Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture and Conjugation
2.2. Bioinformatic Analysis
2.3. Construction of HA-ATG5 and HA-ATG8.2 Strains
2.4. ATG5 Knockdown by RNA Interference
2.5. Labeling of Autophagosomal Structures and Lysosomes
2.6. Indirect Immunofluorescence Analysis
2.7. Western Blot Analysis
2.8. Statistical Analysis
3. Results
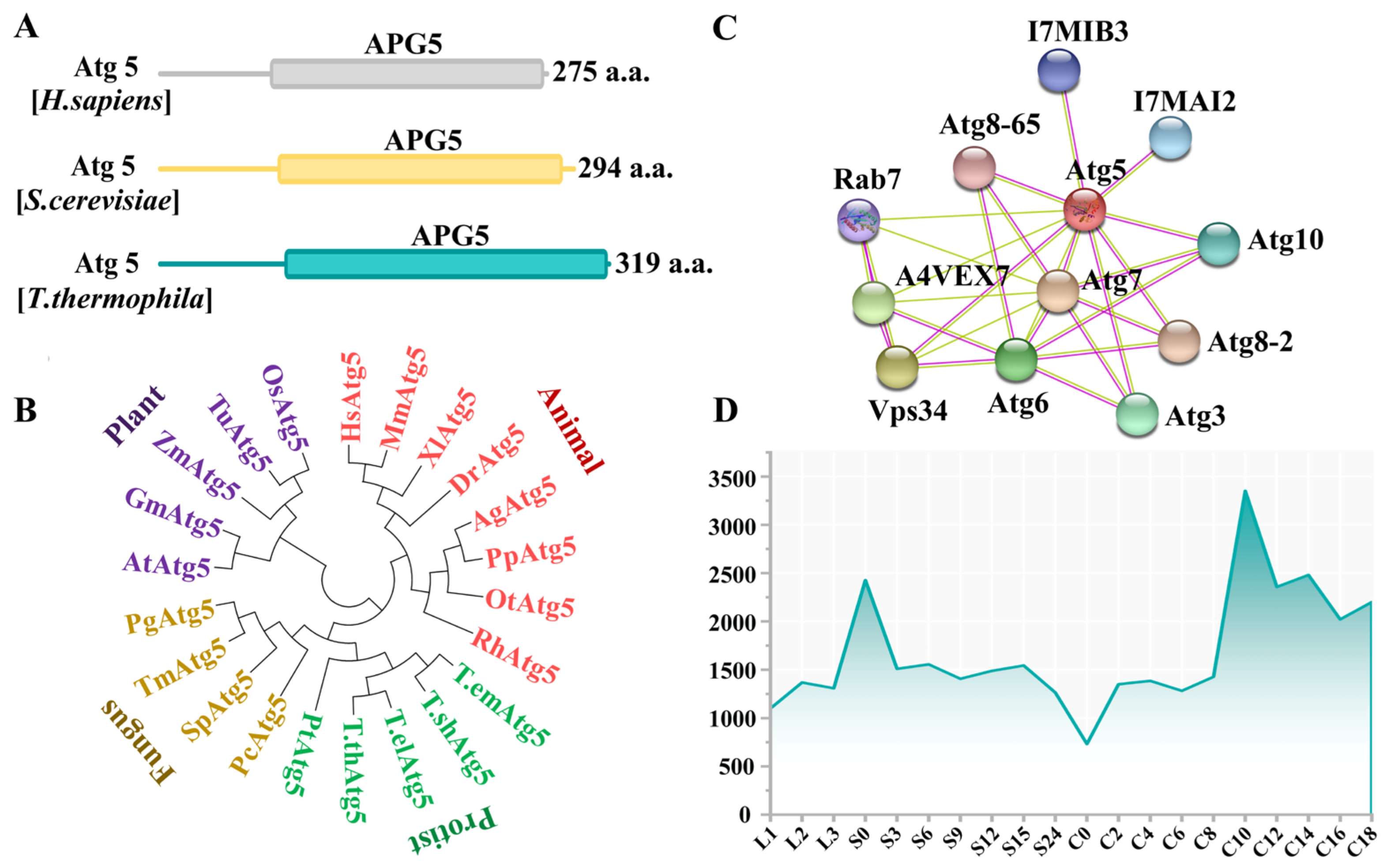
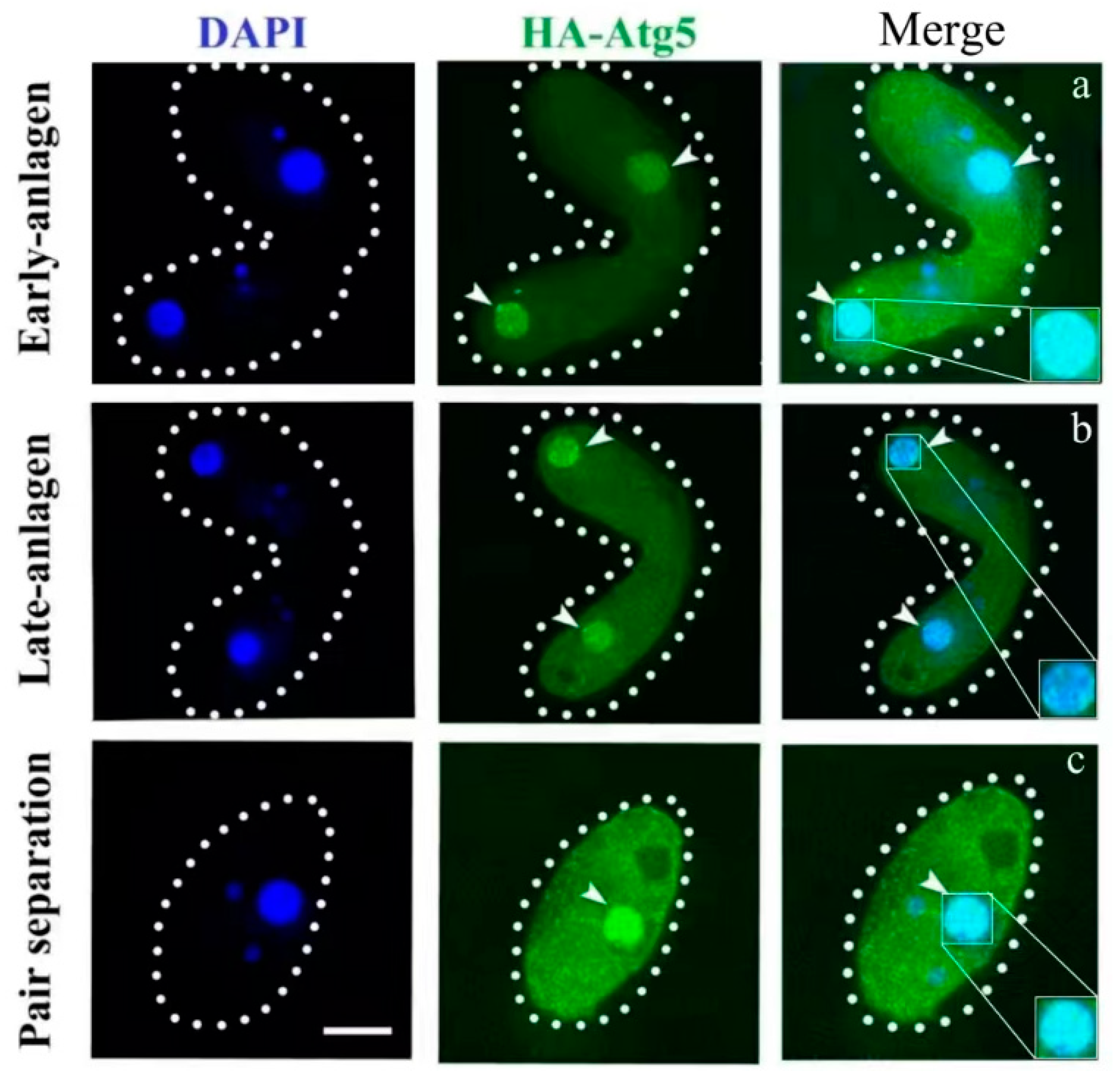
3.1. Characterization of ATG5 in Tetrahymena
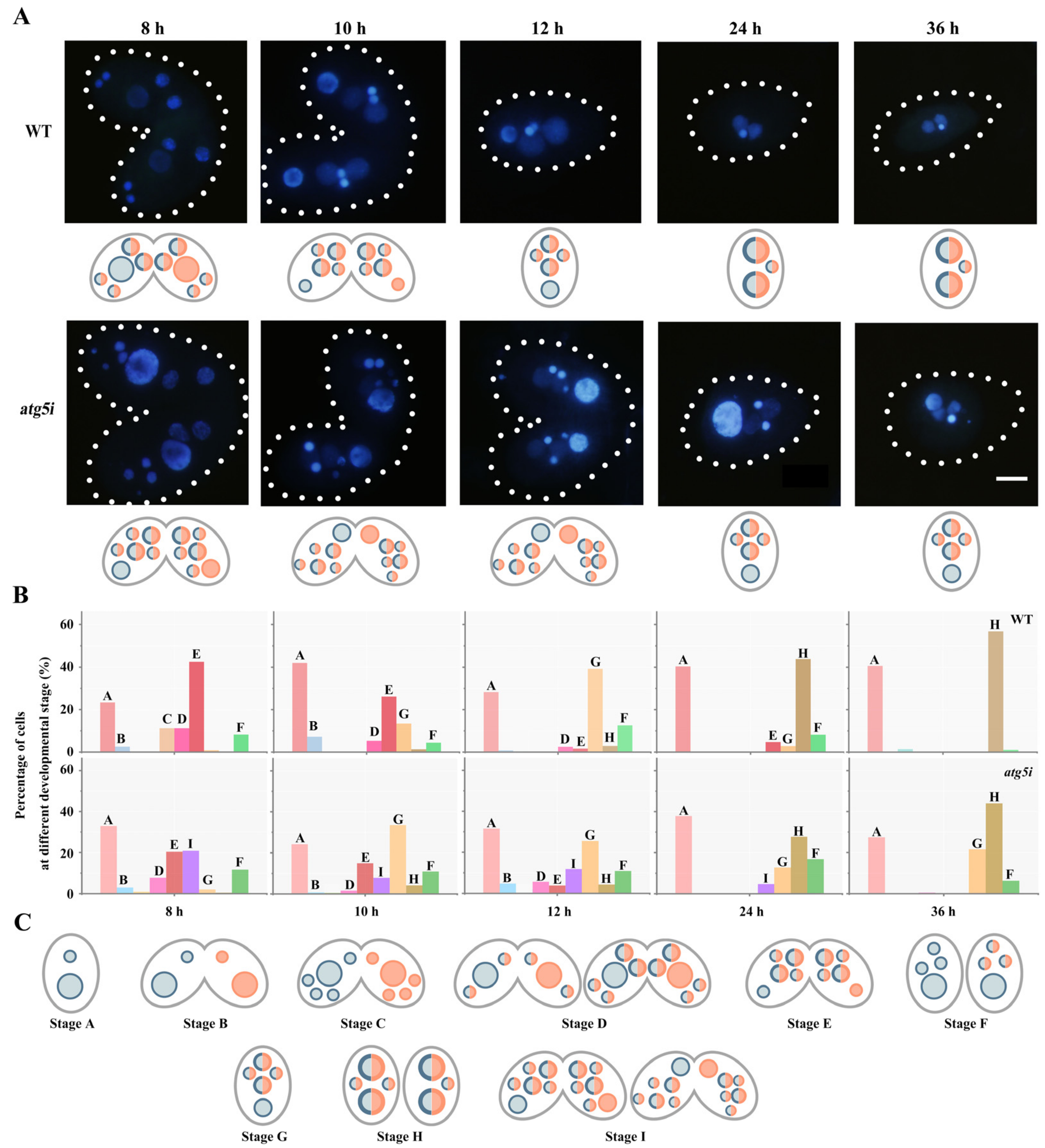
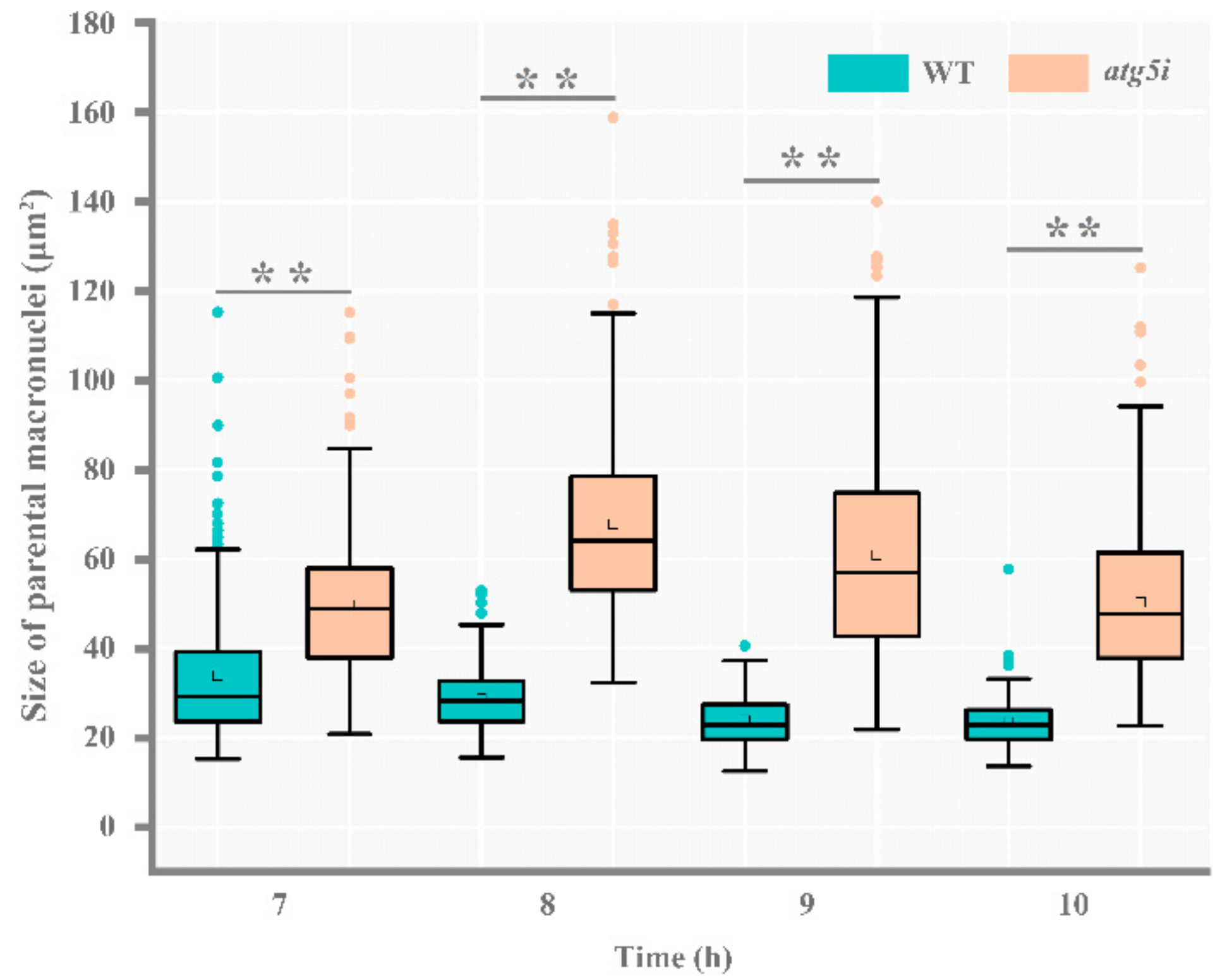
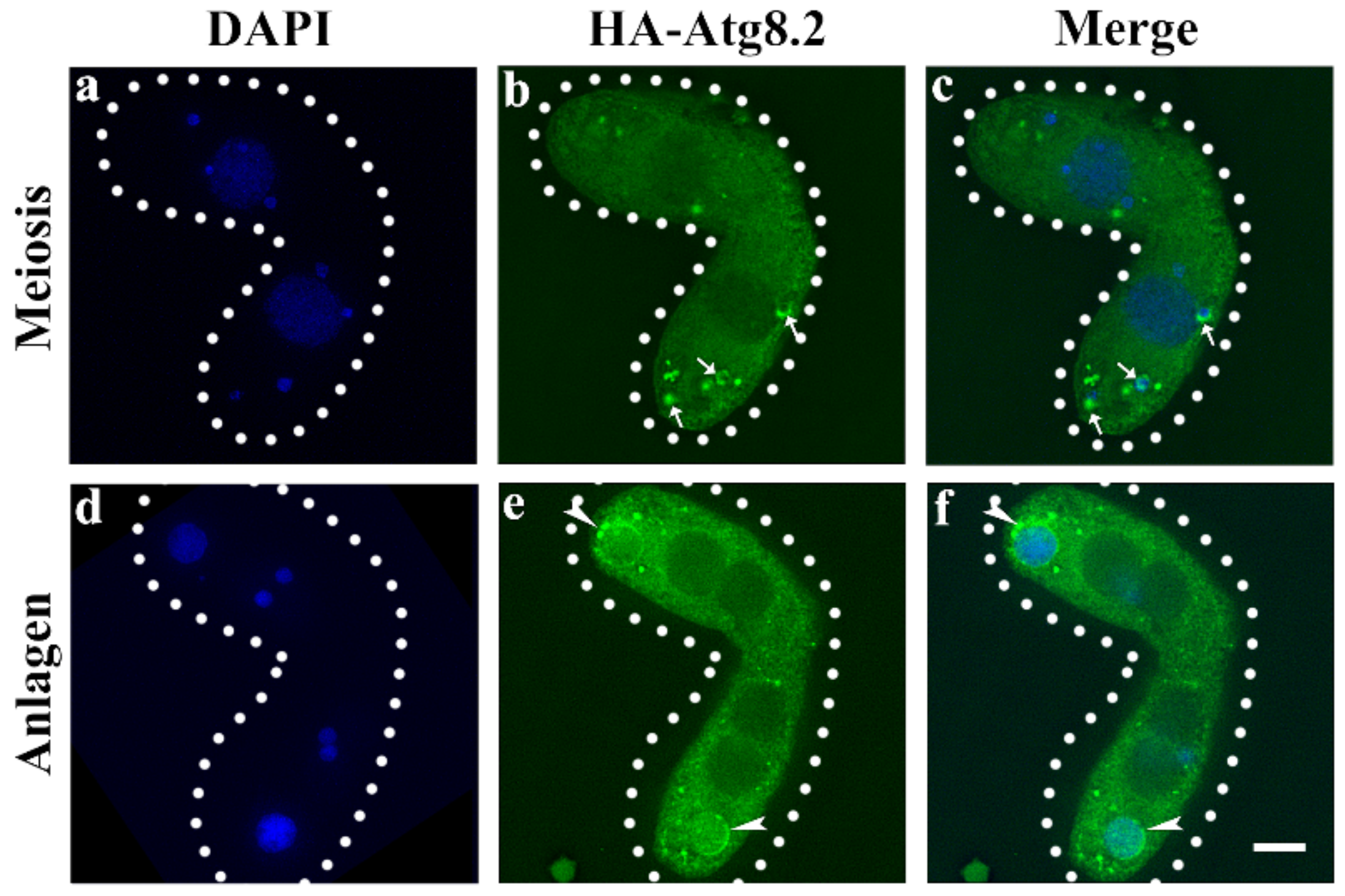
3.2. ATG5 Knockdown Inhibites the PND of paMAC
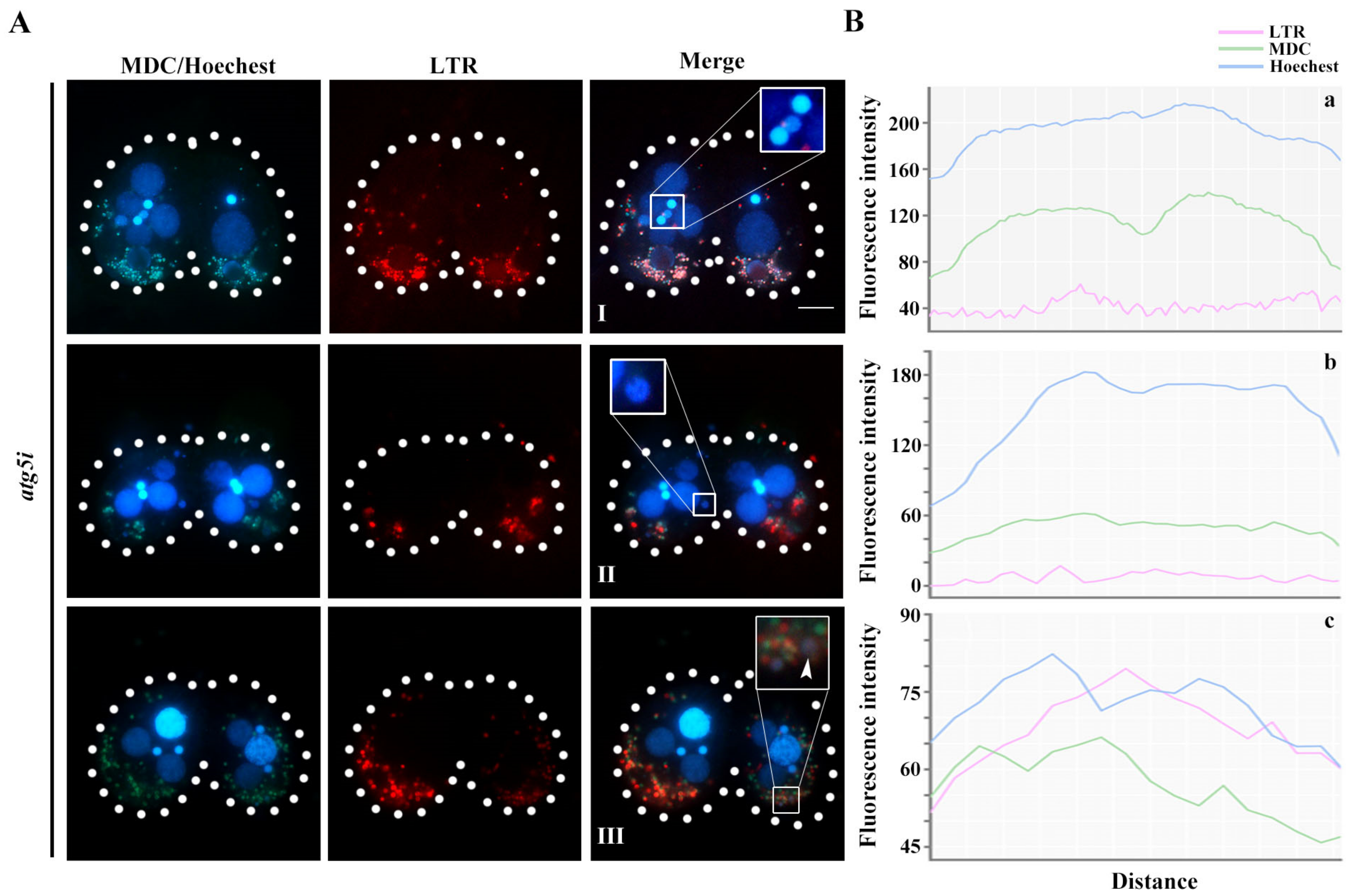
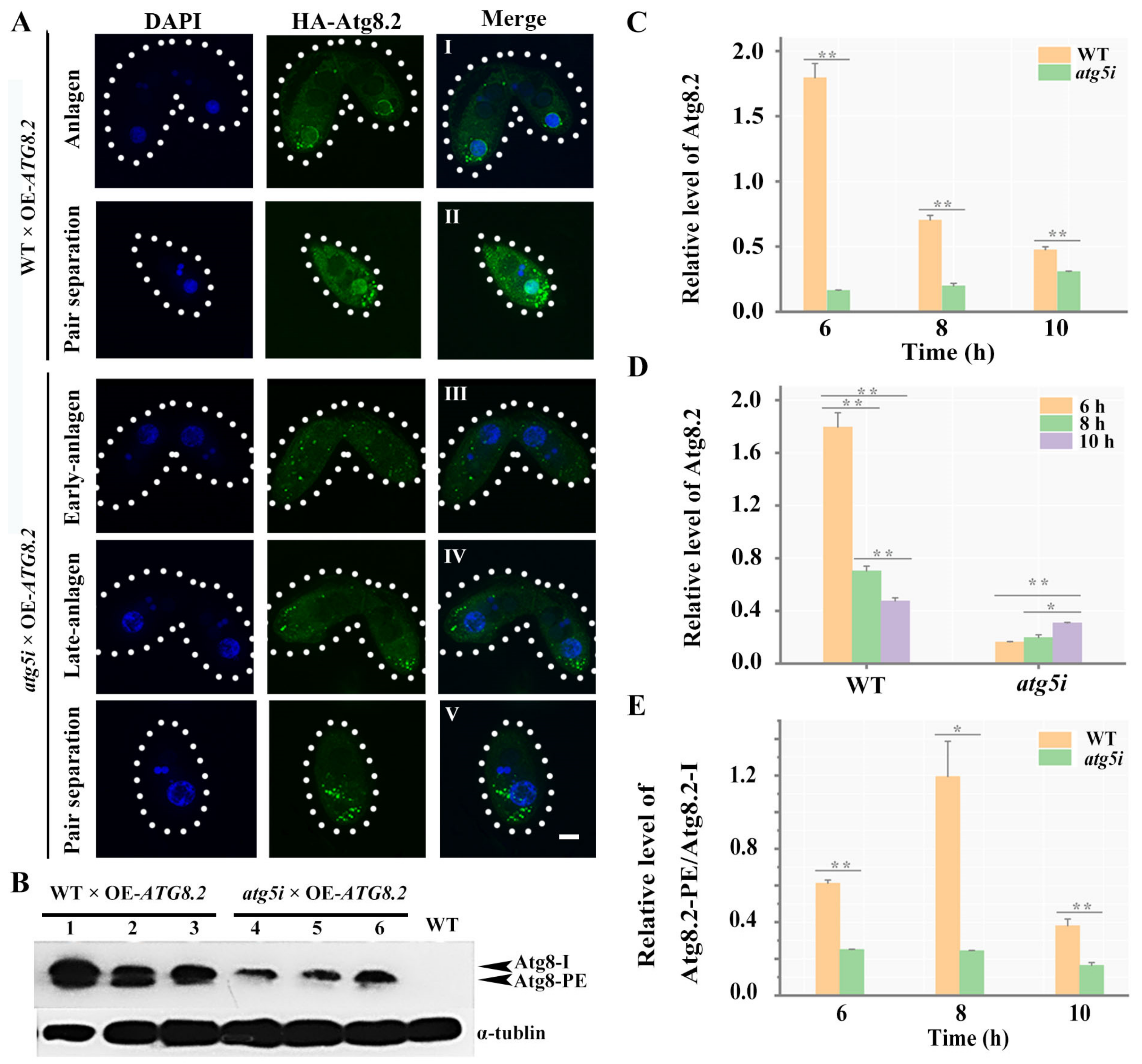
3.3. Atg5 Mediated the PND of paMAC by Regulating the Function of Atg8.2
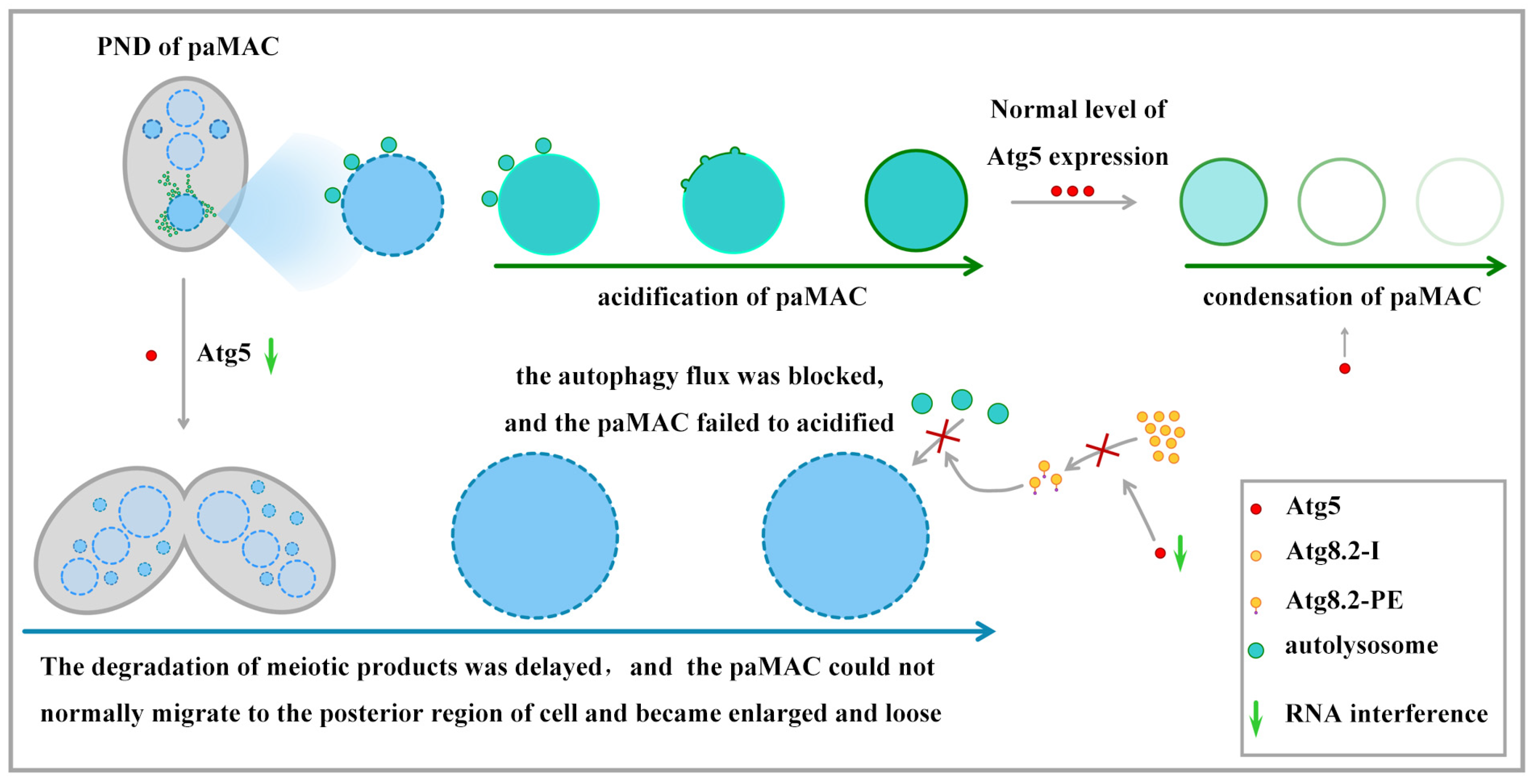
