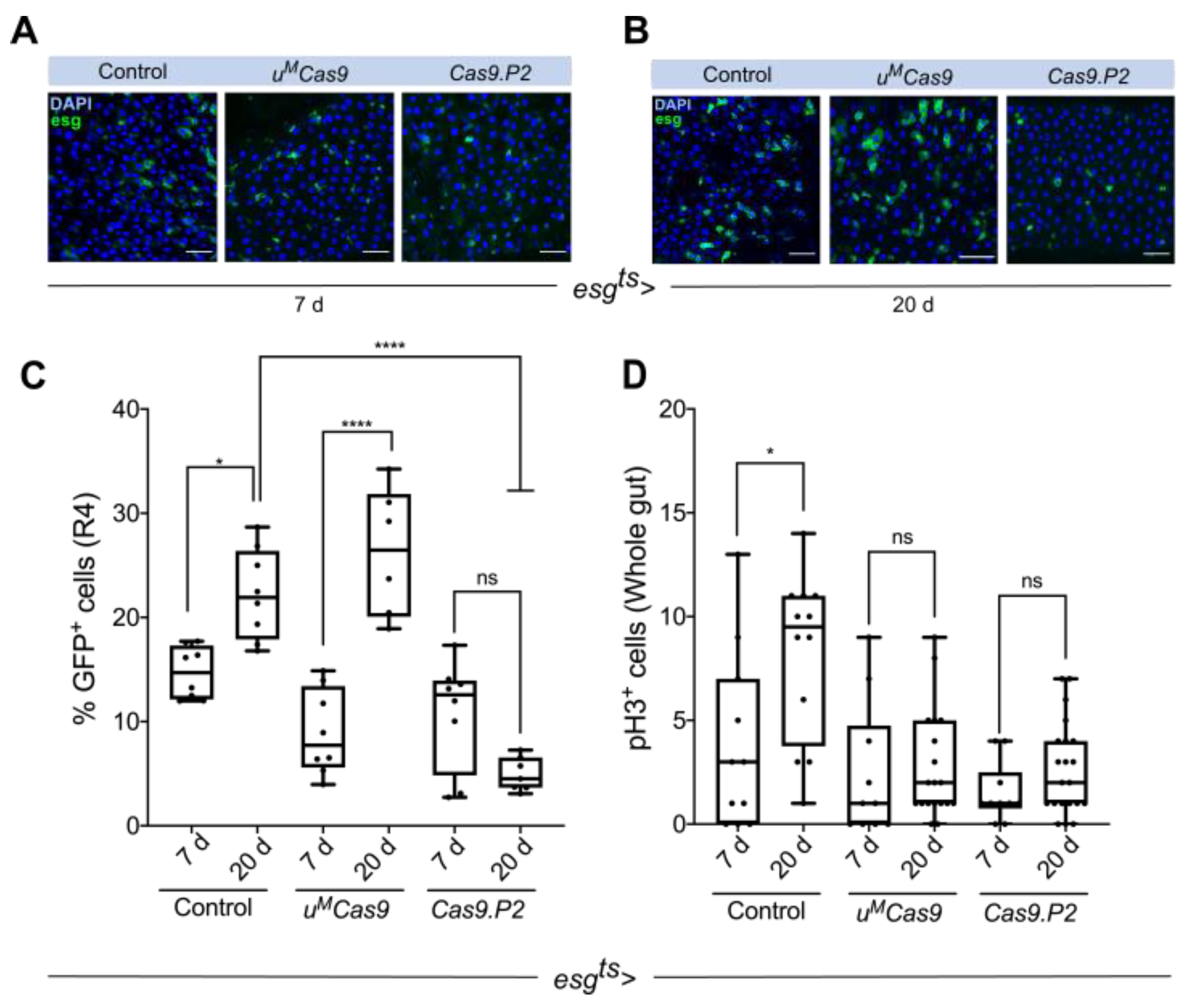
3.1. Cas9 Expression in the Drosophila Midgut
3.2. Defining Temporal Parameters for Cas9 Editing
3.3. Cas9.P2on/off and uMCas9on Are Effective at Inducing Intestinal Tumors
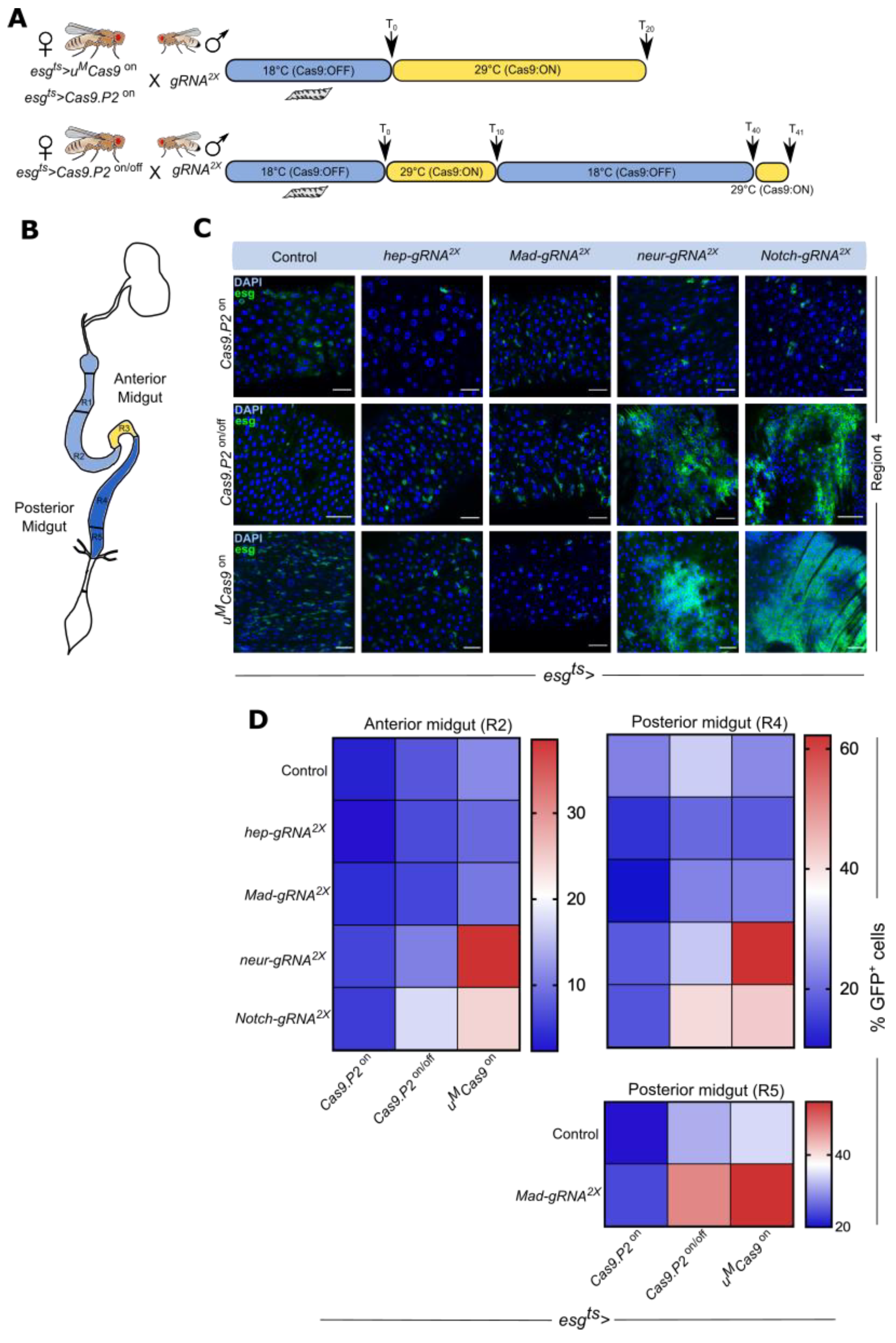
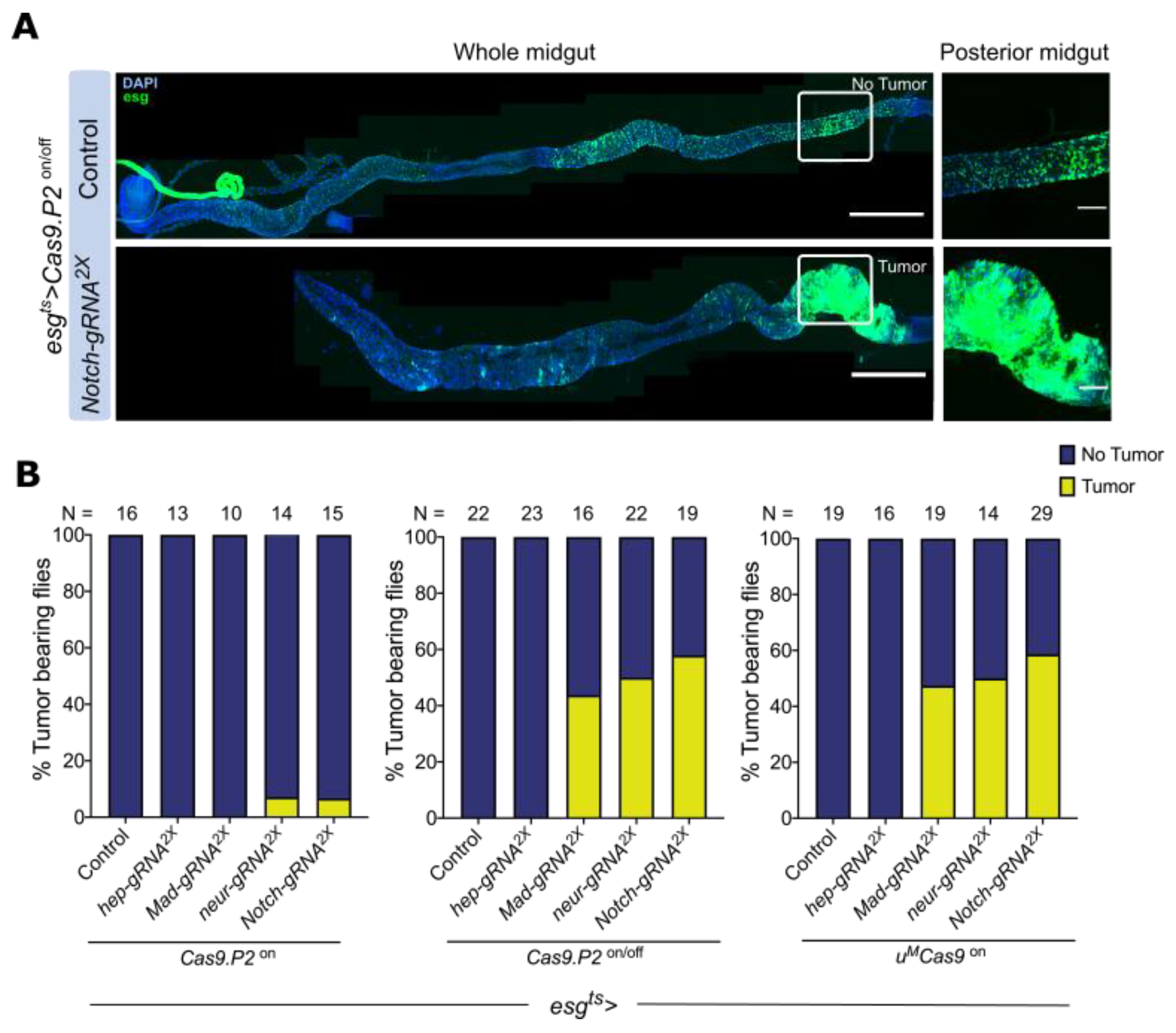
3.4. Mutating Tumor Suppressors Increases Mitotic Cells and Tumor Incidence
3.5. Mutating Tumor Suppressors Changes Intestinal Morphology
4. Discussion
4.1. Comparison of Intestinal-Specific CRISPR-Cas9 Systems
4.2. Effects of High Cas9 Expression and Outlook on Applications of CRISPR Fly Models
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pickar-Oliver, A.; Gersbach, C.A. The next generation of CRISPR-Cas technologies and applications. Nat. Rev. Mol. Cell Biol. 2019, 20, 490–507. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef] [PubMed]
- Adli, M. The CRISPR tool kit for genome editing and beyond. Nat. Commun. 2018, 9, 1911. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Ni, H.; Xu, Y.; Chen, Q.; Jiang, L. CRISPR/Cas9-mediated base-editing system efficiently generates gain-of-function mutations in Arabidopsis. Sci. China Life Sci. 2017, 60, 520–523. [Google Scholar] [CrossRef]
- Zhan, T.; Rindtorff, N.; Betge, J.; Ebert, M.P.; Boutros, M. CRISPR/Cas9 for cancer research and therapy. Semin. Cancer Biol. 2019, 55, 106–119. [Google Scholar] [CrossRef]
- Hales, K.G.; Korey, C.A.; Larracuente, A.M.; Roberts, D.M. Genetics on the Fly: A Primer on the Drosophila Model System. Genetics 2015, 201, 815–842. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duffy, J.B. GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis 2002, 34, 1–15. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef] [PubMed]
- Zeidler, M.P.; Tan, C.; Bellaiche, Y.; Cherry, S.; Hader, S.; Gayko, U.; Perrimon, N. Temperature-sensitive control of protein activity by conditionally splicing inteins. Nat. Biotechnol. 2004, 22, 871–876. [Google Scholar] [CrossRef]
