Abstract
The hypothalamus maintains whole-body homeostasis by integrating information from circulating hormones, nutrients and signaling molecules. Distinct neuronal subpopulations that express and secrete unique neuropeptides execute the individual functions of the hypothalamus, including, but not limited to, the regulation of energy homeostasis, reproduction and circadian rhythms. Alterations at the hypothalamic level can lead to a myriad of diseases, such as type 2 diabetes mellitus, obesity, and infertility. The excessive consumption of saturated fatty acids can induce neuroinflammation, endoplasmic reticulum stress, and resistance to peripheral signals, ultimately leading to hyperphagia, obesity, impaired reproductive function and disturbed circadian rhythms. This review focuses on the how the changes in the underlying molecular mechanisms caused by palmitate exposure, the most commonly consumed saturated fatty acid, and the potential involvement of microRNAs, a class of non-coding RNA molecules that regulate gene expression post-transcriptionally, can result in detrimental alterations in protein expression and content. Studying the involvement of microRNAs in hypothalamic function holds immense potential, as these molecular markers are quickly proving to be valuable tools in the diagnosis and treatment of metabolic disease.
Keywords:
palmitate; hypothalamus; energy homeostasis; reproduction; circadian rhythm; microRNAs; leptin; insulin; NPY; POMC 1. Introduction
1.1. Obesity and Palmitate Consumption
The World Health Organization has estimated that nearly 2 billion people worldwide are obese or overweight, which represents a near tripling of the obesity rate since 1975 [1]. Obesity is accompanied by dangerous comorbidities such as type-2 diabetes mellitus, cardiovascular disease, infertility, and some cancers [2,3]. The primary cause of obesity is positive energy balance wherein individuals consume more energy than is expended. Another important factor in the development or predisposition to obesity is the source of nutrients contributing to the positive energy balance. The increase in obesity rates has coincided with a 50% increase in the consumption of fats [4], and diets high in fat (HFD) induce metabolic syndrome in rodents and mice [5]. There are two general categories of fatty acids: saturated fats, which are commonly called “bad” fats, and unsaturated fats, which are considered “healthy” fats. The increase in dietary fat consumption has been primarily in the form of saturated fatty acids, which make up half of the fatty acid content in palm oil, the most commonly consumed cooking oil in the world.
Palmitate is a 16-carbon saturated fatty acid that is crucial for cellular function, since it is a component in the synthesis of membrane phospholipids, transport lipids, and palmitoylated proteins [6]. Although palmitate is important for cellular function and is synthesized endogenously via de novo lipogenesis, the excessive consumption of palmitate can have adverse consequences [7]. Under normal circumstances, palmitate is converted to triglycerides for long-term energy storage, but excess palmitate leads to increased production of potentially detrimental palmitate metabolites [8,9]. Palmitate and its metabolites can induce several forms of cellular stress, including endoplasmic reticulum (ER) stress, oxidative stress, and inflammation [10,11]. This is particularly a problem for the hypothalamus as obese individuals have higher concentrations of palmitate in their cerebrospinal fluid (CSF) [12] and brain [13]. This review will focus on the disruptive effects of palmitate on the hypothalamic control of basic physiological processes, the mechanisms underlying this dysregulation, and the potential role of microRNAs (miRNA) in these effects.
1.2. The Hypothalamus as a Central Homeostatic Regulator
The hypothalamus is a small region of the brain composed of distinct cell populations that play a crucial role in maintaining whole body homeostasis by regulating energy balance, reproduction, and circadian rhythms [14], among other processes. The hypothalamus, in part, links the central nervous system to the endocrine system, as neurons in the hypothalamus communicate with the pituitary gland by releasing hormones into the hypophyseal portal vein [15]. The secreted hormones will then induce or inhibit the secretion of hormones from the pituitary gland. The position of the hypothalamus allows it to sense nutrients, including palmitate, and peripheral signals that pass through the median eminence, a porous region of the blood brain barrier [15].
The hypothalamus is able to orchestrate multiple processes as a result of the heterogeneous populations of neurons that secrete neuropeptides with distinct functions. This inherent heterogeneity makes studying the direct effects of compounds on specific neuronal subpopulations difficult and identifying the underlying mechanisms nearly impossible. Primary cultures permit the study of direct effects of compounds on neurons, but these cultures have short life spans, limited numbers of surviving neurons, and are still heterogeneous in nature [16]. To resolve this, the Belsham lab generated a bank of clonal immortalized hypothalamic cell lines from murine, embryonic and adult hypothalamii. The immortalization process utilized primary cultures of hypothalamic neurons, the addition of ciliary neurotrophic factor (CNTF) to induce proliferation in adult neurons, and transformation with SV40 T-antigen [17]. The immortalized cells underwent selection with geneticin and were subsequently subcloned to generate genetically identical clonal populations that represent a single subpopulation of hypothalamic neurons. Immortalized heterogeneous neuronal populations of neuropeptide subtypes such as mHypoA-POMC/GFP, mHypoA-NPY/GFP, mHypoA-GnRH/GFP, and mHypoA-Kiss/GFP were also generated from the hypothalamii of transgenic mice containing enhanced green fluorescent protein (eGFP) downstream of the promoters of the genes of interest. The cells were sorted using fluorescent activated cell sorting (FACS), thereby selecting all immortalized neurons expressing GFP and as a result the neuropeptide-expressing neurons of interest [16]. These cell models have been used extensively to characterize neuronal function and the molecular events upon sensing cellular signals, such as palmitate [18].
1.3. Mechanisms and Biogenesis of microRNAs
microRNAs (miRNAs) are short (21–23nt) non-coding strands of RNA that complex with and act as guides for the argonaute (AGO) protein to regulate gene expression post-transcriptionally. They target gene transcripts based on complementarity to a 6-nucleotide seed sequence, which is often found at positions 2–7 from the 5′ end of the miRNA [19]. Upon binding to the targeted messenger RNA (mRNA), miRNAs can either inhibit the translation of the mRNA or lead to its degradation. This mechanism allows for the precise control of gene regulation by the cell and can be drastically altered in disease states [20].
The canonical pathway for miRNA synthesis begins with transcription by RNA polymerase II, which transcribes the primary miRNA (pri-miRNA) containing a stem loop structure with the miRNA sequence. After transcription, the pri-miRNA undergoes further processing within the nucleus by Drosha, a nuclear RNase III, which cleaves the pri-miRNA to release the stem-loop structure, known as the pre-miRNA [21]. The pre-miRNA is then exported to the cytoplasm by exportin 5, a ubiquitously expressed exportin that recognizes the double-stranded RNA portion of the stem-loop structure [22]. Once in the cytoplasm, the pre-miRNA is recognized by Dicer, a cytoplasmic RNase III, which cleaves the miRNA duplex from the stem-loop structure [23]. The miRNA duplex is then combined with AGO to form the RNA-induced silencing complex (RISC) and will concomitantly be used to target mRNAs for inhibition.
In addition to the actions that miRNAs have on the cells that produce them, they are also present in the circulation and can affect distant organs. miRNAs are transported through extracellular fluids within exosomes or bound to proteins. Exosomes are membrane-bound extracellular vesicles with a diameter below 100 nm. Interestingly, the quantity and content of exosomes are disrupted in disease states [24], suggesting miRNAs have potential for use as biomarkers [25]. Furthermore, exosomes enriched with miRNAs linked to the development of glucose intolerance and insulin resistance are detected in the blood of obese individuals [26,27]. The injection of exosomes collected from the adipose tissue macrophages of obese mice into lean mice caused glucose intolerance and insulin resistance, whereas the reverse improved insulin sensitivity in obese mice [28]. Taken together, these results suggest that secreted miRNAs present in exosomes likely mediate disease processes.
miRNAs have been predicted to play a pivotal role in gene regulation, as studies have estimated that as many as 60% of protein coding genes in the human genome may be targeted by miRNAs [29]. Furthermore, miRNAs are important for survival, as the global knockout of Dicer or DGCR8, a cofactor for Drosha function, leads to embryonic mortality [30,31]. The conditional knockout of Dicer in the mouse brain or in hypothalamic POMC neurons leads to hyperphagic obesity [32,33], demonstrating the importance of miRNAs in energy homeostasis. The dysregulation of miRNA expression in disease states indicates distinct miRNA expression profiles for individuals with obesity, type 2 diabetes mellitus, and cancers [34,35,36]. Of interest to this review, multiple miRNA array studies have shown that HFD exposure in mice can induce distinct miRNA expression profiles in the hypothalamus [37,38]. With the emerging potential of miRNAs to be used as biomarkers and therapeutics, studies exploring the physiological effects of miRNAs in the hypothalamus may provide a new class of tools to diagnose and target hypothalamus-associated metabolic disease. Hence, this review will include a perspective on miRNAs involved in hypothalamic function, and how palmitate exposure may alter these mechanistic pathways.
2. Energy Homeostasis
Energy homeostasis is primarily controlled in the arcuate nucleus (ARC) of the hypothalamus by two opposing neuronal populations, the orexigenic, appetite inducing, neuropeptide Y (NPY)/agouti related peptide (AgRP) expressing neurons and the anorexigenic, appetite suppressing, proopiomelanocortin (POMC) expressing neurons. The ARC is situated directly above the median eminence allowing it to sense peripheral signals and nutrients in the circulation. These neuronal populations synthesize and secrete their respective feeding neuropeptides in response to nutritional status communicated via peripheral hormones, nutrients and signaling molecules. The neuropeptides primarily act on second order neurons in the paraventricular nucleus (PVN) and the opposing neuronal population in the ARC to control energy homeostasis. NPY and AgRP induce feeding and inhibit the activity of POMC neurons [39], whereas POMC neurons, secreting α-melanocyte stimulating hormone (α-MSH) have the opposite effect, suppressing food intake and inhibiting NPY/AgRP neurons by activating melanocortin receptors, MC3R and MC4R [39]. This system is drastically altered in obesity as the neurons become resistant to peripheral satiety signals, such as insulin and leptin [40], leading to excessive food intake, weight gain, and metabolic disease [41].
2.1. Hypothalamic Insulin Signaling Is Impaired by Palmitate
2.1.1. Insulin Signaling in the Hypothalamus
Although initially debated, the critical role of hypothalamic insulin action in regulating whole body energy homeostasis is now uncontested [42,43]. Pancreatic beta cells are the primary source of insulin; they secrete insulin in response to elevated blood glucose levels following a meal, allowing peripheral tissues to utilize or store the glucose [44]. Insulin travels through the circulation and is sensed by the hypothalamus at the median eminence [43]. Insulin is an anorexigenic hormone, and its hypothalamic actions lead to reduced food intake, and ultimately body weight, in rodents and primates [45,46]. Besides these anorexigenic effects, central insulin signaling in AgRP neurons is essential for suppression of hepatic gluconeogenesis [47], and in POMC neurons, it is necessary for the insulin-induced suppression of lipolysis in adipose tissue [48]. These actions are mediated by the ability of insulin to suppress orexigenic NPY/AgRP, while promoting anorexigenic POMC/α-MSH [49,50,51], which act on second-order neurons located in the PVN and other hypothalamic nuclei. Ultimately the actions of insulin in the hypothalamus and peripheral tissues achieve the same goal, which is to ensure glucose homeostasis after a meal [44].
Mechanistically, insulin acts via the insulin receptor (InsR), which dimerizes upon insulin binding and undergoes autophosphorylation of tyrosine residues, followed by recruitment and activation of insulin receptor substrate (IRS) proteins. Insulin actions occur primarily through two downstream signaling pathways. The first pathway mediates the effect of insulin on metabolism and begins with the activation of phosphoinositide 3-kinase (PI3K) by IRS. PI3K assists in the synthesis of PIP3, and PIP3 activates Akt via PDK1. Akt then phosphorylates forkhead box protein O1 (FOXO1) causing its expulsion from the nucleus, resulting in the anorexigenic actions of insulin signaling, which is to suppress Npy and Agrp and promote Pomc transcription. Akt also activates the mTOR signaling pathway, leading to the inhibition of AMP-activated protein kinase (AMPK), a protein kinase that is activated in low energy states to replenish ATP levels (Figure 1). The second pathway mediates the effect of insulin on cell proliferation and differentiation and involves mitogen-activated protein kinase kinase(MEK)/extracellular signal-regulated kinase (ERK)1/2 activation [44]. A commonality between insulin and leptin signaling is PI3K, which integrates anorexigenic signals from both pathways [52] and is discussed further later in this review.
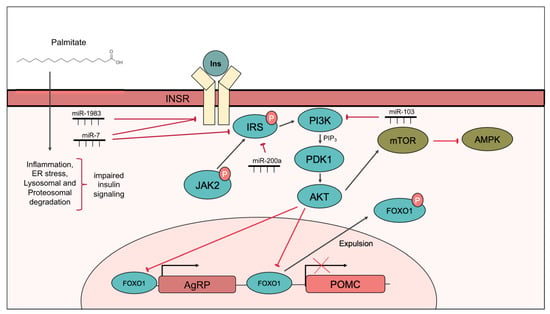
Figure 1.
Hypothalamic insulin signaling. Insulin binds to insulin receptor on the cell membrane, leading to the phosphorylation of IRS, which in turn activates PI3K, to generate PIP3 that activates Akt via PDK1. Akt phosphorylates FOXO1 leading to its expulsion from the nucleus, leading to decreased Npy and Agrp and increased Pomc expression. Akt also activates mTOR signaling. Palmitate impairs insulin signaling by inducing, inflammation, ER stress, and lysosomal and proteasomal degradation of INSR or IRS. Components of the insulin signal transduction pathway are targeted by miRNAs include. INSR is targeted by miR-1983 and miR-7, IRS is targeted by miR-7 and miR-200a, and PI3K is targeted by miR-103.
2.1.2. Hypothalamic Insulin Resistance
A hallmark of pre-diabetes and type II diabetes is an inability to respond appropriately to insulin. This phenomenon, known as insulin resistance, results from chronically elevated insulin levels, termed hyperinsulinemia, and desensitization of the insulin signaling pathway, ultimately resulting in improper energy balance. High calorie diets induce constant insulin secretion and activation of InsR, leading to its downregulation as well as desensitization of downstream signaling molecules [43,44]. Excess insulin-induced insulin resistance has been demonstrated in several hypothalamic neuronal models, including NPY/AgRP- and POMC-expressing neurons [51,52,53]. Specifically, insulin treatment in the mHypoE-46 cell line, an NPY/AgRP-expressing model, downregulated Npy and Agrp expression in the immediate term, as expected [50]. However, high insulin exposure (100 nM) for as little as 8 h downregulated IRβ, IRS1 and diminished subsequent insulin-stimulated Akt phosphorylation [53]. mTOR-S6K1-mediated phosphorylation of IRS-1, and degradation of IRS-1 and InsR via the proteasomal or lysosomal pathway, respectively, were implicated in the mechanism of insulin-induced insulin resistance in these NPY/AgRP-expressing cells [53].
2.1.3. Induction of Central Insulin Resistance by Palmitate
Saturated fats can also directly induce insulin resistance in peripheral and hypothalamic cells in the absence of high insulin levels. Palmitate induces inflammatory responses, facilitated by nuclear factor kappa B (NFκB) activation, which is the primary mediator of HFD-induced insulin resistance in peripheral tissues, including adipocytes, muscle and liver [54]. Given evidence that hypothalamic neuroinflammation occurs within one day of HFD exposure, prior to inflammation in peripheral tissues, [55] and central insulin signaling is required for peripheral insulin action [42,47,48], fat-induced hypothalamic insulin resistance merits attention. In fact, intracerebroventricular (ICV) administration of palmitate impairs hypothalamic insulin signaling and leads to disruptions in hepatic glucose production and peripheral glucose metabolism in rodents [56,57]. Knockdown of protein kinase C theta in the ARC or toll-like receptor 4 (TLR4)-adaptor molecule, MyD88, in the CNS prevented HFD- or ICV palmitate-induced weight gain and insulin resistance [56,57], highlighting some of the mechanisms of palmitate-induced central insulin resistance. In the NPY/AgRP-expressing mHypoE-44 cell line, palmitate pre-treatment diminished the response to an insulin challenge, as demonstrated by reduced pAkt activation [58]. This did not occur in POMC-expressing mHypoA-POMC/GFP-1 cells [59], suggesting a protective mechanism in the POMC-expressing cells. The proinflammatory cytokine tumour necrosis factor alpha (TNFα) also induces insulin resistance in NPY/AgRP neurons [60]. Thus, along with the involvement of MyD88, a molecule that is essential for the generation of pro-inflammatory cytokines [57], these results strongly implicate neuroinflammation as a mediator of palmitate-induced insulin resistance. In contrast to palmitate, Amine et al. recently described that the polyunsaturated fat docosahexaenoic acid (DHA) did not induce pro-inflammatory cytokine production nor lead to hypothalamic insulin resistance in a human neuroblastoma cell line [61]. Our studies in vitro also suggest that DHA is protective against TNFα-induced neuroinflammation through GPR120 [62], implicating DHA as a protective fatty acid against the effects of palmitate.