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.-L. AMPK and mTOR Regulate Autophagy through Direct Phosphorylation of Ulk1. Nature 2011, 13, 132–141. [Google Scholar] [CrossRef]
- Mizushima, N.; Komatsu, M. Autophagy: Renovation of Cells and Tissues. Cell 2011, 147, 728–741. [Google Scholar] [CrossRef] [PubMed]
- Shen, H.M.; Mizushima, N. At the End of the Autophagic Road: An Emerging Understanding of Lysosomal Functions in Autophagy. Trends. Biochem. Sci. 2014, 39, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Tan, S.-H.; Nicolas, N.; Bauvy, C.; Yang, N.-D.; Zhang, J.; Xue, Y.P. Codogno, P.; Shen, H.-M. Activation of Lysosomal Function in the Course of Autophagy via mTORC1 Suppression and Autophagosome-Lysosome Fusion. Cell Res. 2013, 23, 508–523. [Google Scholar] [CrossRef]
- Yordy, B.; Tal, M.; Hayashi, K.; Arojo, O.; Iwasaki, A. Autophagy and Selective Deployment of Atg Proteins in Antiviral Defense. Int. Immunol. 2012, 25, 1–10. [Google Scholar] [CrossRef]
- White, E. The role for autophagy in cancer. J. Clin. Investig. 2015, 125, 42–46. [Google Scholar] [CrossRef]
- Menzies, F.M.; Fleming, A.; Rubinsztein, D.C. Compromised Autophagy and Neurodegenerative Diseases. Nat. Rev. Neurosci. 2015, 16, 345–357. [Google Scholar] [CrossRef]
- Kroemer, G. Autophagy: A Druggable Process that is Deregulated in Aging and Human Disease. J. Clin. Investig. 2015, 125, 1–4. [Google Scholar] [CrossRef]
- Anding, A.L.; Baehrecke, E.H. Cleaning House: Selective Autophagy of Organelles. Dev. Cell 2017, 41, 10–22. [Google Scholar] [CrossRef]
- Léger-Silvestre, I.; Caffrey, J.M.; Dawaliby, R.; Alvarez-Arias, D.A.; Gas, N.; Bertolone, S.J.; Gleizes, P.