- Port, F.; Strein, C.; Stricker, M.; Rauscher, B.; Heigwer, F.; Zhou, J.; Beyersdorffer, C.; Frei, J.; Hess, A.; Kern, K.; et al. A large-scale resource for tissue-specific CRISPR mutagenesis in Drosophila. eLife 2020, 9. [Google Scholar] [CrossRef]
- Meltzer, H.; Marom, E.; Alyagor, I.; Mayseless, O.; Berkun, V.; Segal-Gilboa, N.; Unger, T.; Luginbuhl, D.; Schuldiner, O. Tissue-specific (ts)CRISPR as an efficient strategy for in vivo screening in Drosophila. Nat. Commun. 2019, 10, 2113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Port, F.; Bullock, S.L. Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 2016, 13, 852–854. [Google Scholar] [CrossRef] [Green Version]
- Bier, E.; Harrison, M.M.; O’Connor-Giles, K.M.; Wildonger, J. Advances in Engineering the Fly Genome with the CRISPR-Cas System. Genetics 2018, 208, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, X.; Sun, J.; Housden, B.E.; Hu, Y.; Roesel, C.; Lin, S.; Liu, L.P.; Yang, Z.; Mao, D.; Sun, L.; et al. Optimized gene editing technology for Drosophila melanogaster using germ line-specific Cas9. Proc. Natl. Acad. Sci. USA 2013, 110, 19012–19017. [Google Scholar] [CrossRef] [Green Version]
- Ren, X.; Holsteens, K.; Li, H.; Sun, J.; Zhang, Y.; Liu, L.P.; Liu, Q.; Ni, J.Q. Genome editing in Drosophila melanogaster: From basic genome engineering to the multipurpose CRISPR-Cas9 system. Sci. China Life Sci. 2017, 60, 476–489. [Google Scholar] [CrossRef]
- Port, F.; Chen, H.M.; Lee, T.; Bullock, S.L. Optimized CRISPR/Cas tools for efficient germline and somatic genome engineering in Drosophila. Proc. Natl. Acad. Sci. USA 2014, 111, E2967–E2976. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Port, F.; Muschalik, N.; Bullock, S.L. Systematic Evaluation of Drosophila CRISPR Tools Reveals Safe and Robust Alternatives to Autonomous Gene Drives in Basic Research. G3 (Bethesda) 2015, 5, 1493–1502. [Google Scholar] [CrossRef] [Green Version]
- Miguel-Aliaga, I.; Jasper, H.; Lemaitre, B. Anatomy and Physiology of the Digestive Tract of Drosophila melanogaster. Genetics 2018, 210, 357–396. [Google Scholar] [CrossRef] [Green Version]