Alternative mechanisms by which palmitate can induce insulin resistance include AMPK inhibition and ER stress. Treatment with an AMPK activator, aminoimidazole carboxamide ribonucleotide (AICAR), prevented palmitate-induced phosphorylation of cJun N-terminal kinase (JNK) and restored insulin signaling in the mHypoE-44 cells [58]. Indeed, activation of AMPK prevented hyperglycemia in insulin-resistant, leptin-deficient mice [63], and improved glucose tolerance in insulin-resistant Zucker rats [64]. This protective phenomenon is thought to result from the ability of AMPK to increase fatty acid oxidation, thereby decreasing fatty acid levels [65], or to inhibit mTOR signaling, as overactive mTOR is related to insulin resistance [66]. ER stress, resulting from the accumulation of misfolded proteins in the ER lumen, is also induced with palmitate and may play a role in palmitate-mediated hypothalamic insulin resistance [43,58]. To summarize, neuroinflammation, AMPK inhibition/mTOR activation and ER stress are involved in the mediation of hypothalamic insulin resistance by palmitate [43]. These processes involve signaling proteins that converge on pathways activated by insulin, resulting in modified signaling. As an example, palmitate upregulates SOCS3, as a result of nuclear factor κ B (NFκB) activation, subsequently leading to IRS1 degradation and prevention of InsR auto-phosphorylation. Inhibition of the NFκB pathway with PS1145 in turn reduced food intake and diminished hypothalamic insulin resistance in mice fed a HFD [67], identifying a targetable pathway to restore hypothalamic insulin signaling.
2.1.4. Role of miRNAs in Hypothalamic Insulin Signaling and Resistance
The role of miRNAs in mediating insulin signaling and resistance and their potential ability to therapeutically prevent or reverse insulin resistance is an area of increasing interest. Of note, there is evidence hypothalamic miRNAs play an important role in insulin action and whole-body energy homeostasis. Firstly, hypothalamic knockout of Dicer leads to overactivation of the PI3K/Akt/mTOR pathway, akin to the effects of high insulin levels, suggesting miRNAs may serve to prevent overactivation of insulin signaling [68]. Specifically, miR-103 administration to mice lacking hypothalamic Dicer prevented overeating and obesity [68]. miR-103 targets two components of the insulin signaling pathway: Pik3cg, a catalytic subunit of PI3K, and IRS1 [68,69] (Figure 1). Overexpression of Lin28a in the hypothalamus improved glucose tolerance and insulin sensitivity in HFD-fed mice, suggesting the importance of hypothalamic miRNAs in whole body insulin sensitivity [70]. Components of insulin and leptin signaling often crosstalk and therefore miRNAs can affect both pathways. An example of this is miR-200a, a miRNA upregulated in the hypothalamus of ob/ob mice and targets both IRS2 (Figure 1) and LepR in the hypothalamus [71]. ICV administration of miR-200a antagomir in ob/ob mice increased the expression of insulin receptor in NPY and POMC neurons [71]. Because miRNAs can target hundreds of genes, there resides a potential symphony of intracellular events that target multiple pathways, ultimately leading to metabolic disease.
Two miRNAs, miR-1983 and miR-7, have emerged as miRNAs associated with neuronal insulin resistance that directly target the InsR (Figure 1). Treatment of mHypoE-46 neurons with high levels of insulin for 24 h identified miR-1983 as a candidate that is induced with insulin resistance in hypothalamic neurons. Exposure to miR-1983 mimics downregulated InsR β-subunit protein levels and target analysis identified a binding site for miR-1983 in the 3′UTR of IRβ [72]. miR-1983 levels were also increased in the hypothalamus of MKR mice [72], a non-obese model with impaired insulin signaling and hyperinsulinemia [73], and was positively correlated to insulin and homeostatic model for insulin resistance (HOMA-IR) scores in human serum samples, representing their potential for use as a biomarker. miR-7 is a miRNA that is highly abundant in the hypothalamus, along with let-7c and miR-9 [74]. miR-7 was elevated in mice fed a HFD, and insulin induced the miR-7-expressing intron and its parent gene heterogeneous nuclear ribonucleoprotein K. Interestingly, miR-7 targets multiple components of the insulin signaling pathway, including the InsR, IRS2, and insulin-degrading enzyme (IDE) [75], suggesting potent downregulation of insulin signaling in the presence of this miRNA. However, knockdown of miR-7 in POMC neurons of female mice exacerbated diet-induced obesity [76], suggesting that the neuronal subtype and sex differences play an important role in the function of miR-7. Antagonizing both miR-1983 and miR-7 in specific neuronal subtypes may have therapeutic potential as preventing the downregulation of the InsR is one potential way to combat hypothalamic insulin resistance. Future studies investigating the combined effects of several of these insulin-related miRNAs may provide avenues for combating multiple aspects of central insulin resistance.
2.2. Leptin Signaling Is Impaired by Palmitate
Leptin is an anorexigenic circulating hormone involved in the hypothalamic control of appetite. It is predominately synthesized in adipocytes and secreted into the circulation. The circulating concentration of leptin increases acutely after a meal to suppress appetite and the levels are proportional to the amount of fat mass [77,78]. Mutation of the leptin receptor (LepR), whereby a longer defective variant is expressed, causes hyperphagia, leading to obesity [79]. The restoration of leptin signaling in the central nervous system alone is able to restore the appetite suppressing effects of leptin and reverse the obese phenotype in LepR-deficient mice [80]. Obese individuals typically have chronically elevated levels of circulating leptin as a result of greater fat mass [78]. Despite this, the anorexigenic effects of leptin is lost in obese individuals, a phenomenon known as leptin resistance [81]. The primary cause of leptin resistance has not been identified but has been hypothesized to be caused by either impaired leptin transport across the blood brain barrier or the continuous activation of a negative feedback loop in the leptin signaling pathway [82,83].
2.2.1. The Mechanisms of Hypothalamic Leptin Signaling
Leptin achieves its appetite suppressing effects primarily via LepR signaling in the ventromedial hypothalamus and the ARC, which are involved in the control of feeding [84]. In anorexigenic POMC expressing neurons, leptin induces the transcription of Pomc mRNA and the secretion of α-MSH [85,86]. In contrast, leptin suppresses the transcription and secretion of NPY and AgRP in orexigenic neurons [87,88]. Leptin signaling begins with the phosphorylation of janus kinase 2 (JAK2), which in turn phosphorylates signal transducer and activator of transcription 3 (STAT3), leading to its translocation to the nucleus, where it activates Pomc transcription and inhibits Npy and Agrp transcription [89,90,91,92]. Phosphorylated STAT3 also promotes the transcription of suppressor of cytokine signaling 3 (SOCS3), which in turn inhibits JAK2 activity [93], acting as a negative feedback loop to prevent chronic activation (Figure 2). Thus, a potential mechanism for the development of leptin resistance is the continuous transcription and activation of SOCS3 [93]. Leptin signaling can also crosstalk with the insulin signaling pathway, as JAK2 can lead to the phosphorylation of FOXO1 via IRS1/PI3K/Akt signaling [94]. The induction of insulin resistance in rat- and mouse-derived immortalized neurons also attenuates leptin signaling and its effects on gene expression [52]. Thus, there is a complex relationship between signaling components in neurons that require further investigation and disentangling of their roles in hypothalamic function.

Figure 2.
Hypothalamic leptin signaling. Leptin binds to LepR on the cell membrane, inducing the phosphorylation of JAK2, which in turn phosphorylates STAT3, and causes the translocation of STAT3 to the nucleus. In the nucleus, p-STAT3 induces the expression of Pomc and Socs3 and represses the expression of Npy, miR-488, and miR-384-3p. SOCS3 negatively feeds back to inhibit JAK2 phosphorylation. miR-488 and miR-384-3p target the 3′-UTR of Pomc mRNA and inhibit its expression. miR-200a targets the 3′-UTR of LepR, inhibiting its expression and reducing leptin sensitivity. Socs3 is targeted by miR-19a and can enhance JAK2-STAT3 signaling. Palmitate directly induces leptin resistance by inducing inflammation and ER stress, resulting in the activation of PTP1B and SOCS3. Excess palmitate can also induce leptin resistance by chronically increasing circulating leptin, leading to overactivation of the signaling pathway.
2.2.2. The Induction of Central Leptin Resistance by Palmitate
Leptin resistance is caused primarily by chronically high circulating levels of leptin. This has been demonstrated in the mHypoA-NPY/GFP cell line, an adult murine-derived Npy-expressing neuronal model, as an 8 h leptin exposure attenuates subsequent leptin-induced suppression of NPY secretion [95]. Hypothalamic exposure to saturated fatty acids has also been shown to induce leptin resistance. For example, ICV administration of palmitate in C57BL/6J mice led to central leptin resistance, as the appetite suppressing effects of leptin were lost and the phosphorylation of JAK2 and STAT3 were attenuated [96]. This coincided with an increase in the proinflammatory genes TNFα, interleukin 6 (IL6), and Interleukin 1 beta (IL1-β) [96]. Indeed, neuroinflammation has been implicated in the development of leptin resistance, as the activation of NFκB signaling, the hallmark regulator of proinflammatory cytokines, leads to an induction of SOCS3 and protein tyrosine phosphatase 1B (PTP1B), which are both negative regulators of leptin signaling [97,98]. Similar to neuroinflammation, ER stress induced by palmitate can lead to leptin resistance via PTP1B, and conversely, relieving ER stress with chemical chaperones, increased leptin sensitivity in diet-induced obese mice [99,100,101]. Overall, these findings demonstrate that exposure to palmitate can directly induce central leptin resistance via modifying proteins involved in leptin signaling.
2.2.3. The Role of miRNAs in Leptin Signaling and Resistance
The role of miRNAs in leptin signaling is still a new area of investigation, but the importance of their involvement has been established. The conditional knockout of Dicer in POMC neurons, resulted in increased leptin sensitivity, evident from the greater suppression of food intake by leptin and a reduction in food intake overall [102]. Furthermore, miR-200a and miR-200b have been shown to directly target leptin mRNA in yellow catfish [103]. In mice, miR-200a targets the 3′-UTR of LepR and has been shown to be regulated by leptin itself, as mice with deficient leptin signaling have increased hypothalamic miR-200a expression and central leptin administration decreases it [71]. Therefore, an induction of miR-200a by exogenous compounds, including dietary fats, may lead to reduced leptin sensitivity and blocking mir-200a may serve as a tool to relieve leptin resistance. miRNAs can also target components involved in the leptin signaling pathway, such as SOCS3, which is targeted by miR-19a [104]. Administration of miR-19a can therefore enhance leptin sensitivity by relieving inhibition on JAK-STAT signal transduction and warrants further investigation. A group of conserved miRNAs have been shown to mediate the effects of leptin signaling in POMC neurons. The miRNAs of interest, miR-383, miR-384-3p, and miR-488, target the 3′ UTR of the Pomc mRNA in the mHypoA-POMC/GFP-1 cell line [105] (Figure 2). The expressions of these miRNAs in the hypothalamus were dependent on leptin, as they were downregulated in response to leptin administration and were increased in leptin-deficient ob/ob mice [105]. The potential role of miRNAs in mediating leptin response or resistance in NPY/AgRP-expressing neurons remains to be explored.
2.3. Feeding Neuropeptides
2.3.1. Palmitate-Induced Changes in Neuropeptide Expression
In addition to modifying the response of hypothalamic neurons to peripheral signals, palmitate can also directly affect the expression of feeding neuropeptides. In the mHypoA-POMC/GFP-2 cell line, an adult murine derived Pomc-expressing neuronal model, exposure to palmitate increased Pomc mRNA expression and induced a myriad of neuroinflammatory and ER stress markers [59]. This induction of Pomc mRNA by palmitate was independent of neuroinflammation as neither inhibition of TLR4 nor NF𝜅B signaling were able to block the effects of palmitate, but it was dependent on palmitate metabolism to palmitoyl-coA and activation of MAP kinases, JNK and ERK [59]. Though the effects of palmitate on Pomc and inflammatory marker gene expression were not dependent on each other, the monounsaturated fatty acid oleate, which has been shown to block the effects of palmitate in multiple different tissues, blocked changes in the mRNA expression of Pomc, inflammatory and ER stress markers [59]. In the mHypoE-44 and mHypoE-46 cell lines, which are embryonic-derived Npy-expressing neuronal models, exposure to palmitate increased Npy mRNA expression, which was attenuated by PS1145, an inhibitor of IKK [106,107,108], illustrating a role for neuroinflammation in the alteration of Npy expression. The development of chronic, low-grade neuroinflammation in vivo has been suggested to play a significant role in altering feeding neuropeptide expression [106,109]. Studies in the mHypoE-46 cell line demonstrate that exogenous administration of TNFα and visfatin, proinflammatory cytokines induced by palmitate, induce Npy expression [106,110]. However, the exogenous administration of other palmitate-induced pro-inflammatory cytokines, Macrophage migration inhibitory factor (MIF) and IL-17F, were unable to affect Npy expression in the mHypoE-46 cell line [110]. Whereas PS1145 was only able to partially block the induction of Npy by palmitate [106], inhibition of acyl-coA synthetase completely blocks the increase in the mHypoE-46 cells [110], suggesting that the palmitate-mediated effects of Npy occur primarily through the metabolism of palmitate to ceramides and certain phospholipid species and is only partially dependent on neuroinflammation [110]. Furthermore, Npy dysregulation by palmitate in the mHypoE-44 cell line coincides with disruptions in the cyclic expression of circadian rhythm genes [108], including brain and muscle ARNT-like 1 (BMAL1), an essential mediator of palmitate-induced Npy upregulation [111]. This topic is discussed further in subsequent sections. Agrp mRNA expression is induced with palmitate in the mHypoE-41 cell line at 4 h and was blocked by siRNA-mediated knockdown of autophagy-related gene 5 (Atg5) [112]. In mHypoE-46 cells, palmitate induced Agrp mRNA expression at 16 h, which was blocked by metformin and salicylate treatment [107]. Furthermore, Agrp mRNA expression was unaffected by the proinflammatory cytokine TNFα, demonstrating the differential regulation of Agrp and Npy by pro-inflammatory mediators [107]. Taken together, palmitate is able to directly disrupt the expression of feeding neuropeptides in hypothalamic neurons via the induction of MAP kinases or inflammatory signaling, increasing palmitate metabolites, altering circadian transcription factors, and modifying autophagy.
2.3.2. The Control of Feeding Neuropeptides by miRNAs
Research on miRNAs directly targeting feeding neuropeptides is severely limited. A total of three miRNAs have been identified to directly target the 3′ UTR of Pomc mRNA, miR-383, miR-384-3p, and miR-488 [105]. There are currently no published studies identifying miRNAs that directly target Npy or Agrp. miRNAs do not need to directly target a gene to affect its expression, as they can target components involved in the regulation of the gene, such as transcription factors and signal transduction molecules. Transcription factors involved in the regulation of Npy include cAMP response element binding protein (CREB), octamer transcription factor 1 (OCT1), and BMAL1 [113,114,115,116]. CREB, a transcription factor that positively regulates Npy expression, is targeted by several miRNAs, including miR-22-3p, miR-26a-5p, miR-27a-3p, miR-221-3p, miR-4474-3p, and miR-4717-3p [117,118]. A miRNA-induced decrease in CREB would likely lead to a downstream decrease in Npy expression. miR-155 has two binding sites in the 3′UTR of Bmal1, a transcription factor crucial for the rhythmic expression of Npy [119]. Furthermore, miR-155 is induced by inflammatory signaling and is increased in the adipose of obese humans [119,120]. A miRNA-induced decrease in Bmal1 expression could result in the loss of rhythmic Npy expression, similar to what is seen in BMAL1-KO mice. Our lab recently identified miR-708-5p as a miRNA involved in Npy regulation, as transfection of the mHypoA-59 cell line with miR-708-5p mimic increased Npy mRNA [121]. This increase in Npy may be the result of miR-708-5p-mediated downregulation of its target neuronatin (NNAT), a putative sarco/endoplasmic reticulum Ca2+ inhibitor [122]. This hypothesis is supported by the fact that knockout of NNAT resulted in increased Npy expression in the ARC and an increased propensity to develop obesity in mice [123]. Transcription factors involved in the regulation of Agrp include Kruppel-like factor 4 (KLF4), activating transcription factor 3 (ATF3), STAT3, and FOXO1 [90,124,125,126]. KLF4, a transcription factor crucial for development and a positive regulator of Agrp, is targeted by two miRNAs, miR-206 and miR-145 [127,128]. The expression of miR-206 is increased 6-fold in the brain after a 20-week HFD [129], suggesting the potential involvement of miR-206 and KLF4 in palmitate-mediated Agrp induction. ATF3 is a stress induced transcription factor that induces Agrp transcription [130] and is targeted by at least three miRNAs, miR-27a-3p, miR-488, and miR-222 [131,132,133]. miR-222 is found in serum exosomes primarily produced by the gonadal white adipose tissue and is elevated in the serum of obese patients [134], implying a far-reaching mechanism by which palmitate can act. As with all miRNAs that target a positive regulator of a gene of interest, the miRNA-induced downregulation would cascade to a downregulation of the gene of interest. Thus, although a variety of miRNAs that target components involved in feeding neuropeptide regulation have been identified, there is still much to be done with respect to miRNAs that directly target Npy, Agrp, and Pomc, as many have been predicted to do so with in silico analysis.