-E.; Ellis, S.R. Specific Role for Yeast Homologs of the Diamond Blackfan Anemia-associated Rps19 Protein in Ribosome Synthesis. J. Biol. Chem. 2005, 280, 38177–38185. [Google Scholar] [CrossRef]
- Chen, J.-H.; Zhang, P.; Chen, W.-D.; Li, D.-D.; Wu, X.-Q.; Deng, R.; Jiao, L.; Li, X.; Ji, J.; Feng, G.-K.; et al. ATM-mediated PTEN Phosphorylation Promotes PTEN Nuclear Translocation and Autophagy in Response to DNA-Damaging Agents in Cancer Cells. Autophagy 2015, 11, 239–252. [Google Scholar] [CrossRef]
- Krick, R.; Muehe, Y.; Prick, T.; Bremer, S.; Schlotterhose, P.; Eskelinen, E.-L.; Millen, J.; Goldfarb, D.S.; Thumm, M. Piecemeal Microautophagy of the Nucleus Requires the Core Macroautophagy Genes. Mol. Biol. Cell 2008, 19, 4492–4505. [Google Scholar] [CrossRef]
- Kvam, E.; Goldfarb, D.S. Nucleus-Vacuole Junctions and Piecemeal Microautophagy of the Nucleus in S. Cerevisiae. Autophagy 2007, 3, 85–92. [Google Scholar] [CrossRef]