- Liu, Q.; Jin, L.H. Tissue-resident stem cell activity: A view from the adult Drosophila gastrointestinal tract. Cell Commun. Signal. 2017, 15, 33. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Boutros, M. JNK-dependent intestinal barrier failure disrupts host-microbe homeostasis during tumorigenesis. Proc. Natl. Acad. Sci. USA 2020, 117, 9401–9412. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Valentini, E.; Boutros, M. Microenvironmental innate immune signaling and cell mechanical responses promote tumor growth. Dev. Cell 2021, 56, 1884–1899 e1885. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Patel, P.H.; Kohlmaier, A.; Grenley, M.O.; McEwen, D.G.; Edgar, B.A. Cytokine/Jak/Stat signaling mediates regeneration and homeostasis in the Drosophila midgut. Cell 2009, 137, 1343–1355. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Biteau, B.; Hochmuth, C.E.; Jasper, H. JNK activity in somatic stem cells causes loss of tissue homeostasis in the aging Drosophila gut. Cell Stem Cell 2008, 3, 442–455. [Google Scholar] [CrossRef] [Green Version]
- Patel, P.H.; Dutta, D.; Edgar, B.A. Niche appropriation by Drosophila intestinal stem cell tumours. Nat. Cell Biol. 2015, 17, 1182–1192. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Driver, I.; Ohlstein, B. Injury-induced BMP signaling negatively regulates Drosophila midgut homeostasis. J. Cell Biol. 2013, 201, 945–961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Massironi, S.; Cavalcoli, F.; Rausa, E.; Invernizzi, P.; Braga, M.; Vecchi, M. Understanding short bowel syndrome: Current status and future perspectives. Dig. Liver Dis. 2020, 52, 253–261. [Google Scholar] [CrossRef] [Green Version]
- Dietzl, G.; Chen, D.; Schnorrer, F.; Su, K.C.; Barinova, Y.; Fellner, M.; Gasser, B.; Kinsey, K.; Oppel, S.; Scheiblauer, S.; et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 2007, 448, 151–156. [Google Scholar] [CrossRef]