3. Reproduction
Infertility occurs in 10% of women worldwide and often accompanies obesity [135]. Specifically, overweight and obese women are three times more likely to experience infertility [136,137] and the odds ratio of infertility increases with increasing body mass index (BMI) in men [138]. Even in cases of successful pregnancies, overweight and obesity is a main risk factor for gestational diabetes, occurring in up to 20% of pregnant women in Canada, the complications not only affecting the mother, but reaching adult offspring [139]. As such, consumption of an HFD impacts reproductive and offspring health, with evidence of excess fats disrupting multiple aspects of the hypothalamic-pituitary-gonadal (HPG) axis [136,140], the master regulator of reproductive function. In this section, we describe the effects of palmitate on the hypothalamic cells that initiate the HPG axis.
3.1. Hypothalamic-Pituitary-Gonadal Axis
Gonadotropin releasing hormone (GnRH) neurons of the hypothalamus are the main regulators of reproductive function, and disruptions to these neurons leads to infertility and improper development [141]. These neurons are primarily located within the medial preoptic nucleus of the hypothalamus but receive inputs from surrounding neuronal populations. At the anterior pituitary, GnRH induces the release of luteinizing hormone (LH) and follicle stimulating hormone (FSH) into circulation. LH and FSH then travel to the gonads, triggering the production and secretion of sex steroid hormones, estrogen, testosterone, and progesterone. The expression and secretion of GnRH are tightly regulated by afferent neuropeptides, satiety signals, hormones, and stress.
Kisspeptin (KISS1) is a key reproductive peptide that regulates GnRH expression. The importance of KISS1 in reproduction cannot be understated as whole-body knockouts of KISS1 or its receptor, GPR54/KISS1r, lead to hypogonadotropic hypogonadism, a condition where little to no sex hormones are produced, resulting in infertility and the loss of puberty [142,143]. The role of KISS1 in reproduction can be traced to the hypothalamus as the conditional knockout of KISS1r in GnRH neurons or KISS1 in hypothalamic neurons is enough to induce an infertile phenotype [144,145]. KISS1 neurons are primarily located in the ARC and anteroventral periventricular nucleus (AVPV) of the hypothalamus allowing integration of nutrient signals into their actions [146]. ARC KISS1 induces the secretion of GnRH (Figure 3), leading to pulsatile LH and FSH secretion required for the onset of puberty and maintenance of reproductive function [147,148,149], while AVPV KISS1 evokes the pre-ovulatory GnRH and LH surge [141]. Phoenixin (PNX) is another recently discovered hypothalamic peptide that afferently controls GnRH expression. siRNA knockdown of Pnx in rats delayed the start of the estrous cycle by 2.3 days [150], highlighting its role in maintaining proper reproductive function. In hypothalamic neuronal models, PNX bound to GPR173 receptors and increased Gnrh mRNA, GnRH secretion, Gnrh receptor (Gnrh-r) mRNA and Kiss1 mRNA [151]. GnRH-, KISS1- and PNX-expressing neurons are all subject to HFD- or palmitate-induced dysregulation as described below.
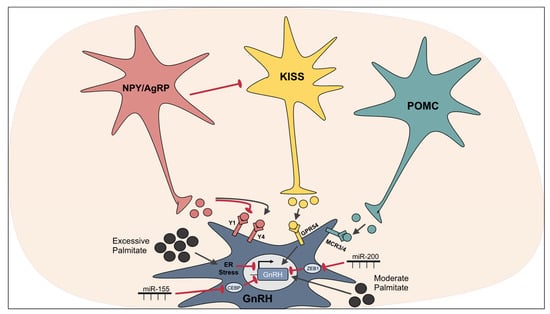
Figure 3.
Regulation of GnRH neurons. NPY differentially affects GnRH neuron excitability depending on receptor variant availability; activation of Y1 is inhibitory and Y4 is excitatory. AgRP indirectly affects GnRH neurons by inhibiting afferent KISS1 neurons. KISS1 neurons secrete KISS1 which activates GPR54/KISS1r on GnRH neurons, leading to increased excitability and GnRH transcription. POMC neurons secrete α-MSH, which acts via MC3R and MC4R to increase GnRH neuron excitability. Moderate concentrations of palmitate induce Gnrh transcription, whereas high concentrations of palmitate induce ER stress and inhibit Gnrh transcription. miR-155 and miR-200 increase Gnrh mRNA by targeting Gnrh repressors CEBP and ZEB1, respectively.
3.2. The Integration of Energy Homeostasis in the Control of Reproduction
Reproduction is a costly process that requires an appropriate nutritional status to succeed. As such, it is imperative that reproductive neurons integrate satiety signals from feeding neurons, including NPY/AgRP and POMC neurons. This requirement for proper energy homeostasis is emphasized by the fact that both malnourished and obese individuals often suffer from reproductive dysfunction [137,152].
Orexigenic NPY and AgRP signals are able to directly and indirectly affect GnRH neurons. NPY also suppresses KISS1 neuron activity in the ARC, thereby acting indirectly on GnRH through its afferents [153]. The overall directionality and magnitude of the effects of NPY on GnRH neurons is highly variable and dependent on the receptor subtypes present, NPY can activate or inhibit the reproductive axis. In contrast to the effects of NPY, the direct effects of AgRP on GnRH neuron excitability are minimal [154]. Instead, AgRP neurons may act on GnRH neurons indirectly via KISS1 neurons. Padilla et al. demonstrated that overactivation of AgRP neurons decreased fertility through a γ-aminobutyric acid (GABA)-dependent inhibition of KISS1 neurons [155]. The anorexigenic POMC neurons release α-MSH, which exerts an excitatory effect on GnRH neurons via MC3R and MC4R [154,156]. This excitatory effect of α-MSH on GnRH neurons may be a signal indicating sufficient nutrients are available for reproduction (Figure 3). Taken together, whether direct or indirect, communication between GnRH and feeding neurons is essential for proper reproductive function, and disruptions to feeding neurons as described previously may also impair reproductive function.
In addition to receiving input from feeding neurons, GnRH neurons also contain receptors for peripheral satiety signals, such as insulin and leptin, and as a result can become insulin- or leptin-resistant. However, the presence of insulin signaling in GnRH neurons seems to be dispensable as mice with conditional knockout of the InsR in GnRH neurons develop normally and are fertile [157]. This is similar for leptin signaling, as mice with conditional knockout of LepR in GnRH neurons are fertile but do have a slight delay in the onset of puberty [158]. Thus, these peripheral satiety signals may relay their signals primarily through feeding-related neurons that project onto GnRH neurons as discussed above. KISS1 neurons may also relay this information as circulating leptin levels are correlated with circulating KISS1 levels and leptin is able to increase Kiss1 mRNA expression [159,160]. In addition to sensing peripheral hormones, GnRH neurons can also directly sense nutrients such as glucose [161] and fatty acids [9]. Overall, these studies demonstrate the ability of reproductive neurons to integrate energy status and draws attention to probable mechanisms of HFD- and palmitate-induced disruption of hypothalamic reproductive neurons.
3.3. Palmitate-Mediated Disruption of Hypothalamic Reproductive Control
As discussed, obesity in both men and women is associated with a greater incidence of infertility [136,138], however this association can be confounded by genetic and other lifestyle factors. Fertility complications in rodents consuming an HFD have delineated direct effects of an HFD on reproductive function. For example, female rats fed an HFD had increased incidences of irregular estrous cycles after just 4 months on the diet and showed accelerated follicle loss [162]. HFD-fed male mice had functional sperm but reduced sperm counts [163]. At the pituitary, the effects of HFD on LH secretion are highly variable, with some studies observing an increase in LH, whereas others reported a decrease or no change [164,165,166,167]. Nevertheless, the majority of studies identify some dysregulation of the HPG axis, resulting in detrimental effects on reproductive function.
GnRH neurons can directly sense fatty acids as illustrated by in vitro studies. In the mHypoA-GnRH/GFP cell line, exposure to 50 to 100 µM palmitate increased Gnrh mRNA, which was mechanistically linked to palmitoyl-coA synthesis and increased PI3K signaling [9]. In the GT1-7 cell line, treatment with 500 or 1000 µM palmitate decreased Gnrh mRNA and this effect was seemingly the result of ER stress, which is commonly observed among obese individuals [168]. These dose-dependent effects of palmitate demonstrate the fatty acid sensing capabilities of GnRH neurons: lower concentrations of palmitate likely signals that sufficient nutrients are available for reproduction, thereby increasing Gnrh, but higher concentrations cause cellular stress and dysfunction and dampen Gnrh as may be seen in obesity. Inflammatory cytokines have been reported to decrease Gnrh expression and may represent a mechanism of HFD-mediated impairment of the HPG axis [169]. For example, HFD-fed rabbits had decreased GnRH and KISS1r immunopositivity alongside increased microglial activation and hypothalamic inflammatory cytokine expression [170]. Furthermore, ovariectomized female C57BL/6J mice become obese, but are protected against changes in the HPG axis. This is remarkably accompanied by a lack of microglial activation and inflammatory cytokine production as well as an increase in anti-inflammatory IL-10 levels in the hypothalamii of these female mice despite the lack of ovarian estrogen. These studies strongly tie together hypothalamic inflammation and reduced Gnrh expression [171].
In addition to the direct effects of palmitate on GnRH neurons, palmitate can also alter the function of neurons that afferently regulate GnRH neurons, including feeding neuropeptide expressing- and PNX-expressing neurons. Treatment of the mHypoE-46 cell line with palmitate increased the mRNA expression of Pnx, which has been shown to positively regulate GnRH expression and function [172]. However, palmitate also reduced the expression of the PNX receptor, Gpr173, via a p38-mediated mechanism and prevented PNX-induced upregulation of pCREB [173], suggesting potential dampening of the HPG axis. Although Kiss1 and Kiss1r expression is reduced in the hypothalamus of male mice fed a HFD for 19 weeks [174], the direct effects and mechanisms of palmitate action on KISS1 neurons remain to be studied. Overall, palmitate alters hypothalamic reproductive neurons via direct actions on Gnrh expression or through the regulation of its afferents. How these effects can be combatted to prevent fertility complications in obesity remains an important question.
3.4. miRNAs Involved in the Hypothalamic Control of Reproduction
miRNAs may become useful tools in the study of palmitate-induced dysregulation of reproduction as they are already being investigated as biomarkers of infertility and pregnancy outcomes [175,176]. The literature surrounding miRNAs specifically involved in the hypothalamic control of reproduction is still very limited, but their importance is evident as conditional knockout of Dicer in GnRH neurons or in the pituitary gland leads to infertility or severely reduced fertility, respectively [162,177]. A miRNA “switch” has been identified in GnRH neurons involving miR-155, which targets CCAAT-enhancer binding protein beta (CEBPß), a transcription factor that negatively regulates GnRH, and miR-200, which targets Zinc finger E-box binding homeobox 1 (Zeb1), another repressor of GnRH and its transcriptional activators [177]. Prior to the onset of puberty, these miRNAs are increased, leading to the downregulation of the targeted GnRH repressors, and ultimately lead to increased GnRH [177]. In addition, 16 other miRNAs have been identified by in silico analysis to directly target the 3′ UTR of GnRH but none have been experimentally validated [178]. Another miRNA that has been associated with the onset of puberty in the hypothalamus is miR-30b. miR-30b targets the 3′-UTR of makorin RING-finger protein-3 (MKRN3), a protein associated with the inhibition of the onset of puberty [179]. Central administration of target site blockers prevented the binding of miR-30b to the 3′-UTR of MKRN3 and delayed the onset of puberty in female mice [179]. As for KISS1, a recent abstract described a miRNA that directly targets the 3′ UTR of Kiss1 and is increased in obese individuals with hypogonadism, but due to intellectual property reasons has not been disclosed [180]. Overall, investigation into miRNAs involved in the hypothalamic control of reproduction and how they are altered by palmitate holds potential for future intervention studies.
4. Circadian Rhythms
Circadian rhythms are 24-h patterns of biological activity that occur to synchronize homeostatic functions, including feeding, reproduction, stress, temperature control, blood pressure and hormone production, with the outside environment [181]. Disruptions to these rhythms in humans through shift work or through genetic polymorphisms of the genes responsible for maintaining these rhythms are linked to increased rates of obesity, metabolic syndrome and diabetes [182,183,184]. As such, disrupting circadian rhythms may promote obesity by altering metabolism of nutrients; however, dietary factors can themselves disrupt circadian rhythms [185]. This section will explore how circadian rhythms in the feeding centers of the hypothalamus are disrupted by palmitate and how these changes mediate the other downstream actions of palmitate, including neuroinflammation, neuropeptide dysregulation and potentially miRNA alterations.
4.1. Control of Circadian Rhythms by the SCN and other Hypothalamic Nuclei
Circadian rhythms are generated internally by the suprachiasmatic nucleus (SCN) of the hypothalamus and are entrained or synchronized by zeitgebers or “time givers”, including light, food, body temperature and social cues [185]. The SCN is then able to control the rhythms of all other cells in the body via projections to other hypothalamic nuclei and brain regions [181,186]. At the molecular level, this occurs as a result of transcriptional-translational feedback loops involving clock genes, Bmal1 and circadian locomotor output cycles protein kaput (Clock) that are transcribed and translated and act as transcription factors to positively regulate period (Per1-3), cryptochrome (Cry1-2), nuclear receptor subfamily 1,group D, member 1 (Rev-erbs) and retinoic acid-related orphan receptor (Rors) by binding to promoter elements called E-boxes. PER and CRY proteins then inhibit the action of BMAL1:CLOCK to prevent their own transcription, and REV-ERBs, RORs and peroxisome proliferator-activated receptors (PPARs) directly influence Bmal1 transcription. This feedback loop creates a 24 h period where Per and Cry oscillate in an antiphasic manner to Bmal1 expression [187]. These genes are subject to post-translational as well as post-transcriptional modifications (i.e., miRNA regulation) that contributes the length of the 24 h period [188,189,190] (Figure 4).

Figure 4.
Circadian rhythm. The rhythmic expression of genes within a cell is controlled by a transcriptional-translational feedback loop primarily involving BMAL1, CLOCK, PER, and CRY proteins. The BMAL1 and CLOCK dimer binds to E-box sites in promoters to induce the expression of the downstream genes, which include Per and Cry. These negatively feedback onto the BMAL1:CLOCK dimer, thereby inhibiting their own transcription. Other circadian proteins involved in regulating the core elements of the loop include ROR, PPAR, and REV-ERB. Palmitate disrupts the rhythmicity of the loop, altering the amplitude or phase of rhythmic genes, including Bmal1, Per2 and Npy. Core clock genes are also regulated by miRNAs. For example, Bmal1 is targeted by miR-155 and miR-142-3p, Clock is targeted by miR-17-5p and miR-183, and Pers are targeted by miR-24, miR-29, miR-30a, and miR-96.
SCN rhythms are essential to maintain proper energy regulation and reproduction, as rodents with SCN lesions exhibit increased body weight, disrupted timing of food intake and activity [191], and a dysregulated LH surge and estrous cycle [192,193]. However, the SCN is not the sole player in circadian rhythms; the ARC and the PVN of the hypothalamus are core nuclei involved in food intake and energy expenditure [17], and synchronization of rhythms in these nuclei is critical to metabolic homeostasis. For example, SCN-ARC connections play a crucial role as microcuts removing this interconnectivity abolished rhythms in locomotor activity, corticosterone levels and body temperature in Wistar rats despite uninterrupted SCN rhythms [186]. Furthermore, deletion of BMAL1 in the PVN disrupts diurnal rhythms in metabolism and decreases neuronal response to refeeding, leading to obesity [194]. This dysregulation was attributed to perturbation of the rhythmic expression of the GABA-A receptor y2 subunit in the PVN neurons [194]. ARC neuropeptides NPY/AgRP and POMC show rhythmicity in vivo, with a peak in Pomc expression at 4 h after the dark phase [195], in Agrp expression in the transition between the light and dark phases, and in Npy expression once in the dark and once in the light phase [196]. These rhythms are also studied in vitro, where mechanisms of rhythmicity can be elucidated. Remarkably, the non-SCN, NPY/AgRP-expressing mHypoE-44 cell line demonstrated rhythmic expression of clock genes with an approximate period of 24 h [197]. Rhythmic binding of BMAL1 to the promoter region of Npy in these cells was associated with the rhythmic expression of Npy [197]. This ability of non-SCN hypothalamic cell lines to endogenously express rhythmicity has been recapitulated in murine adult-derived cell lines [111]. The importance of the molecular clock in NPY/AgRP-expressing neurons of the ARC is further demonstrated with the knockout of BMAL1 in AgRP-expressing neurons. These knockout mice demonstrated increased food intake with a trend towards increased body weight, a higher respiratory exchange ratio, and elevated hepatic glucose production [198]. Furthermore, coinciding with the transcriptional regulatory activity of BMAL1, the expressions of several genes were altered in AgRP-specific BMAL1-KO mice, including those involved in protein folding, secretory pathways, glucagon signaling, spliceosome activity and transcriptional response to leptin [198]. The observation that the major functions of AgRP-neurons were disrupted in these KO mice highlights the significance of BMAL1 and the circadian clock in NPY/AgRP neuron activity.