- Roberts, P.; Moshitch-Moshkovitz, S.; Kvam, E.; O’Toole, E.; Winey, M.; Goldfarb, D.S. Piecemeal Microautophagy of Nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 2003, 14, 129–141. [Google Scholar] [CrossRef]
- Luo, M.; Zhao, X.; Song, Y.; Cheng, H.; Zhou, R. Nuclear Autophagy: An evolutionarily Conserved Mechanism of Nuclear Degradation in the Cytoplasm. Autophagy 2016, 12, 1973–1983. [Google Scholar] [CrossRef]
- Park, Y.-E.; Hayashi, Y.K.; Bonne, G.; Arimura, T.; Noguchi, S.; Nonaka, I.; Nishino, I. Autophagic Degradation of Nuclear Components in Mammalian Cells. Autophagy 2009, 5, 795–804. [Google Scholar] [CrossRef]
- Kovács, A.L.; Réz, G.; Pálfia, Z.; Kovács, J. Autophagy in the Epithelial Cells of Murine Seminal Vesicle In Vitro. Formation of Large Sheets of Nascent Isolation Membranes, Sequestration of the Nucleus and Inhibition by Wortmannin and 3-ethyladenine. Cell Tissue Res. 2000, 302, 253–261. [Google Scholar]
- Akinduro, O.; Sully, K.; Patel, A.; Robinson, D.J.; Chikh, A.; McPhail, G.; Braun, K.M.; Philpott, M.P.; Harwood, C.; Byrne, C.; et al. Constitutive Autophagy and Nucleophagy during Epidermal Differentiation. J. Investig. Dermatol. 2016, 136, 1460–1470. [Google Scholar] [CrossRef]
- Nishimura, T.; Tooze, S.A. Emerging Roles of ATG Proteins and Membrane Lipids in Autophagosome Formation. Cell Discov. 2020, 6, 1–18. [Google Scholar] [CrossRef]
- Ye, X.; Zhou, X.-J.; Zhang, H. Exploring the Role of Autophagy-Related Gene 5 (ATG5) Yields Important Insights into Autophagy in Autoimmune/Autoinflammatory Diseases. Front. Immunol. 2018, 9, 2334. [Google Scholar] [CrossRef]