- Horn, T.; Arziman, Z.; Berger, J.; Boutros, M. GenomeRNAi: A database for cell-based RNAi phenotypes. Nucleic Acids Res. 2007, 35, D492–D497. [Google Scholar] [CrossRef] [Green Version]
- Horn, T.; Boutros, M. E-RNAi: A web application for the multi-species design of RNAi reagents--2010 update. Nucleic Acids Res. 2010, 38, W332–W339. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.Q.; Liu, L.P.; Binari, R.; Hardy, R.; Shim, H.S.; Cavallaro, A.; Booker, M.; Pfeiffer, B.D.; Markstein, M.; Wang, H.; et al. A Drosophila resource of transgenic RNAi lines for neurogenetics. Genetics 2009, 182, 1089–1100. [Google Scholar] [CrossRef] [Green Version]
- Ni, J.Q.; Markstein, M.; Binari, R.; Pfeiffer, B.; Liu, L.P.; Villalta, C.; Booker, M.; Perkins, L.; Perrimon, N. Vector and parameters for targeted transgenic RNA interference in Drosophila melanogaster. Nat. Methods 2008, 5, 49–51. [Google Scholar] [CrossRef]
- Green, E.W.; Fedele, G.; Giorgini, F.; Kyriacou, C.P. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat. Methods 2014, 11, 222–223. [Google Scholar] [CrossRef] [PubMed]
- Larkin, A.; Marygold, S.J.; Antonazzo, G.; Attrill, H.; Dos Santos, G.; Garapati, P.V.; Goodman, J.L.; Gramates, L.S.; Millburn, G.; Strelets, V.B.; et al. FlyBase: Updates to the Drosophila melanogaster knowledge base. Nucleic Acids Res. 2021, 49, D899–D907. [Google Scholar] [CrossRef]
- Whitworth, C. The Bloomington Drosophila Stock Center: Management, Maintenance, Distribution, and Research. In The Biological Resources of Model Organisms; Jarret, R.L., McCluskey, K., Eds.; Taylor & Francis Group: London, UK, 2019; pp. 145–162. [Google Scholar]
- Jiang, W.; Brueggeman, A.J.; Horken, K.M.; Plucinak, T.M.; Weeks, D.P. Successful transient expression of Cas9 and single guide RNA genes in Chlamydomonas reinhardtii. Eukaryot. Cell 2014, 13, 1465–1469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.; Li, S.; Li, X.J. Shortening the Half-Life of Cas9 Maintains Its Gene Editing Ability and Reduces Neuronal Toxicity. Cell Rep. 2018, 25, 2653–2659 e2653. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Zhao, R.; Huang, P.; Yang, F.; Quan, Z.; Xu, N.; Xi, R. APC loss-induced intestinal tumorigenesis in Drosophila: Roles of Ras in Wnt signaling activation and tumor progression. Dev. Biol. 2013, 378, 122–140. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, M.; Petkau, K.; Shin, M.; Galenza, A.; Fast, D.; Foley, E. Differential effects of commensal bacteria on progenitor cell adhesion, division symmetry and tumorigenesis in the Drosophila intestine. Development 2021, 148. [Google Scholar] [CrossRef]
- Suijkerbuijk, S.J.; Kolahgar, G.; Kucinski, I.; Piddini, E. Cell Competition Drives the Growth of Intestinal Adenomas in Drosophila. Curr. Biol. 2016, 26, 428–438. [Google Scholar] [CrossRef] [Green Version]
- Markstein, M.; Dettorre, S.; Cho, J.; Neumuller, R.A.; Craig-Muller, S.; Perrimon, N. Systematic screen of chemotherapeutics in Drosophila stem cell tumors. Proc. Natl. Acad. Sci. USA 2014, 111, 4530–4535. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ngo, S.; Liang, J.; Su, Y.H.; O’Brien, L.E. Disruption of EGF Feedback by Intestinal Tumors and Neighboring Cells in Drosophila. Curr. Biol. 2020, 30, 1537–1546 e1533. [Google Scholar] [CrossRef]
- Cordero, J.B.; Stefanatos, R.K.; Myant, K.; Vidal, M.; Sansom, O.J. Non-autonomous crosstalk between the Jak/Stat and Egfr pathways mediates Apc1-driven intestinal stem cell hyperplasia in the Drosophila adult midgut. Development 2012, 139, 4524–4535. [Google Scholar] [CrossRef] [Green Version]
- Zhai, W.; Lim, T.K.; Zhang, T.; Phang, S.T.; Tiang, Z.; Guan, P.; Ng, M.H.; Lim, J.Q.; Yao, F.; Li, Z.; et al. The spatial organization of intra-tumour heterogeneity and evolutionary trajectories of metastases in hepatocellular carcinoma. Nat. Commun. 2017, 8, 4565. [Google Scholar] [CrossRef] [Green Version]
- Bilder, D.; Ong, K.; Hsi, T.C.; Adiga, K.; Kim, J. Tumour-host interactions through the lens of Drosophila. Nat. Rev. Cancer 2021, 21, 687–700. [Google Scholar] [CrossRef] [PubMed]