4.2. Palmitate-Induced Dysregulation of Circadian Rhythms in Hypothalamic Neurons
A major way in which the ARC and PVN neurons can desynchronize from the SCN is through exposure to exogenous factors, including high levels of fats and sugars [108,111,185,199,200,201]. Several in vivo studies have described hypothalamic neuropeptide expression and disruption of the rhythm of associated homeostatic processes, including body weight, food intake, and locomotion as a result of high fat diet exposure [199]. Consumption of a 45% HFD for 6 weeks dampened the diurnal pattern of food intake in C57BL6/J mice and although the expression profiles of hypothalamic clock genes were not altered, these mice displayed drastic changes in rhythmic Npy, Agrp and Pomc expression, indicative of HFD-induced disruptions in rhythmic feeding regulation at the level of the hypothalamus [199]. Moreover, although combined high-sugar, high-fat diets (termed a Western Diet (WD)) lead to obesity and disrupt circadian rhythms in food intake, the effects of a WD on hypothalamic neuropeptides or circadian proteins was not investigated until recently by our lab. Specifically, C57BL/6J mice exposed to a WD for 16 weeks showed altered diurnal feeding patterns of mice, with decreased food intake during the dark period and increased energy consumption during the light period [111]. These mice also displayed loss of rhythmic Agrp expression, a phase-shift in Pomc expression, and an overall reduction in Npy and Agrp expression in the hypothalamus [111]. In vitro experiments where neurons have been directly exposed to palmitate have indicated a causal relationship between palmitate, clock gene, and neuropeptide changes. For instance, palmitate-treated mHypoE-44 and mHypoE-37 neurons showed increased Bmal1 expression and decreased Per2 expression [108,200]. Likewise, palmitate-exposure in the mHypoE-44 neurons increased the amplitude of rhythmic Npy expression, which was mechanistically linked to AMPK activation by palmitate [108]. Thus, although causal links remained to be established, these studies strengthened the link between disruption of circadian clock genes, disruption of neuropeptides and disruption of whole-body homeostatic processes.
Remarkably, DHA, a ω-3 polyunsaturated fatty acid, prevented the palmitate-mediated upregulation in the amplitude of Bmal1 expression in the mHypoE-37 cells [200]. DHA also led to a phase advance in Bmal1 expression, however, DHA pre-treatment for palmitate-treated cells allowed normalization of the phase changed caused by both fatty acids. These results suggest potential protective actions of DHA against the saturated fatty acid palmitate in neurons and is corroborated by the recent finding of DHA-induced protection against fatty liver disease in HFD-fed mice through mechanisms involving circadian genes, RORα and REV-ERBα [202]. The monounsaturated fatty acid oleate has demonstrated protective effects against palmitate-induced changes in inflammatory markers and neuropeptides [9,59]. Interestingly, Tal et al. recently reported in 3T3-L1 adipocytes that palmitate increased the amplitude of clock gene expression, but decreased their overall expression after 24 h of treatment [203]. Oleate, in contrast, did not alter the amplitude of rhythmic expression nor lead to decreased expression. In fact, oleate increased the expression of Clock and Cry1 [203]. Mechanistically, these differences were attributed to differential enzyme activation by oleate (AMPK) and palmitate (acetyl-coA carboxylase (ACC)). Palmitate also induced the activation of Akt and GS3Kβ, which both phosphorylate BMAL1 for exclusion from the nucleus of for ubiquitination [203,204,205]. Whether oleate has protective effects against palmitate-induced circadian dysregulation in the hypothalamus warrants investigation given the considerable ability of oleate to protect against neuropeptide and inflammatory dysregulation. These studies also highlight the potential mechanistic basis of palmitate-induced circadian dysregulation, where palmitate acts by disrupting proteins and enzymes that post-transcriptionally modify BMAL1 and the other circadian regulators to maintain proper rhythmicity.
To further establish the molecular clock as mediating the disruptive effects of an HFD or palmitate, peripheral as well as brain-specific BMA1-KO models have become useful tools. For instance, mice with BMAL1-KO in skeletal muscle resisted HFD-induced obesity due to increased oxidative capacity [206]. Microglial-specific BMAL1-KO also prevented diet-induced obesity due to increased microglial phagocytic capacity [207]. This ability led to increased retention of POMC neurons in the hypothalamus, which are typically selectively lost with exposure to an HFD [207,208]. Thus, brain-specific BMAL1 and circadian control are implicated as mediators of the effects of HFD-exposure. To establish links between BMAL1 and the direct effects of palmitate on hypothalamic neurons, our lab generated immortalized hypothalamic cell lines from male or female BMAL1-KO mice and their wildtype littermates, which served as appropriate controls [116]. Palmitate treatment in wild-type mixed population neurons (mHypoA-BMAL1-WT/F) or clonal NPY-expressing neurons (mHypoA-BMAL1-WT/8) increases Npy expression, while the respective BMAL1-KO counterparts failed to show increased Npy expression [111]. Mechanistically, palmitate treatment increases binding of BMAL1 to the promoter of Npy, indicating a causal link between palmitate-mediated Npy dysregulation and BMAL1 [111]. Furthermore, BMAL1 may play a role in the inflammatory actions of palmitate. In mHypoA-BMAL1-KO/F cells, palmitate increased basal Il6 expression, but decreased basal Nf𝜅b expression. Moreover, the absence of BMAL1 altered the Il6 and Nf𝜅b response to palmitate treatment [209]. To further highlight this inflammatory role, BMAL1 knockdown in microglial BV-2 cells led to an increased anti-inflammatory phenotype, and increased protection from palmitate-induced neuroinflammation [207]. As neuroinflammation has been established as a critical mediator of palmitate actions [55,59,107,210], the role of the circadian system in influencing this inflammation should not be ignored, particularly considering that in the mHypoE-37 neurons, circadian gene dysregulation with palmitate occurred prior to changes in inflammatory markers [200]. In contrast to knockdown of BMAL1, knockdown of REV-ERBα and REV-ERBβ specifically in tuberal hypothalamic nuclei (non-SCN) led to exacerbated weight gain and day-time food intake in male mice, but not female mice, fed an HFD; leptin insensitivity was implicated as the mediator [211]. These results coincide with the negative regulatory role of REV-ERBs on BMAL1 [185].
4.3. Role of miRNAs in Palmitate-Mediated Circadian Dysregulation
The role of miRNAs in the maintenance of normal circadian rhythms is demonstrated by whole-body knockdown of Dicer in mice. Dicer knockdown shortened the circadian period by 2 h as a result of faster translation of PER1 and PER2, due to the absence of three miRNAs that target PER: miR-24, miR-29 and miR-30a [212]. The role of miRNAs in palmitate-mediated circadian rhythm dysregulation can be thought of as two-fold. Firstly, miRNAs that target and control clock gene expression can be altered by palmitate, thereby altering the rhythmic expression of these genes and the transcriptional targets of clock genes. Secondly, miRNAs themselves can be rhythmically expressed, leading to specific fine-tuning and control of their targets. Palmitate-mediated alterations in these rhythmic miRNAs can influence a wide variety of gene products and this miRNA influence may underlie many of the palmitate-mediated changes in gene expression mentioned above. Palmitate-specific examples of both of these scenarios remain to be investigated in the hypothalamus; however, examples of both scenarios exist from other models [189,190]. For example, miR-142-3p in the SCN and miR-155 in bone marrow cells target BMAL1, both with two independent binding sites in the BMAL1 3′UTR [119,213,214]. Given that HFD-fed mice show reduced expression of miR-142-3p in muscle [215], it remains plausible that palmitate or an HFD may alter the expression of this miRNA in the hypothalamus. Furthermore, deletion of miR-155 protected HFD-fed female mice from developing obesity, accumulating white adipose tissue (WAT) and glucose tolerance, while it decreased WAT accumulation in males without any reduction in body weight [216]. Although this protection was associated with increased expression of brown adipose tissue promoting genes, the role of BMAL1 cannot be excluded [216]. Lastly, in a recent screen to identify rhythm-controlling miRNAs, 120 out of 989 miRNAs were identified to alter the period of Bmal1-dluc or Per2-dluc reporter containing cells. From this, miR-183/96/182 was followed up to be a conserved cluster and maintains the period and amplitude in human cells containing the Per2-dluc reporter, controls rhythmic locomotor activity, and targets Per2 (miR-96) or Clock (miR-183) [217]. Interestingly, miR-96 was upregulated in the liver cells with palmitate exposure or with HFD exposure, targets IRS1 and insulin receptor and may contribute to fatty acid induced hepatic insulin resistance [218]. Whether its circadian function is also affected warrants investigation in these mice. Other miRNAs that have been reported to play a role in the murine SCN include miR-219, which is a target of CLOCK:BMAL1 [219], miR-132, which is responsive to light and fine tunes the clock by targeting chromatin remodeling [220], miR-17-5p, which itself displays a rhythm in the SCN via regulating CLOCK and is reciprocally regulated by CLOCK [221] (Figure 4). Any disruption to the rhythmicity of these miRNAs by palmitate has potential to alter the regulation of their targets. Dicer mRNA itself shows a diurnal rhythm of expression in the SCN, retina and bone marrow of mice, whereas this rhythm was phase-advanced in the SCN by aging and levels were decreased in the retina and bone marrow in diabetic animals [222]. Thus, the importance of rhythmic miRNAs in metabolic disorders warrants closer analysis and may prove therapeutically beneficial to fine-tune the expression of clock genes and their downstream targets.
5. Conclusions
Neurons in the hypothalamus control appetite, reproduction, and circadian rhythms and disruptions to these neurons have detrimental consequences for whole body health. Ultimately, these processes are interconnected and often undergo concurrent dysregulation. Thus, investigating the mechanisms of factors that dysregulate several of these functions will provide tools to combat their broad effects. Excessive exposure to saturated fatty acids such as palmitate, which are commonly found in HFDs, can disturb hypothalamic neurons resulting in resistance to insulin and leptin, dysregulation of feeding and reproductive neuropeptides, and disruptions of circadian rhythms that maintain energy homeostasis, and in turn reproduction. The effects of palmitate have been primarily linked to the induction of cellular stress, including neuroinflammation and ER stress, but avenues remain yet to be fully explored, including the actions of palmitate metabolites [110]. miRNAs play a key role in hypothalamic function and are dysregulated in disease states, and as a result, have become a novel and exciting area of study. Extracellular miRNAs can act as biomarkers to identify patients predisposed to certain metabolic conditions [25]. Furthermore, with the advent of technologies that enable packaging and therapeutic delivery of miRNAs to patients [223], they hold promise as an innovative tool to target key regulatory pathways in palmitate- and HFD-induced obesity and type 2 diabetes mellitus.
Author Contributions
Writing—original draft preparation, C.V.L. and N.L.; writing—review and editing, C.V.L., N.L. and D.D.B.; funding acquisition, D.D.B. All authors have read and agreed to the published version of the manuscript.
Funding
Our funding sources include the Canadian Institutes for Health Research (CIHR), Natural Sciences and Engineering Research Council (NSERC), Canada Foundation for Innovation and Canada Research Chairs Program (DDB). C.V.L. was supported by an NSERC CGS M Studentship and a Banting and Best Diabetes Centre Studentship. N.L. was supported by an Alexander Graham Bell Graduate (CGS D), NSERC (CGS D), and Banting and Best Diabetes Centre Studentship.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Emma McIlwraith for reading and providing suggestions for the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
| ACC | Acetyl-CoA carboxylase |
| AGO | Argonaute |
| AgRP | Agouti related peptide |
| AICAR | Aminoimidazole carboxamide ribonucleotide |
| AMPK | AMP-activated protein kinase |
| Arc | Arcuate nucleus |
| ATF3 | Activating transcription factor 3 |
| AVPV | Anteroventral periventricular nucleus |
| BMAL1 | Brain and muscle ARNT-like 1 |
| BMI | Body mass index |
| CEBPß | CCAAT-enhancer binding protein beta |
| CLOCK | Circadian locomotor output cycles protein kaput |
| CNTF | Ciliary neurotrophic factor |
| CREB | cAMP response element binding protein |
| CRY(1-2) | Cryptochrome (1-2) |
| CSF | Cerebrospinal fluid |
| DHA | Docosahexaenoic acid |
| eGFP | Enhanced green fluorescent protein |
| ER ERK | Endoplasmic reticulum Extracellular signal-regulated kinase |
| FACS | Fluorescent activated cell sorting |
| FOXO1 | Forkhead box protein O1 |
| FSH GABA | Follicle stimulating hormone γ-aminobutyric acid |
| GnRH | Gonadotropin releasing hormone |
| GNRH-R | Gonadotropin releasing hormone receptor |
| HFD | High fat diet |
| HOMA-IR | Homeostatic model for insulin resistance |
| HPG | Hypothalamus-pituitary-gonadal |
| ICV | Intracerebroventricular |
| IL1-β | Interleukin 1 beta |
| IL6 | Interleukin 6 |
| InsR | Insulin receptor |
| IRS | Insulin receptor substrate |
| IRβ | Insulin receptor beta |
| JAK2 | Janus kinase 2 |
| JNK | cJun N-terminal kinases |
| KISS1 | Kisspeptin |
| KISS1r/GPR54 | Kisspeptin receptor |
| KLF4 | Kruppel-like factor 4 |
| LepR | Leptin receptor |
| LH | Luteinizing hormone |
| MAPK | Mitogen activated protein kinase |
| MC3R | Melanocortin 3 receptor |
| MC4R | Melanocortin 4 receptor |
| MEK | Mitogen-activated protein kinase kinase |
| MIF | Macrophage migration inhibitory factor |
| miRNA | microRNA |
| MKRN3 | Makorin RING-finger protein 3 |
| mRNA | Messenger RNA |
| NF𝜅B | Nuclear factor kappa B |
| NNAT | Neuronatin |
| NPY | Neuropeptide Y |
| OCT1 | Octamer transcription factor 1 |
| pCREB | Phosphorylated cAMP response element binding protein |
| PER (1-3) | Period (1-3) |
| PI3K | phosphoinositide 3-kinase |
| PIP3 | Phosphatidylinositol 3-phosphate |
| PKA | Protein Kinase A |
| PNX | Phoenexin |
| POMC | Proopiomelanocortin |
| PPAR | Peroxisome proliferator-activated receptor |
| pri-miRNA | Primary miRNA |
| PTP1B | Protein tyrosine phosphatase 1B |
| PVN | Paraventricular nucleus |
| Rev-erb | Nuclear receptor subfamily 1, group D, member 1 |
| RISC | RNA induced silencing complex |
| ROR | Retinoic acid-related orphan receptor |
| SCN | Suprachiasmatic nucleus |
| SOCS3 | Suppressor of cytokine signaling 3 |
| STAT3 | signal transducer and activator of transcription 3 |
| TLR4 | Toll like receptor 4 |
| TNFα | Tumour necrosis factor alpha |
| UTR | Untranslated region |
| WAT | White adipose tissue |
| WD | Western diet |
| ZEB1 | Zinc finger E-box binding homeobox 1 |
| α-MSH | α-melanocyte stimulating hormone |
References
- World Health Organization. Obesity and Overweight. Available online: https://www.who.int/news-room/fact-sheets/detail/obesity-and-overweight (accessed on 5 October 2021).
- Bhupathiraju, S.N.; Hu, F.B. Epidemiology of obesity and diabetes and their cardiovascular complications. Circ. Res. 2016, 118, 1723–1735. [Google Scholar] [CrossRef] [PubMed]
- Weihrauch-Bluher, S.; Schwarz, P.; Klusmann, J.H. Childhood obesity: Increased risk for cardiometabolic disease and cancer in adulthood. Metabolism 2019, 92, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Ritchie, H.; Roser, M. Diet Compositions by Macronutrient. Daily Caloric Supply Derived from Carbohydrates, Protein and Fat, United States, 1961 to 2013. Available online: https://ourworldindata.org/diet-compositions (accessed on 5 October 2021).
- Rodriguez-Correa, E.; Gonzalez-Perez, I.; Clavel-Perez, P.I.; Contreras-Vargas, Y.; Carvajal, K. Biochemical and nutritional overview of diet-induced metabolic syndrome models in rats: What is the best choice? Nutr. Diabetes 2020, 10, 24. [Google Scholar] [CrossRef] [PubMed]
- Carta, G.; Murru, E.; Banni, S.; Manca, C. Palmitic acid: Physiological role, metabolism and nutritional implications. Front. Physiol. 2017, 8, 902. [Google Scholar] [CrossRef] [PubMed]
- Martínez de Morentin, P.B.; Varela, L.; Fernø, J.; Nogueiras, R.; Diéguez, C.; López, M. Hypothalamic lipotoxicity and the metabolic syndrome. Biochim. Biophys. Acta 2010, 1801, 350–361. [Google Scholar] [CrossRef] [PubMed]
- Aon, M.A.; Bhatt, N.; Cortassa, S.C. Mitochondrial and cellular mechanisms for managing lipid excess. Front. Physiol. 2014, 5, 282. [Google Scholar] [CrossRef]
- Tran, D.Q.; Ramos, E.H.; Belsham, D.D. Induction of Gnrh mRNA expression by the omega-3 polyunsaturated fatty acid docosahexaenoic acid and the saturated fatty acid palmitate in a GnRH-synthesizing neuronal cell model, mHypoA-GnRH/GFP. Mol. Cell. Endocrinol. 2016, 426, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Karaskov, E.; Scott, C.; Zhang, L.; Teodoro, T.; Ravazzola, M.; Volchuk, A. Chronic palmitate but not oleate exposure induces endoplasmic reticulum stress, which may contribute to INS-1 pancreatic beta-cell apoptosis. Endocrinology 2006, 147, 3398–3407. [Google Scholar] [CrossRef]
- McFadden, J.W.; Aja, S.; Li, Q.; Bandaru, V.V.; Kim, E.K.; Haughey, N.J.; Kuhajda, F.P.; Ronnett, G.V. Increasing fatty acid oxidation remodels the hypothalamic neurometabolome to mitigate stress and inflammation. PLoS ONE 2014, 9, e115642. [Google Scholar] [CrossRef]
- Melo, H.M.; Seixas da Silva, G.D.S.; Sant’Ana, M.R.; Teixeira, C.V.L.; Clarke, J.R.; Miya Coreixas, V.S.; de Melo, B.C.; Fortuna, J.T.S.; Forny-Germano, L.; Ledo, J.H.; et al. Palmitate Is increased in the cerebrospinal fluid of humans with obesity and induces memory impairment in mice via pro-inflammatory TNF-α. Cell. Rep. 2020, 30, 2180–2194. [Google Scholar] [CrossRef]
- Karmi, A.; Iozzo, P.; Viljanen, A.; Hirvonen, J.; Fielding, B.A.; Virtanen, K.; Oikonen, V.; Kemppainen, J.; Viljanen, T.; Guiducci, L.; et al. Increased brain fatty acid uptake in metabolic syndrome. Diabetes 2010, 59, 2171–2177. [Google Scholar] [CrossRef] [PubMed]
- Saper, C.B.; Lowell, B.B. The hypothalamus. Curr. Biol. 2014, 24, R1111–R1116. [Google Scholar] [CrossRef] [PubMed]
- Flament-Durand, J. The hypothalamus: Anatomy and functions. Acta Psychiatr. Belg. 1980, 80, 364–375. [Google Scholar] [PubMed]
- Wellhauser, L.; Gojska, N.M.; Belsham, D.D. Delineating the regulation of energy homeostasis using hypothalamic cell models. Front. Neuroendocrinol. 2015, 36, 130–149. [Google Scholar] [CrossRef] [PubMed]
- Dalvi, P.S.; Nazarians-Armavil, A.; Tung, S.; Belsham, D.D. Immortalized neurons for the study of hypothalamic function. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011, 300, R1030–R1052. [Google Scholar] [CrossRef] [PubMed]
- Dalvi, P.S.; Loganathan, N.; McIlwraith, E.K.; Tran, A.; Belsham, D.D. (Eds.) Chapter 2—Hypothalamic Cell Models, 2nd ed.; Academic Press: Cambridge, MA, USA, 2021; pp. 27–77. [Google Scholar]
- Ellwanger, D.C.; Büttner, F.A.; Mewes, H.W.; Stümpflen, V. The sufficient minimal set of miRNA seed types. Bioinformatics 2011, 27, 1346–1350. [Google Scholar] [CrossRef] [PubMed]
- Pratt, A.J.; MacRae, I.J. The RNA-induced silencing complex: A versatile gene-silencing machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S.; et al. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Yi, R.; Qin, Y.; Macara, I.G.; Cullen, B.R. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003, 17, 3011–3016. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, E.; Caudy, A.A.; Hammond, S.M.; Hannon, G.J. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 2001, 409, 363–366. [Google Scholar] [CrossRef]
- Ludwig, N.; Yerneni, S.S.; Razzo, B.M.; Whiteside, T.L. Exosomes from HNSCC Promote Angiogenesis through reprogramming of endothelial cells. Mol. Cancer Res. 2018, 16, 1798–1808. [Google Scholar] [CrossRef] [PubMed]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandao, B.B.; Kahn, C.R. Extracellular miRNAs: From biomarkers to mediators of physiology and disease. Cell. Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed]
- Deiuliis, J.A. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose tissue macrophage-derived Exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105. [Google Scholar] [CrossRef]
- Bernstein, E.; Kim, S.Y.; Carmell, M.A.; Murchison, E.P.; Alcorn, H.; Li, M.Z.; Mills, A.A.; Elledge, S.J.; Anderson, K.V.; Hannon, G.J. Dicer is essential for mouse development. Nat. Genet. 2003, 35, 215–217. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, J.; Yang, C.; Fan, P.; Balazs, L.; Jiao, Y.; Lu, M.; Gu, W.; Li, C.; Pfeffer, L.M.; et al. DiGeorge syndrome critical region 8 (DGCR8) protein-mediated microRNA biogenesis is essential for vascular smooth muscle cell development in mice. J. Biol. Chem. 2012, 287, 19018–19028. [Google Scholar] [CrossRef]
- Mang, G.M.; Pradervand, S.; Du, N.H.; Arpat, A.B.; Preitner, F.; Wigger, L.; Gatfield, D.; Franken, P. A neuron-specific deletion of the microRNA-processing enzyme DICER induces severe but transient obesity in mice. PLoS ONE 2015, 10, e0116760. [Google Scholar] [CrossRef]
- Greenman, Y.; Kuperman, Y.; Drori, Y.; Asa, S.L.; Navon, I.; Forkosh, O.; Gil, S.; Stern, N.; Chen, A. Postnatal ablation of POMC neurons induces an obese phenotype characterized by decreased food intake and enhanced anxiety-like behavior. Mol. Endocrinol. 2013, 27, 1091–1102. [Google Scholar] [CrossRef]
- Ouyang, S.; Tang, R.; Liu, Z.; Ma, F.; Li, Y.; Wu, J. Characterization and predicted role of microRNA expression profiles associated with early childhood obesity. Mol. Med. Rep. 2017, 16, 3799–3806. [Google Scholar] [CrossRef] [PubMed][Green Version]
- de Candia, P.; Spinetti, G.; Specchia, C.; Sangalli, E.; La Sala, L.; Uccellatore, A.; Lupini, S.; Genovese, S.; Matarese, G.; Ceriello, A. A unique plasma microRNA profile defines type 2 diabetes progression. PLoS ONE 2017, 12, e0188980. [Google Scholar] [CrossRef] [PubMed]
- Di Leva, G.; Croce, C.M. miRNA profiling of cancer. Curr. Opin. Genet. Dev. 2013, 23, 3–11. [Google Scholar] [CrossRef] [PubMed]
- Sangiao-Alvarellos, S.; Pena-Bello, L.; Manfredi-Lozano, M.; Tena-Sempere, M.; Cordido, F. Perturbation of hypothalamic microRNA expression patterns in male rats after metabolic distress: Impact of obesity and conditions of negative energy balance. Endocrinology 2014, 155, 1838–1850. [Google Scholar] [CrossRef]
- Benoit, C.; Doubi-Kadmiri, S.; Benigni, X.; Crepin, D.; Riffault, L.; Poizat, G.; Vacher, C.M.; Taouis, M.; Baroin-Tourancheau, A.; Amar, L. miRNA Long-term response to early metabolic environmental challenge in hypothalamic arcuate nucleus. Front. Mol. Neurosci. 2018, 11, 90. [Google Scholar] [CrossRef]
- Joly-Amado, A.; Cansell, C.; Denis, R.G.; Delbes, A.S.; Castel, J.; Martinez, S.; Luquet, S. The hypothalamic arcuate nucleus and the control of peripheral substrates. Best Pract. Res. Clin. Endocrinol. Metab. 2014, 28, 725–737. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Obici, S.; Morgan, K.; Barzilai, N.; Feng, Z.; Rossetti, L. Overfeeding rapidly induces leptin and insulin resistance. Diabetes 2001, 50, 2786–2791. [Google Scholar] [CrossRef]
- Darbre, P.D. Endocrine Disruptors and Obesity. Curr. Obes. Rep. 2017, 6, 18–27. [Google Scholar] [CrossRef]
- Ruud, J.; Steculorum, S.M.; Bruning, J.C. Neuronal control of peripheral insulin sensitivity and glucose metabolism. Nat. Commun. 2017, 8, 15259. [Google Scholar] [CrossRef]
- Belsham, D.D.; Dalvi, P.S. Insulin signalling in hypothalamic neurones. J. Neuroendocrinol. 2020, 33, e12919. [Google Scholar] [CrossRef] [PubMed]
- Petersen, M.C.; Shulman, G.I. Mechanisms of insulin action and insulin resistance. Physiol. Rev. 2018, 98, 2133–2223. [Google Scholar] [CrossRef]
- Loh, K.; Zhang, L.; Brandon, A.; Wang, Q.; Begg, D.; Qi, Y.; Fu, M.; Kulkarni, R.; Teo, J.; Baldock, P.; et al. Insulin controls food intake and energy balance via NPY neurons. Mol. Metab. 2017, 6, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Hallschmid, M.; Benedict, C.; Schultes, B.; Fehm, H.L.; Born, J.; Kern, W. Intranasal insulin reduces body fat in men but not in women. Diabetes 2004, 53, 3024–3029. [Google Scholar] [CrossRef]
- Konner, A.C.; Janoschek, R.; Plum, L.; Jordan, S.D.; Rother, E.; Ma, X.; Xu, C.; Enriori, P.; Hampel, B.; Barsh, G.S.; et al. Insulin action in AgRP-expressing neurons is required for suppression of hepatic glucose production. Cell. Metab. 2007, 5, 438–449. [Google Scholar] [CrossRef]
- Shin, A.C.; Filatova, N.; Lindtner, C.; Chi, T.; Degann, S.; Oberlin, D.; Buettner, C. Insulin receptor signaling in POMC, but not AgRP, neurons controls adipose tissue insulin action. Diabetes 2017, 66, 1560–1571. [Google Scholar] [CrossRef] [PubMed]
- Qiu, J.; Zhang, C.; Borgquist, A.; Nestor, C.C.; Smith, A.W.; Bosch, M.A.; Ku, S.; Wagner, E.J.; Ronnekleiv, O.K.; Kelly, M.J. Insulin excites anorexigenic proopiomelanocortin neurons via activation of canonical transient receptor potential channels. Cell. Metab. 2014, 19, 682–693. [Google Scholar] [CrossRef]
- Mayer, C.M.; Belsham, D.D. Insulin directly regulates NPY and AgRP gene expression via the MAPK MEK/ERK signal transduction pathway in mHypoE-46 hypothalamic neurons. Mol. Cell. Endocrinol. 2009, 307, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Nazarians-Armavil, A.; Chalmers, J.A.; Lee, C.B.; Ye, W.; Belsham, D.D. Cellular insulin resistance disrupts hypothalamic mHypoA-POMC/GFP neuronal signaling pathways. J. Endocrinol. 2014, 220, 13–24. [Google Scholar] [CrossRef]
- Nazarians-Armavil, A.; Menchella, J.A.; Belsham, D.D. Cellular insulin resistance disrupts leptin-mediated control of neuronal signaling and transcription. Mol. Endocrinol. 2013, 27, 990–1003. [Google Scholar] [CrossRef]
- Mayer, C.M.; Belsham, D.D. Central insulin signaling is attenuated by long-term insulin exposure via insulin receptor substrate-1 serine phosphorylation, proteasomal degradation, and lysosomal insulin receptor degradation. Endocrinology 2010, 151, 75–84. [Google Scholar] [CrossRef]
- de Luca, C.; Olefsky, J.M. Inflammation and insulin resistance. FEBS Lett. 2008, 582, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Thaler, J.P.; Yi, C.X.; Schur, E.A.; Guyenet, S.J.; Hwang, B.H.; Dietrich, M.O.; Zhao, X.L.; Sarruf, D.A.; Izgur, V.; Maravilla, K.R.; et al. Obesity is associated with hypothalamic injury in rodents and humans. J. Clin. Investig. 2012, 122, 153–162. [Google Scholar] [CrossRef]
- Benoit, S.C.; Kemp, C.J.; Elias, C.F.; Abplanalp, W.; Herman, J.P.; Migrenne, S.; Lefevre, A.L.; Cruciani-Guglielmacci, C.; Magnan, C.; Yu, F.; et al. Palmitic acid mediates hypothalamic insulin resistance by altering PKC-theta subcellular localization in rodents. J. Clin. Investing. 2009, 119, 2577–2589. [Google Scholar] [CrossRef] [PubMed]
- Kleinridders, A.; Schenten, D.; Konner, A.C.; Belgardt, B.F.; Mauer, J.; Okamura, T.; Wunderlich, F.T.; Medzhitov, R.; Bruning, J.C. MyD88 signaling in the CNS is required for development of fatty acid-induced leptin resistance and diet-induced obesity. Cell. Metab. 2009, 10, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.M.; Belsham, D.D. Palmitate attenuates insulin signaling and induces endoplasmic reticulum stress and apoptosis in hypothalamic neurons: Rescue of resistance and apoptosis through adenosine 5′ monophosphate-activated protein kinase activation. Endocrinology 2010, 151, 576–585. [Google Scholar] [CrossRef] [PubMed]
- Tse, E.K.; Belsham, D.D. Palmitate induces neuroinflammation, ER stress, and Pomc mRNA expression in hypothalamic mHypoA-POMC/GFP neurons through novel mechanisms that are prevented by oleate. Mol. Cell. Endocrinol. 2018, 472, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Clemenzi, M.N.; Wellhauser, L.; Aljghami, M.E.; Belsham, D.D. Tumour necrosis factor alpha induces neuroinflammation and insulin resistance in immortalised hypothalamic neurones through independent pathways. J. Neuroendocr. 2019, 31, e12678. [Google Scholar] [CrossRef]
- Amine, H.; Benomar, Y.; Taouis, M. Palmitic acid promotes resistin-induced insulin resistance and inflammation in SH-SY5Y human neuroblastoma. Sci. Rep. 2021, 11, 5427. [Google Scholar] [CrossRef]
- Wellhauser, L.; Belsham, D.D. Activation of the omega-3 fatty acid receptor GPR120 mediates anti-inflammatory actions in immortalized hypothalamic neurons. J. Neuroinflammation 2014, 11, 60. [Google Scholar] [CrossRef] [PubMed]
- Halseth, A.E.; Ensor, N.J.; White, T.A.; Ross, S.A.; Gulve, E.A. Acute and chronic treatment of ob/ob and db/db mice with AICAR decreases blood glucose concentrations. Biochem. Biophys. Res. Commun. 2002, 294, 798–805. [Google Scholar] [CrossRef]
- Buhl, E.S.; Jessen, N.; Pold, R.; Ledet, T.; Flyvbjerg, A.; Pedersen, S.B.; Pedersen, O.; Schmitz, O.; Lund, S. Long-term AICAR administration reduces metabolic disturbances and lowers blood pressure in rats displaying features of the insulin resistance syndrome. Diabetes 2002, 51, 2199–2206. [Google Scholar] [CrossRef]
- Yamauchi, T.; Kamon, J.; Minokoshi, Y.; Ito, Y.; Waki, H.; Uchida, S.; Yamashita, S.; Noda, M.; Kita, S.; Ueki, K.; et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002, 8, 1288–1295. [Google Scholar] [CrossRef] [PubMed]