- Pandey, U.B.; Nichols, C.D. Human disease models in Drosophila melanogaster and the role of the fly in therapeutic drug discovery. Pharm. Rev. 2011, 63, 411–436. [Google Scholar] [CrossRef] [Green Version]
- Ugur, B.; Chen, K.; Bellen, H.J. Drosophila tools and assays for the study of human diseases. Dis. Model. Mech. 2016, 9, 235–244. [Google Scholar] [CrossRef] [Green Version]
- Bangi, E.; Murgia, C.; Teague, A.G.; Sansom, O.J.; Cagan, R.L. Functional exploration of colorectal cancer genomes using Drosophila. Nat. Commun. 2016, 7, 13615. [Google Scholar] [CrossRef] [PubMed]
- Bangi, E.; Ang, C.; Smibert, P.; Uzilov, A.V.; Teague, A.G.; Antipin, Y.; Chen, R.; Hecht, C.; Gruszczynski, N.; Yon, W.J.; et al. A personalized platform identifies trametinib plus zoledronate for a patient with KRAS-mutant metastatic colorectal cancer. Sci. Adv. 2019, 5, eaav6528. [Google Scholar] [CrossRef] [Green Version]
- Redhai, S.; Boutros, M. The Role of Organelles in Intestinal Function, Physiology, and Disease. Trends Cell Biol. 2021, 31, 485–499. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bahuguna, S.; Redhai, S.; Zhou, J.; Wang, T.; Port, F.; Boutros, M. Conditional CRISPR-Cas Genome Editing in Drosophila to Generate Intestinal Tumors. Cells 2021, 10, 3156. https://doi.org/10.3390/cells10113156
Bahuguna S, Redhai S, Zhou J, Wang T, Port F, Boutros M. Conditional CRISPR-Cas Genome Editing in Drosophila to Generate Intestinal Tumors. Cells. 2021; 10(11):3156. https://doi.org/10.3390/cells10113156
Chicago/Turabian StyleBahuguna, Shivohum, Siamak Redhai, Jun Zhou, Tianyu Wang, Fillip Port, and Michael Boutros. 2021. "Conditional CRISPR-Cas Genome Editing in Drosophila to Generate Intestinal Tumors" Cells 10, no. 11: 3156. https://doi.org/10.3390/cells10113156
APA StyleBahuguna, S., Redhai, S., Zhou, J., Wang, T., Port, F., & Boutros, M. (2021). Conditional CRISPR-Cas Genome Editing in Drosophila to Generate Intestinal Tumors. Cells, 10(11), 3156. https://doi.org/10.3390/cells10113156