- Ong, P.S.; Wang, L.Z.; Dai, X.; Tseng, S.H.; Loo, S.J.; Sethi, G. Judicious toggling of mTOR activity to combat insulin resistance and cancer: Current evidence and perspectives. Front. Pharmacol. 2016, 7, 395. [Google Scholar] [CrossRef] [PubMed]
- Posey, K.A.; Clegg, D.J.; Printz, R.L.; Byun, J.; Morton, G.J.; Vivekanandan-Giri, A.; Pennathur, S.; Baskin, D.G.; Heinecke, J.W.; Woods, S.C.; et al. Hypothalamic proinflammatory lipid accumulation, inflammation, and insulin resistance in rats fed a high-fat diet. Am. J. Physiol. Endocrinol. Metab. 2009, 296, E1003–E1012. [Google Scholar] [CrossRef] [PubMed]
- Vinnikov, I.A.; Hajdukiewicz, K.; Reymann, J.; Beneke, J.; Czajkowski, R.; Roth, L.C.; Novak, M.; Roller, A.; Dorner, N.; Starkuviene, V.; et al. Hypothalamic miR-103 protects from hyperphagic obesity in mice. J. Neurosci. 2014, 34, 10659–10674. [Google Scholar] [CrossRef]
- Mu, J.; Yu, P.; Li, Q. microRNA-103 Contributes to progression of polycystic ovary syndrome through modulating the IRS1/PI3K/AKT signal axis. Arch. Med. Res. 2021, 52, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.D.; Toda, C.; Ramirez, C.M.; Fernandez-Hernando, C.; Diano, S. Hypothalamic ventromedial Lin28a enhances glucose metabolism in diet-induced obesity. Diabetes 2017, 66, 2102–2111. [Google Scholar] [CrossRef]
- Crepin, D.; Benomar, Y.; Riffault, L.; Amine, H.; Gertler, A.; Taouis, M. The over-expression of miR-200a in the hypothalamus of ob/ob mice is linked to leptin and insulin signaling impairment. Mol. Cell. Endocrinol. 2014, 384, 1–11. [Google Scholar] [CrossRef]
- Chalmers, J.A.; Dalvi, P.S.; Wellhauser, L.; Nazarians-Armavil, A.; Eversley, J.A.; Mohan, H.; Wheeler, M.B.; Belsham, D.D. Hypothalamic miR-1983 targets insulin receptor beta to induce insulin resistance, and the insulin-mediated increase in miR-1983 is blocked by metformin. Endocrinology 2021. in revision. [Google Scholar]
- Vaitheesvaran, B.; LeRoith, D.; Kurland, I.J. MKR mice have increased dynamic glucose disposal despite metabolic inflexibility, and hepatic and peripheral insulin insensitivity. Diabetologia 2010, 53, 2224–2232. [Google Scholar] [CrossRef]
- Amar, L.; Benoit, C.; Beaumont, G.; Vacher, C.M.; Crepin, D.; Taouis, M.; Baroin-Tourancheau, A. MicroRNA expression profiling of hypothalamic arcuate and paraventricular nuclei from single rats using Illumina sequencing technology. J. Neurosci. Methods 2012, 209, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-de Frutos, M.; Galan-Chilet, I.; Goedeke, L.; Kim, B.; Pardo-Marques, V.; Perez-Garcia, A.; Herrero, J.I.; Fernandez-Hernando, C.; Kim, J.; Ramirez, C.M. MicroRNA 7 impairs insulin signaling and regulates abeta levels through posttranscriptional regulation of the Insulin receptor substrate 2, insulin receptor, insulin-degrading enzyme, and liver X receptor pathway. Mol. Cell. Biol. 2019, 39, e00170-19. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Li, J.; Zhang, Z.; Zhang, R.; Pollock, A.; Sun, T. MicroRNA miR-7 and miR-17-92 in the arcuate nucleus of Mouse Hypothalamus regulate sex-specific diet-induced obesity. Mol. Neurobiol. 2019, 56, 7508–7521. [Google Scholar] [CrossRef] [PubMed]
- Romon, M.; Lebel, P.; Velly, C.; Marecaux, N.; Fruchart, J.C.; Dallongeville, J. Leptin response to carbohydrate or fat meal and association with subsequent satiety and energy intake. Am. J. Physiol. 1999, 277, E855–E861. [Google Scholar] [CrossRef] [PubMed]
- Ostlund, R.E.; Yang, J.W.; Klein, S.; Gingerich, R. Relation between plasma leptin concentration and body fat, gender, diet, age, and metabolic covariates. J. Clin. Endocrinol. Metab. 1996, 81, 3909–3913. [Google Scholar] [CrossRef]
- Clement, K.; Vaisse, C.; Lahlou, N.; Cabrol, S.; Pelloux, V.; Cassuto, D.; Gourmelen, M.; Dina, C.; Chambaz, J.; Lacorte, J.M.; et al. A mutation in the human leptin receptor gene causes obesity and pituitary dysfunction. Nature 1998, 392, 398–401. [Google Scholar] [CrossRef]
- de Luca, C.; Kowalski, T.J.; Zhang, Y.; Elmquist, J.K.; Lee, C.; Kilimann, M.W.; Ludwig, T.; Liu, S.M.; Chua, S.C., Jr. Complete rescue of obesity, diabetes, and infertility in db/db mice by neuron-specific LEPR-B transgenes. J. Clin. Investig. 2005, 115, 3484–3493. [Google Scholar] [CrossRef]
- Havel, P.J.; Kasim-Karakas, S.; Mueller, W.; Johnson, P.R.; Gingerich, R.L.; Stern, J.S. Relationship of plasma leptin to plasma insulin and adiposity in normal weight and overweight women: Effects of dietary fat content and sustained weight loss. J. Clin. Endocrinol. Metab. 1996, 81, 4406–4413. [Google Scholar] [CrossRef][Green Version]
- Banks, W.A.; Farrell, C.L. Impaired transport of leptin across the blood-brain barrier in obesity is acquired and reversible. Am. J. Physiol. Endocrinol. Metab. 2003, 285, E10–E15. [Google Scholar] [CrossRef]
- Zhao, S.; Zhu, Y.; Schultz, R.D.; Li, N.; He, Z.; Zhang, Z.; Caron, A.; Zhu, Q.; Sun, K.; Xiong, W.; et al. Partial Leptin Reduction as an Insulin Sensitization and Weight Loss Strategy. Cell. Metab. 2019, 30, 706–719. [Google Scholar] [CrossRef]
- Donato, J., Jr.; Cravo, R.M.; Frazao, R.; Elias, C.F. Hypothalamic sites of leptin action linking metabolism and reproduction. Neuroendocrinology 2011, 93, 9–18. [Google Scholar] [CrossRef]
- Schwartz, M.W.; Seeley, R.J.; Woods, S.C.; Weigle, D.S.; Campfield, L.A.; Burn, P.; Baskin, D.G. Leptin increases hypothalamic pro-opiomelanocortin mRNA expression in the rostral arcuate nucleus. Diabetes 1997, 46, 2119–2123. [Google Scholar] [CrossRef]
- Kim, Y.B.; Uotani, S.; Pierroz, D.D.; Flier, J.S.; Kahn, B.B. In vivo administration of leptin activates signal transduction directly in insulin-sensitive tissues: Overlapping but distinct pathways from insulin. Endocrinology 2000, 141, 2328–2339. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bing, C.; Al-Barazanji, K.; Mossakowaska, D.E.; Wang, X.M.; McBay, D.L.; Neville, W.A.; Taddayon, M.; Pickavance, L.; Dryden, S.; et al. Interactions between leptin and hypothalamic neuropeptide Y neurons in the control of food intake and energy homeostasis in the rat. Diabetes 1997, 46, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Korner, J.; Savontaus, E.; Chua, S.C., Jr.; Leibel, R.L.; Wardlaw, S.L. Leptin regulation of Agrp and Npy mRNA in the rat hypothalamus. J. Neuroendocrinol. 2001, 13, 959–966. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Li, Z.; Rui, L. Leptin stimulates both JAK2-dependent and JAK2-independent signaling pathways. J. Biol. Chem. 2008, 283, 28066–28073. [Google Scholar] [CrossRef]
- Gong, L.; Yao, F.; Hockman, K.; Heng, H.H.; Morton, G.J.; Takeda, K.; Akira, S.; Low, M.J.; Rubinstein, M.; MacKenzie, R.G. Signal transducer and activator of transcription-3 is required in hypothalamic agouti-related protein/neuropeptide Y neurons for normal energy homeostasis. Endocrinology 2008, 149, 3346–3354. [Google Scholar] [CrossRef]
- Ernst, M.B.; Wunderlich, C.M.; Hess, S.; Paehler, M.; Mesaros, A.; Koralov, S.B.; Kleinridders, A.; Husch, A.; Munzberg, H.; Hampel, B.; et al. Enhanced Stat3 activation in POMC neurons provokes negative feedback inhibition of leptin and insulin signaling in obesity. J. Neurosci. 2009, 29, 11582–11593. [Google Scholar] [CrossRef]
- Ma, W.; Fuentes, G.; Shi, X.; Verma, C.; Radda, G.K.; Han, W. FoxO1 negatively regulates leptin-induced POMC transcription through its direct interaction with STAT3. Biochem. J. 2015, 466, 291–298. [Google Scholar] [CrossRef]
- Lubis, A.R.; Widia, F.; Soegondo, S.; Setiawati, A. The role of SOCS-3 protein in leptin resistance and obesity. Acta Med. Indones. 2008, 40, 89–95. [Google Scholar]
- Carvalheira, J.B.; Torsoni, M.A.; Ueno, M.; Amaral, M.E.; Araujo, E.P.; Velloso, L.A.; Gontijo, J.A.; Saad, M.J. Cross-talk between the insulin and leptin signaling systems in rat hypothalamus. Obes. Res. 2005, 13, 48–57. [Google Scholar] [CrossRef]
- Dhillon, S.S.; McFadden, S.A.; Chalmers, J.A.; Centeno, M.L.; Kim, G.L.; Belsham, D.D. Cellular leptin resistance impairs the leptin-mediated suppression of neuropeptide Y secretion in hypothalamic neurons. Endocrinology 2011, 152, 4138–4147. [Google Scholar] [CrossRef] [PubMed]
- Cheng, L.; Yu, Y.; Szabo, A.; Wu, Y.; Wang, H.; Camer, D.; Huang, X.F. Palmitic acid induces central leptin resistance and impairs hepatic glucose and lipid metabolism in male mice. J. Nutr. Biochem. 2015, 26, 541–548. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Dhillon, H.; Yin, H.; Yoshimura, A.; Lowell, B.B.; Maratos-Flier, E.; Flier, J.S. Selective inactivation of Socs3 in SF1 neurons improves glucose homeostasis without affecting body weight. Endocrinology 2008, 149, 5654–5661. [Google Scholar] [CrossRef] [PubMed]
- Zabolotny, J.M.; Kim, Y.B.; Welsh, L.A.; Kershaw, E.E.; Neel, B.G.; Kahn, B.B. Protein-tyrosine phosphatase 1B expression is induced by inflammation in vivo. J. Biol. Chem. 2008, 283, 14230–14241. [Google Scholar] [CrossRef]
- Hakim, F.; Wang, Y.; Carreras, A.; Hirotsu, C.; Zhang, J.; Peris, E.; Gozal, D. Chronic sleep fragmentation during the sleep period induces hypothalamic endoplasmic reticulum stress and PTP1b-mediated leptin resistance in male mice. Sleep 2015, 38, 31–40. [Google Scholar] [CrossRef]
- Ozcan, L.; Ergin, A.; Lu, A.; Chung, J.; Sarkar, S.; Nie, D.; Myers, M.G.; Ozcan, U. Endoplasmic reticulum stress plays a central role in development of leptin resistance. Cell. Metab. 2009, 9, 35–51. [Google Scholar] [CrossRef]
- Marwarha, G.; Claycombe, K.; Schommer, J.; Collins, D.; Ghribi, O. Palmitate-induced endoplasmic reticulum stress and subsequent C/EBPalpha homologous protein activation attenuates leptin and insulin-like growth factor 1 expression in the brain. Cell. Signal 2016, 28, 1789–1805. [Google Scholar] [CrossRef]
- Derghal, A.; Djelloul, M.; Azzarelli, M.; Degonon, S.; Tourniaire, F.; Landrier, J.F.; Mounien, L. MicroRNAs are involved in the hypothalamic leptin sensitivity. Epigenetics 2018, 13, 1127–1140. [Google Scholar] [CrossRef]
- Zhang, J.; Ma, W.; He, Y.; Dawar, F.U.; Xiong, S.; Mei, J. Potential Contributions of miR-200a/-200b and their target gene-leptin to the sexual Size dimorphism in Yellow Catfish. Front. Physiol. 2017, 8, 970. [Google Scholar] [CrossRef]
- Collins, A.S.; McCoy, C.E.; Lloyd, A.T.; O’Farrelly, C.; Stevenson, N.J. miR-19a: An effective regulator of SOCS3 and enhancer of JAK-STAT signalling. PLoS ONE 2013, 8, e69090. [Google Scholar] [CrossRef] [PubMed]
- Derghal, A.; Astier, J.; Sicard, F.; Couturier, C.; Landrier, J.F.; Mounien, L. Leptin modulates the expression of miRNAs-targeting POMC mRNA by the JAK2-STAT3 and PI3K-Akt pathways. J. Clin. Med. 2019, 8, 2213. [Google Scholar] [CrossRef] [PubMed]
- Dalvi, P.S.; Chalmers, J.A.; Luo, V.; Han, D.-Y.; Wellhauser, L.; Liu, Y.; Tran, D.Q.; Castel, J.; Luquet, S.; Wheeler, M.B.; et al. High fat induces acute and chronic inflammation in the hypothalamus: Effect of high-fat diet, palmitate and TNF-α on appetite-regulating NPY neurons. Int. J. Obes. 2017, 41, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Ye, W.; Ramos, E.H.; Wong, B.C.; Belsham, D.D. Beneficial effects of metformin and/or salicylate on palmitate- or TNFalpha-induced neuroinflammatory marker and neuropeptide gene regulation in immortalized NPY/AgRP neurons. PLoS ONE 2016, 11, e0166973. [Google Scholar] [CrossRef]
- Fick, L.J.; Fick, G.H.; Belsham, D.D. Palmitate alters the rhythmic expression of molecular clock genes and orexigenic neuropeptide Y mRNA levels within immortalized, hypothalamic neurons. Biochem. Biophys. Res. Commun. 2011, 413, 414–419. [Google Scholar] [CrossRef]
- Jais, A.; Bruning, J.C. Hypothalamic inflammation in obesity and metabolic disease. J. Clin. Investig. 2017, 127, 24–32. [Google Scholar] [CrossRef] [PubMed]
- Tran, A.; He, W.; Chen, J.T.C.; Wellhauser, L.; Hopperton, K.E.; Bazinet, R.P.; Belsham, D.D. Palmitate-mediated induction of neuropeptide Y expression occurs through intracellular metabolites and not direct exposure to proinflammatory cytokines. J. Neurochem. 2021, 159, 574–589. [Google Scholar] [CrossRef]
- Clemenzi, M.N.; Martchenko, A.; Loganathan, N.; Tse, E.K.; Brubaker, P.L.; Belsham, D.D. Analysis of Western diet, palmitate and BMAL1 regulation of neuropeptide Y expression in the murine hypothalamus and BMAL1 knockout cell models. Mol. Cell. Endocrinol. 2020, 507, 110773. [Google Scholar] [CrossRef]
- Park, S.; Oh, T.S.; Kim, S.; Kim, E.K. Palmitate-induced autophagy liberates monounsaturated fatty acids and increases Agrp expression in hypothalamic cells. Anim. Cells. Syst. 2019, 23, 384–391. [Google Scholar] [CrossRef]
- Titolo, D.; Mayer, C.M.; Dhillon, S.S.; Cai, F.; Belsham, D.D. Estrogen facilitates both phosphatidylinositol 3-kinase/Akt and ERK1/2 mitogen-activated protein kinase membrane signaling required for long-term neuropeptide Y transcriptional regulation in clonal, immortalized neurons. J. Neurosci. 2008, 28, 6473–6482. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, Y.-S.; Yang, S.-F.; Kuo, D.-Y. Intracerebral administration of protein kinase A or cAMP response element-binding protein antisense oligonucleotide can modulate amphetamine-mediated appetite suppression in free-moving rats. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E123–E131. [Google Scholar] [CrossRef] [PubMed]
- Mayer, C.M.; Cai, F.; Cui, H.; Gillespie, J.M.; MacMillan, M.; Belsham, D.D. Analysis of a repressor region in the human neuropeptide Y gene that binds Oct-1 and Pbx-1 in GT1-7 neurons. Biochem. Biophys. Res. Commun. 2003, 307, 847–854. [Google Scholar] [CrossRef]
- Loganathan, N.; Salehi, A.; Chalmers, J.A.; Belsham, D.D. Bisphenol A alters Bmal1, Per2, and Rev-Erba mRNA and requires Bmal1 to increase neuropeptide Y expression in hypothalamic neurons. Endocrinology 2019, 160, 181–192. [Google Scholar] [CrossRef]
- Friedrich, M.; Heimer, N.; Stoehr, C.; Steven, A.; Wach, S.; Taubert, H.; Hartmann, A.; Seliger, B. CREB1 is affected by the microRNAs miR-22-3p, miR-26a-5p, miR-27a-3p, and miR-221-3p and correlates with adverse clinicopathological features in renal cell carcinoma. Sci. Rep. 2020, 10, 6499. [Google Scholar] [CrossRef]
- Feng, Y.Y.; Zeng, D.Z.; Tong, Y.N.; Lu, X.X.; Dun, G.D.; Tang, B.; Zhang, Z.J.; Ye, X.L.; Li, Q.; Xie, J.P.; et al. Alteration of microRNA-4474/4717 expression and CREB-binding protein in human colorectal cancer tissues infected with Fusobacterium nucleatum. PLoS ONE 2019, 14, e0215088. [Google Scholar] [CrossRef] [PubMed]
- Curtis, A.M.; Fagundes, C.T.; Yang, G.; Palsson-McDermott, E.M.; Wochal, P.; McGettrick, A.F.; Foley, N.H.; Early, J.O.; Chen, L.; Zhang, H.; et al. Circadian control of innate immunity in macrophages by miR-155 targeting Bmal1. Proc. Nat. Acad. Sci. USA 2015, 112, 7231–7236. [Google Scholar] [CrossRef]
- Karkeni, E.; Astier, J.; Tourniaire, F.; El Abed, M.; Romier, B.; Gouranton, E.; Wan, L.; Borel, P.; Salles, J.; Walrand, S.; et al. Obesity-associated inflammation induces microRNA-155 expression in adipocytes and adipose tissue: Outcome on adipocyte function. J. Clin. Endocrinol. Metab. 2016, 101, 1615–1626. [Google Scholar] [CrossRef] [PubMed]
- McIlwraith, E.K.; Lieu, C.V.; Belsham, D.D. Bisphenol A induces miR-708-5p through an ER stress-mediated mechanism achieving its obesogenic effects by altering neuronatin and neuropeptide Y expression in hypothalamic neuronal models. Mol. Cell. Endocrinol. 2021, in press. [Google Scholar]
- Lin, H.H.; Bell, E.; Uwanogho, D.; Perfect, L.W.; Noristani, H.; Bates, T.J.; Snetkov, V.; Price, J.; Sun, Y.M. Neuronatin promotes neural lineage in ESCs via Ca(2+) signaling. Stem Cells 2010, 28, 1950–1960. [Google Scholar] [CrossRef] [PubMed]
- Cimino, I.; Rimmington, D.; Tung, Y.C.L.; Lawler, K.; Larraufie, P.; Kay, R.G.; Virtue, S.; Lam, B.Y.H.; Fagnocchi, L.; Ma, M.K.L.; et al. Murine neuronatin deficiency is associated with a hypervariable food intake and bimodal obesity. Sci. Rep. 2021, 11, 17571. [Google Scholar] [CrossRef]
- Kitamura, T.; Feng, Y.; Kitamura, Y.I.; Chua, S.C., Jr.; Xu, A.W.; Barsh, G.S.; Rossetti, L.; Accili, D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat. Med. 2006, 12, 534–540. [Google Scholar] [CrossRef] [PubMed]
- Imbernon, M.; Sanchez-Rebordelo, E.; Gallego, R.; Gandara, M.; Lear, P.; Lopez, M.; Dieguez, C.; Nogueiras, R. Hypothalamic KLF4 mediates leptin’s effects on food intake via AgRP. Mol. Metab. 2014, 3, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Loganathan, N.; McIlwraith, E.K.; Belsham, D.D. Bisphenol A Induces Agrp Gene Expression in Hypothalamic Neurons through a Mechanism Involving ATF3. Neuroendocrinology 2021, 111, 678–695. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.C.; Liu, L.Z.; Addison, J.B.; Wonderlin, W.F.; Ivanov, A.V.; Ruppert, J.M. A KLF4-miRNA-206 autoregulatory feedback loop can promote or inhibit protein translation depending upon cell context. Mol. Cell. Biol. 2011, 31, 2513–2527. [Google Scholar] [CrossRef]
- Xu, N.; Papagiannakopoulos, T.; Pan, G.; Thomson, J.A.; Kosik, K.S. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 2009, 137, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Vinod, M.; Patankar, J.V.; Sachdev, V.; Frank, S.; Graier, W.F.; Kratky, D.; Kostner, G.M. MiR-206 is expressed in pancreatic islets and regulates glucokinase activity. Am. J. Physiol. Endocrinol. Metab. 2016, 311, E175–E185. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.S.; Sasaki, T.; Kobayashi, M.; Kikuchi, O.; Kim, H.J.; Yokota-Hashimoto, H.; Shimpuku, M.; Susanti, V.Y.; Ido-Kitamura, Y.; Kimura, K.; et al. Hypothalamic ATF3 is involved in regulating glucose and energy metabolism in mice. Diabetologia 2013, 56, 1383–1393. [Google Scholar] [CrossRef] [PubMed]
- Choe, N.; Kwon, D.H.; Ryu, J.; Shin, S.; Cho, H.J.; Joung, H.; Eom, G.H.; Ahn, Y.; Park, W.J.; Nam, K.I.; et al. Targets ATF3 to Reduce Calcium Deposition in Vascular Smooth Muscle Cells. Mol. Ther. Nucleic Acids 2020, 22, 627–639. [Google Scholar] [CrossRef]
- Shi, B.; Yan, W.; Liu, G.; Guo, Y. MicroRNA-488 inhibits tongue squamous carcinoma cell invasion and EMT by directly targeting ATF3. Cell. Mol. Biol. Lett. 2018, 23, 28. [Google Scholar] [CrossRef]
- Li, S.; Yan, G.; Yue, M.; Wang, L. Extracellular vesicles-derived microRNA-222 promotes immune escape via interacting with ATF3 to regulate AKT1 transcription in colorectal cancer. BMC Cancer 2021, 21, 349. [Google Scholar] [CrossRef]
- Li, D.; Song, H.; Shuo, L.; Wang, L.; Xie, P.; Li, W.; Liu, J.; Tong, Y.; Zhang, C.Y.; Jiang, X.; et al. Gonadal white adipose tissue-derived exosomal MiR-222 promotes obesity-associated insulin resistance. Aging 2020, 12, 22719–22743. [Google Scholar] [CrossRef] [PubMed]
- World Health Organization. Sexual and Reproductive Health: Infertility Is a Global Public Health Issue. Available online: https://www.who.int/reproductivehealth/topics/infertility/perspective/en/ (accessed on 20 September 2020).
- Silvestris, E.; de Pergola, G.; Rosania, R.; Loverro, G. Obesity as disruptor of the female fertility. Reprod. Biol. Endocrinol. 2018, 16, 22. [Google Scholar] [CrossRef] [PubMed]
- Rich-Edwards, J.W.; Goldman, M.B.; Willett, W.C.; Hunter, D.J.; Stampfer, M.J.; Colditz, G.A.; Manson, J.E. Adolescent body mass index and infertility caused by ovulatory disorder. Am. J. Obstet. Gynecol. 1994, 171, 171–177. [Google Scholar] [CrossRef]
- Sallmen, M.; Sandler, D.P.; Hoppin, J.A.; Blair, A.; Baird, D.D. Reduced fertility among overweight and obese men. Epidemiology 2006, 17, 520–523. [Google Scholar] [CrossRef] [PubMed]
- Diabetes Canada Clinical Practice Guidelines Expert Committee; Feig, D.S.; Berger, H.; Donovan, L.; Godbout, A.; Kader, T.; Keely, E.; Sanghera, R. Diabetes and Pregnancy. Can. J. Diabetes 2018, 42, S255–S282. [Google Scholar] [CrossRef]
- Katib, A. Mechanisms linking obesity to male infertility. Cent. European J. Urol. 2015, 68, 79–85. [Google Scholar] [CrossRef]
- Skorupskaite, K.; George, J.T.; Anderson, R.A. The kisspeptin-GnRH pathway in human reproductive health and disease. Hum. Reprod. Update 2014, 20, 485–500. [Google Scholar] [CrossRef]
- Lapatto, R.; Pallais, J.C.; Zhang, D.; Chan, Y.M.; Mahan, A.; Cerrato, F.; Le, W.W.; Hoffman, G.E.; Seminara, S.B. Kiss1-/- mice exhibit more variable hypogonadism than Gpr54-/- mice. Endocrinology 2007, 148, 4927–4936. [Google Scholar] [CrossRef] [PubMed]
- Seminara, S.B.; Messager, S.; Chatzidaki, E.E.; Thresher, R.R.; Acierno, J.S., Jr.; Shagoury, J.K.; Bo-Abbas, Y.; Kuohung, W.; Schwinof, K.M.; Hendrick, A.G.; et al. The GPR54 gene as a regulator of puberty. N. Engl. J. Med. 2003, 349, 1614–1627. [Google Scholar] [CrossRef]
- Kirilov, M.; Clarkson, J.; Liu, X.; Roa, J.; Campos, P.; Porteous, R.; Schutz, G.; Herbison, A.E. Dependence of fertility on kisspeptin-Gpr54 signaling at the GnRH neuron. Nat. Commun. 2013, 4, 2492. [Google Scholar] [CrossRef]
- Ikegami, K.; Goto, T.; Nakamura, S.; Watanabe, Y.; Sugimoto, A.; Majarune, S.; Horihata, K.; Nagae, M.; Tomikawa, J.; Imamura, T.; et al. Conditional kisspeptin neuron-specific Kiss1 knockout with newly generated Kiss1-floxed and Kiss1-Cre mice replicates a hypogonadal phenotype of global Kiss1 knockout mice. J. Reprod. Dev. 2020, 66, 359–367. [Google Scholar] [CrossRef]
- De Bond, J.A.; Smith, J.T. Kisspeptin and energy balance in reproduction. Reproduction 2014, 147, R53–R63. [Google Scholar] [CrossRef] [PubMed]
- Messager, S.; Chatzidaki, E.E.; Ma, D.; Hendrick, A.G.; Zahn, D.; Dixon, J.; Thresher, R.R.; Malinge, I.; Lomet, D.; Carlton, M.B.; et al. Kisspeptin directly stimulates gonadotropin-releasing hormone release via G protein-coupled receptor 54. Proc. Nat. Acad. Sci. USA 2005, 102, 1761–1766. [Google Scholar] [CrossRef]
- Gottsch, M.L.; Cunningham, M.J.; Smith, J.T.; Popa, S.M.; Acohido, B.V.; Crowley, W.F.; Seminara, S.; Clifton, D.K.; Steiner, R.A. A role for kisspeptins in the regulation of gonadotropin secretion in the mouse. Endocrinology 2004, 145, 4073–4077. [Google Scholar] [CrossRef] [PubMed]
- Matsui, H.; Takatsu, Y.; Kumano, S.; Matsumoto, H.; Ohtaki, T. Peripheral administration of metastin induces marked gonadotropin release and ovulation in the rat. Biochem. Biophys. Res. Commun. 2004, 320, 383–388. [Google Scholar] [CrossRef]
- Yosten, G.L.; Lyu, R.M.; Hsueh, A.J.; Avsian-Kretchmer, O.; Chang, J.K.; Tullock, C.W.; Dun, S.L.; Dun, N.; Samson, W.K. A novel reproductive peptide, phoenixin. J. Neuroendocrinol. 2013, 25, 206–215. [Google Scholar] [CrossRef]
- Treen, A.K.; Luo, V.; Belsham, D.D. Phoenixin activates immortalized GnRH and kisspeptin neurons through the novel receptor GPR173. Mol. Endocrinol. 2016, 30, 872–888. [Google Scholar] [CrossRef] [PubMed]
- Elias, S.G.; van Noord, P.A.; Peeters, P.H.; den Tonkelaar, I.; Grobbee, D.E. Childhood exposure to the 1944–1945 Dutch famine and subsequent female reproductive function. Hum. Reprod. 2005, 20, 2483–2488. [Google Scholar] [CrossRef] [PubMed]
- Hessler, S.; Liu, X.; Herbison, A.E. Direct inhibition of arcuate kisspeptin neurones by neuropeptide Y in the male and female mouse. J. Neuroendocrinol. 2020, 32, e12849. [Google Scholar] [CrossRef]
- Roa, J.; Herbison, A.E. Direct regulation of GnRH neuron excitability by arcuate nucleus POMC and NPY neuron neuropeptides in female mice. Endocrinology 2012, 153, 5587–5599. [Google Scholar] [CrossRef] [PubMed]
- Padilla, S.L.; Qiu, J.; Nestor, C.C.; Zhang, C.; Smith, A.W.; Whiddon, B.B.; Ronnekleiv, O.K.; Kelly, M.J.; Palmiter, R.D. AgRP to Kiss1 neuron signaling links nutritional state and fertility. Proc. Nat. Acad. Sci. USA 2017, 114, 2413–2418. [Google Scholar] [CrossRef] [PubMed]
- Matsuyama, S.; Ohkura, S.; Sakurai, K.; Tsukamura, H.; Maeda, K.; Okamura, H. Activation of melanocortin receptors accelerates the gonadotropin-releasing hormone pulse generator activity in goats. Neurosci. Lett. 2005, 383, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Divall, S.A.; Williams, T.R.; Carver, S.E.; Koch, L.; Bruning, J.C.; Kahn, C.R.; Wondisford, F.; Radovick, S.; Wolfe, A. Divergent roles of growth factors in the GnRH regulation of puberty in mice. J. Clin. Investig. 2010, 120, 2900–2909. [Google Scholar] [CrossRef] [PubMed]
- Quennell, J.H.; Mulligan, A.C.; Tups, A.; Liu, X.; Phipps, S.J.; Kemp, C.J.; Herbison, A.E.; Grattan, D.R.; Anderson, G.M. Leptin indirectly regulates gonadotropin-releasing hormone neuronal function. Endocrinology 2009, 150, 2805–2812. [Google Scholar] [CrossRef]
- Quennell, J.H.; Howell, C.S.; Roa, J.; Augustine, R.A.; Grattan, D.R.; Anderson, G.M. Leptin deficiency and diet-induced obesity reduce hypothalamic kisspeptin expression in mice. Endocrinology 2011, 152, 1541–1550. [Google Scholar] [CrossRef] [PubMed]
- Smith, J.T.; Acohido, B.V.; Clifton, D.K.; Steiner, R.A. KiSS-1 neurones are direct targets for leptin in the ob/ob mouse. J. Neuroendocrinol. 2006, 18, 298–303. [Google Scholar] [CrossRef]
- McFadden, S.A.; Menchella, J.A.; Chalmers, J.A.; Centeno, M.L.; Belsham, D.D. Glucose responsiveness in a novel adult-derived GnRH cell line, mHypoA-GnRH/GFP: Involvement of AMP-activated protein kinase. Mol. Cell. Endocrinol. 2013, 377, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Graham, I.; Hastings, R.; Gunewardena, S.; Brinkmeier, M.L.; Conn, P.M.; Camper, S.A.; Kumar, T.R. Gonadotrope-specific deletion of Dicer results in severely suppressed gonadotropins and fertility defects. J. Biol. Chem. 2015, 290, 2699–2714. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Elías, M.D.; Rainero Cáceres, T.S.; Giaccagli, M.M.; Guazzone, V.A.; Dalton, G.N.; De Siervi, A.; Cuasnicú, P.S.; Cohen, D.J.; Da Ros, V.G. Association between high-fat diet feeding and male fertility in high reproductive performance mice. Sci. Rep. 2019, 9, 18546. [Google Scholar] [CrossRef]
- Soulis, G.; Kitraki, E.; Gerozissis, K. Early neuroendocrine alterations in female rats following a diet moderately enriched in fat. Cell. Mol. Neurobiol. 2005, 25, 869–880. [Google Scholar] [CrossRef]
- DiVall, S.A.; Herrera, D.; Sklar, B.; Wu, S.; Wondisford, F.; Radovick, S.; Wolfe, A. Insulin receptor signaling in the GnRH neuron plays a role in the abnormal GnRH pulsatility of obese female mice. PLoS ONE 2015, 10, e0119995. [Google Scholar] [CrossRef]
- Balasubramanian, P.; Jagannathan, L.; Mahaley, R.E.; Subramanian, M.; Gilbreath, E.T.; Mohankumar, P.S.; Mohankumar, S.M. High fat diet affects reproductive functions in female diet-induced obese and dietary resistant rats. J. Neuroendocrinol. 2012, 24, 748–755. [Google Scholar] [CrossRef]
- Hussain, M.A.; Abogresha, N.M.; Hassan, R.; Tamany, D.A.; Lotfy, M. Effect of feeding a high-fat diet independently of caloric intake on reproductive function in diet-induced obese female rats. Arch. Med. Sci. 2016, 12, 906–914. [Google Scholar] [CrossRef] [PubMed]
- Levi, N.J.; Wilson, C.W.; Redweik, G.A.J.; Gray, N.W.; Grzybowski, C.W.; Lenkey, J.A.; Moseman, A.W.; Bertsch, A.D.; Dao, N.; Walsh, H.E. Obesity-related cellular stressors regulate gonadotropin releasing hormone gene expression via c-Fos/AP-1. Mol. Cell. Endocrinol. 2018, 478, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Barabas, K.; Szabo-Meleg, E.; Abraham, I.M. Effect of inflammation on female gonadotropin-releasing hormone (GnRH) neurons: Mechanisms and consequences. Int. J. Mol. Sci. 2020, 21, 529. [Google Scholar] [CrossRef] [PubMed]
- Morelli, A.; Sarchielli, E.; Comeglio, P.; Filippi, S.; Vignozzi, L.; Marini, M.; Rastrelli, G.; Maneschi, E.; Cellai, I.; Persani, L.; et al. Metabolic syndrome induces inflammation and impairs gonadotropin-releasing hormone neurons in the preoptic area of the hypothalamus in rabbits. Mol. Cell. Endocrinol. 2014, 382, 107–119. [Google Scholar] [CrossRef]
- Lainez, N.M.; Jonak, C.R.; Nair, M.G.; Ethell, I.M.; Wilson, E.H.; Carson, M.J.; Coss, D. Diet-induced obesity elicits macrophage infiltration and reduction in spine density in the hypothalami of male but not female mice. Front. Immunol. 2018, 9, 1992. [Google Scholar] [CrossRef] [PubMed]
- McIlwraith, E.K.; Loganathan, N.; Belsham, D.D. Phoenixin expression is Regulated by the fatty acids palmitate, docosahexaenoic acid and oleate, and the endocrine disrupting chemical bisphenol a in immortalized hypothalamic neurons. Front. Neurosci. 2018, 12, 838. [Google Scholar] [CrossRef]
- McIlwraith, E.K.; Loganathan, N.; Belsham, D.D. Regulation of Gpr173 expression, a putative phoenixin receptor, by saturated fatty acid palmitate and endocrine-disrupting chemical bisphenol A through a p38-mediated mechanism in immortalized hypothalamic neurons. Mol. Cell. Endocrinol. 2019, 485, 54–60. [Google Scholar] [CrossRef]
- Zhai, L.; Zhao, J.; Zhu, Y.; Liu, Q.; Niu, W.; Liu, C.; Wang, Y. Downregulation of leptin receptor and kisspeptin/GPR54 in the murine hypothalamus contributes to male hypogonadism caused by high-fat diet-induced obesity. Endocrine 2018, 62, 195–206. [Google Scholar] [CrossRef]
- Barchitta, M.; Maugeri, A.; Quattrocchi, A.; Agrifoglio, O.; Agodi, A. The role of miRNAs as biomarkers for pregnancy outcomes: A comprehensive review. Int. J. Genomics 2017, 2017, 8067972. [Google Scholar] [CrossRef] [PubMed]
- Vashisht, A.; Gahlay, G.K. Using miRNAs as diagnostic biomarkers for male infertility: Opportunities and challenges. Mol. Hum. Reprod. 2020, 26, 199–214. [Google Scholar] [CrossRef] [PubMed]
- Messina, A.; Langlet, F.; Chachlaki, K.; Roa, J.; Rasika, S.; Jouy, N.; Gallet, S.; Gaytan, F.; Parkash, J.; Tena-Sempere, M.; et al. A microRNA switch regulates the rise in hypothalamic GnRH production before puberty. Nat. Neurosci. 2016, 19, 835–844. [Google Scholar] [CrossRef] [PubMed]
- Odle, A.K.; MacNicol, M.C.; Childs, G.V.; MacNicol, A.M. Post-transcriptional regulation of Gnrhr: A checkpoint for metabolic control of female reproduction. Int. J. Mol. Sci. 2021, 22, 3312. [Google Scholar] [CrossRef] [PubMed]
- Heras, V.; Sangiao-Alvarellos, S.; Manfredi-Lozano, M.; Sanchez-Tapia, M.J.; Ruiz-Pino, F.; Roa, J.; Lara-Chica, M.; Morrugares-Carmona, R.; Jouy, N.; Abreu, A.P.; et al. Hypothalamic miR-30 regulates puberty onset via repression of the puberty-suppressing factor, Mkrn3. PLoS Biol. 2019, 17, e3000532. [Google Scholar] [CrossRef] [PubMed]
- Perdices López, C.M.; Avendaño Herrador, M.S.; Vázquez Villar, M.S.; Romero, A.B.; Ruiz-Pino, F.; Morrugares-Carmona, R.; Sáez, A.C.; Calzado, M.A.; Rodríguez, J.M.C.; Alonso, A.M.B.; et al. Identification of a novel hypothalamic miRNA/Kisspeptin pathway as pathophysiological mechanism and putative target for management of obesity-induced hypogonadism. In Proceedings of the 22nd European Congress of Endocrinology, Online, 5–9 September 2020. [Google Scholar]
- Saper, C.B.; Lu, J.; Chou, T.C.; Gooley, J. The hypothalamic integrator for circadian rhythms. Trends Neurosci. 2005, 28, 152–157. [Google Scholar] [CrossRef]
- Valenzuela, F.J.; Vera, J.; Venegas, C.; Munoz, S.; Oyarce, S.; Munoz, K.; Lagunas, C. Evidences of polymorphism associated with circadian system and risk of pathologies: A review of the literature. Int. J. Endocrinol. 2016, 2016, 2746909. [Google Scholar] [CrossRef]
- Di Lorenzo, L.; De Pergola, G.; Zocchetti, C.; L’Abbate, N.; Basso, A.; Pannacciulli, N.; Cignarelli, M.; Giorgino, R.; Soleo, L. Effect of shift work on body mass index: Results of a study performed in 319 glucose-tolerant men working in a Southern Italian industry. Int. J. Obes. Relat. Metab. Disord. 2003, 27, 1353–1358. [Google Scholar] [CrossRef]
- Karlsson, B.; Knutsson, A.; Lindahl, B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occup. Environ. Med. 2001, 58, 747–752. [Google Scholar] [CrossRef] [PubMed]
- Oosterman, J.E.; Kalsbeek, A.; la Fleur, S.E.; Belsham, D.D. Impact of nutrients on circadian rhythmicity. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2015, 308, R337–R350. [Google Scholar] [CrossRef]
- Buijs, F.N.; Guzman-Ruiz, M.; Leon-Mercado, L.; Basualdo, M.C.; Escobar, C.; Kalsbeek, A.; Buijs, R.M. Suprachiasmatic nucleus interaction with the arcuate nucleus; essential for organizing physiological rhythms. eNeuro 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Buhr, E.D.; Takahashi, J.S. Molecular components of the Mammalian circadian clock. Handb. Exp. Pharmacol. 2013, 3–27. [Google Scholar] [CrossRef]
- Kinoshita, C.; Aoyama, K.; Nakaki, T. Neuroprotection afforded by circadian regulation of intracellular glutathione levels: A key role for miRNAs. Free Radic. Biol. Med. 2018, 119, 17–33. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Mo, G.; Tan, Y. Micro RNAs and the biological clock: A target for diseases associated with a loss of circadian regulation. Afr. Health Sci. 2020, 20, 1887–1894. [Google Scholar] [CrossRef] [PubMed]
- Taouis, M. MicroRNAs in the hypothalamus. Best Pract. Res. Clin. Endocrinol. Metab. 2016, 30, 641–651. [Google Scholar] [CrossRef] [PubMed]
- Arble, D.M.; Bass, J.; Laposky, A.D.; Vitaterna, M.H.; Turek, F.W. Circadian timing of food intake contributes to weight gain. Obesity 2009, 17, 2100–2102. [Google Scholar] [CrossRef] [PubMed]
- Brown-Grant, K.; Raisman, G. Abnormalities in reproductive function associated with the destruction of the suprachiasmatic nuclei in female rats. Proc. R. Soc. Lond. B. Biol. Sci. 1977, 198, 279–296. [Google Scholar] [CrossRef]
- Simonneaux, V. A Kiss to drive rhythms in reproduction. Eur. J. Neurosci. 2018, 51, 509–530. [Google Scholar] [CrossRef]
- Kim, E.R.; Xu, Y.; Cassidy, R.M.; Lu, Y.; Yang, Y.; Tian, J.; Li, D.P.; Van Drunen, R.; Ribas-Latre, A.; Cai, Z.L.; et al. Paraventricular hypothalamus mediates diurnal rhythm of metabolism. Nat. Commun. 2020, 11, 3794. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, J.A.; Lin, S.Y.; Martino, T.A.; Arab, S.; Liu, P.; Husain, M.; Sole, M.J.; Belsham, D.D. Diurnal profiling of neuroendocrine genes in murine heart, and shift in proopiomelanocortin gene expression with pressure-overload cardiac hypertrophy. J. Mol. Endocrinol. 2008, 41, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Stutz, A.M.; Staszkiewicz, J.; Ptitsyn, A.; Argyropoulos, G. Circadian expression of genes regulating food intake. Obesity 2007, 15, 607–615. [Google Scholar] [CrossRef] [PubMed]
- Fick, L.J.; Fick, G.H.; Belsham, D.D. Rhythmic clock and neuropeptide gene expression in hypothalamic mHypoE-44 neurons. Mol.Cell. Endocrinol. 2010, 323, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Cedernaes, J.; Huang, W.; Ramsey, K.M.; Waldeck, N.; Cheng, L.; Marcheva, B.; Omura, C.; Kobayashi, Y.; Peek, C.B.; Levine, D.C.; et al. Transcriptional basis for rhythmic control of hunger and metabolism within the AgRP neuron. Cell. Metab. 2019, 29, 1078–1091.e5. [Google Scholar] [CrossRef] [PubMed]
- Kohsaka, A.; Laposky, A.D.; Ramsey, K.M.; Estrada, C.; Joshu, C.; Kobayashi, Y.; Turek, F.W.; Bass, J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell. Metab. 2007, 6, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Greco, J.A.; Oosterman, J.E.; Belsham, D.D. Differential effects of omega-3 fatty acid docosahexaenoic acid and palmitate on the circadian transcriptional profile of clock genes in immortalized hypothalamic neurons. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2014, 307, R1049–R1060. [Google Scholar] [CrossRef]
- Oosterman, J.E.; Belsham, D.D. Glucose Alters Per2 Rhythmicity Independent of AMPK, Whereas AMPK Inhibitor Compound C Causes Profound Repression of Clock Genes and AgRP in mHypoE-37 Hypothalamic Neurons. PLoS ONE 2016, 11, e0146969. [Google Scholar] [CrossRef]
- Chen, R.; Zuo, Z.; Li, Q.; Wang, H.; Li, N.; Zhang, H.; Yu, X.; Liu, Z. DHA substitution overcomes high-fat diet-induced disturbance in the circadian rhythm of lipid metabolism. Food Funct. 2020, 11, 3621–3631. [Google Scholar] [CrossRef]
- Tal, Y.; Chapnik, N.; Froy, O. Non-obesogenic doses of palmitate disrupt circadian metabolism in adipocytes. Adipocyte 2019, 8, 392–400. [Google Scholar] [CrossRef] [PubMed]
- Sahar, S.; Zocchi, L.; Kinoshita, C.; Borrelli, E.; Sassone-Corsi, P. Regulation of BMAL1 protein stability and circadian function by GSK3beta-mediated phosphorylation. PLoS ONE 2010, 5, e8561. [Google Scholar] [CrossRef] [PubMed]
- Dang, F.; Sun, X.; Ma, X.; Wu, R.; Zhang, D.; Chen, Y.; Xu, Q.; Wu, Y.; Liu, Y. Insulin post-transcriptionally modulates Bmal1 protein to affect the hepatic circadian clock. Nat. Commun. 2016, 7, 12696. [Google Scholar] [CrossRef]
- Wada, T.; Ichihashi, Y.; Suzuki, E.; Kosuge, Y.; Ishige, K.; Uchiyama, T.; Makishima, M.; Nakao, R.; Oishi, K.; Shimba, S. Deletion of Bmal1 prevents diet-induced ectopic fat accumulation by controlling oxidative capacity in the skeletal muscle. Int. J. Mol. Sci. 2018, 19, 2813. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.L.; Kooijman, S.; Gao, Y.; Tzeplaeff, L.; Cosquer, B.; Milanova, I.; Wolff, S.E.C.; Korpel, N.; Champy, M.F.; Petit-Demoulière, B.; et al. Microglia-specific knock-down of Bmal1 improves memory and protects mice from high fat diet-induced obesity. Mol. Psychiatry 2021. [Google Scholar] [CrossRef] [PubMed]
- Moraes, J.C.; Coope, A.; Morari, J.; Cintra, D.E.; Roman, E.A.; Pauli, J.R.; Romanatto, T.; Carvalheira, J.B.; Oliveira, A.L.; Saad, M.J.; et al. High-fat diet induces apoptosis of hypothalamic neurons. PLoS ONE 2009, 4, e5045. [Google Scholar] [CrossRef]
- Tran, A.; He, W.; Jiang, N.; Chen, J.T.C.; Belsham, D.D. NAMPT and BMAL1 are independently involved in the palmitate-mediated induction of neuroinflammation in hypothalamic neurons. Front. Endocrinol. 2020, 11, 351. [Google Scholar] [CrossRef] [PubMed]
- Sergi, D.; Morris, A.C.; Kahn, D.E.; McLean, F.H.; Hay, E.A.; Kubitz, P.; MacKenzie, A.; Martinoli, M.G.; Drew, J.E.; Williams, L.M. Palmitic acid triggers inflammatory responses in N42 cultured hypothalamic cells partially via ceramide synthesis but not via TLR4. Nutr. Neurosci. 2020, 23, 321–334. [Google Scholar] [CrossRef]
- Adlanmerini, M.; Nguyen, H.C.; Krusen, B.M.; Teng, C.W.; Geisler, C.E.; Peed, L.C.; Carpenter, B.J.; Hayes, M.R.; Lazar, M.A. Hypothalamic REV-ERB nuclear receptors control diurnal food intake and leptin sensitivity in diet-induced obese mice. J. Clin. Investig. 2021, 131, e140424. [Google Scholar] [CrossRef]
- Chen, R.; D’Alessandro, M.; Lee, C. miRNAs are required for generating a time delay critical for the circadian oscillator. Curr. Biol. 2013, 23, 1959–1968. [Google Scholar] [CrossRef]
- Shende, V.R.; Goldrick, M.M.; Ramani, S.; Earnest, D.J. Expression and rhythmic modulation of circulating microRNAs targeting the clock gene Bmal1 in mice. PLoS ONE 2011, 6, e22586. [Google Scholar] [CrossRef]
- Shende, V.R.; Neuendorff, N.; Earnest, D.J. Role of miR-142-3p in the post-transcriptional regulation of the clock gene Bmal1 in the mouse SCN. PLoS ONE 2013, 8, e65300. [Google Scholar] [CrossRef]
- Chemello, F.; Grespi, F.; Zulian, A.; Cancellara, P.; Hebert-Chatelain, E.; Martini, P.; Bean, C.; Alessio, E.; Buson, L.; Bazzega, M.; et al. Transcriptomic analysis of single isolated myofibers identifies miR-27a-3p and miR-142-3p as regulators of metabolism in skeletal muscle. Cell. Rep. 2019, 26, 3784–3797.e8. [Google Scholar] [CrossRef]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in female mice prevents diet-induced obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef]
- Zhou, L.; Miller, C.; Miraglia, L.J.; Romero, A.; Mure, L.S.; Panda, S.; Kay, S.A. A genome-wide microRNA screen identifies the microRNA-183/96/182 cluster as a modulator of circadian rhythms. Proc. Natl. Acad. Sci. USA 2021, 118. [Google Scholar] [CrossRef]
- Yang, W.M.; Min, K.H.; Lee, W. Induction of miR-96 by dietary saturated fatty acids exacerbates hepatic insulin resistance through the suppression of INSR and IRS-1. PLoS ONE 2016, 11, e0169039. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.Y.; Papp, J.W.; Varlamova, O.; Dziema, H.; Russell, B.; Curfman, J.P.; Nakazawa, T.; Shimizu, K.; Okamura, H.; Impey, S.; et al. microRNA modulation of circadian-clock period and entrainment. Neuron 2007, 54, 813–829. [Google Scholar] [CrossRef]
- Alvarez-Saavedra, M.; Antoun, G.; Yanagiya, A.; Oliva-Hernandez, R.; Cornejo-Palma, D.; Perez-Iratxeta, C.; Sonenberg, N.; Cheng, H.Y. miRNA-132 orchestrates chromatin remodeling and translational control of the circadian clock. Hum. Mol. Genet. 2011, 20, 731–751. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Casals, N.; Keung, W.; Moran, T.H.; Lopaschuk, G.D. Differential effects of central ghrelin on fatty acid metabolism in hypothalamic ventral medial and arcuate nuclei. Physiol. Behav. 2013, 118, 165–170. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Salazar, T.E.; Dominguez, J.M.; Nguyen, D.V.; Li Calzi, S.; Bhatwadekar, A.D.; Qi, X.; Busik, J.V.; Boulton, M.E.; Grant, M.B. Dicer expression exhibits a tissue-specific diurnal pattern that is lost during aging and in diabetes. PLoS ONE 2013, 8, e80029. [Google Scholar] [CrossRef]
- Dasgupta, I.; Chatterjee, A. Recent advances in miRNA delivery systems. Methods Protoc. 2021, 4, 10. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).