- Romanov, J.; Walczak, M.; Ibiricu, I.; Schüchner, S.; Ogris, E.; Kraft, C.; Martens, S. Mechanism and Functions of Membrane Binding by the Atg5-Atg12/Atg16 Complex during Autophagosome Formation. EMBO J. 2012, 31, 4304–4317. [Google Scholar] [CrossRef] [PubMed]
- Kharaziha, P.; Panaretakis, T. Dynamics of Atg5-Atg12-Atg16L1 Aggregation and Deaggregation. Methods Enzymol. 2017, 587, 247–255. [Google Scholar]
- Filimonenko, M.; Isakson, P.; Finley, K.D.; Anderson, M.; Jeong, H.; Melia, T.J.; Bartlett, B.J.; Myers, K.M.; Birkeland, H.C.; Lamark, T.; et al. The Selective Macroautophagic Degradation of Aggregated Proteins Requires the PI3P-Binding Protein Alfy. Mol. Cell 2010, 38, 265–279. [Google Scholar] [CrossRef]
- Fracchiolla, D.; Sawa-Makarska, J.; Zens, B.; de Ruiter, A.; Zaffagnini, G.; Brezovich, A.; Romanov, J.; Runggatscher, K.; Kraft, C.; Zagrovic, B.; et al. Mechanism of Cargo-Directed Atg8 Conjugation during Selective Autophagy. eLife 2016, 5, e18544. [Google Scholar] [CrossRef]
- Kirkin, V. History of the Selective Autophagy Research: How did it Begin and Where Does It Stand Today? J. Mol. Biol. 2020, 432, 3–27. [Google Scholar] [CrossRef]
- Collins, K.; Gorovsky, M.A. Tetrahymena thermophila. Curr. Biol. 2005, 15, R317–R318. [Google Scholar] [CrossRef]
- Xu, J.; Yuan, Y.; Liang, A.; Wang, W. Chromodomain Protein Tcd1 is Required for Macronuclear Genome Rearrangement and repair in Tetrahymena. Sci. Rep. 2015, 5, 10243. [Google Scholar] [CrossRef]
- Orias, E.; Cervantes, M.D.; Hamilton, E.P. Tetrahymena thermophila, a unicellular eukaryote with separate germline and somatic genomes. Res. Microbiol. 2011, 162, 578–586. [Google Scholar] [CrossRef]
- Liang, H.; Xu, J.; Wang, W. Ran1 is Essential for Parental Macronuclear Import of Apoptosis-Inducing Factor and Programmed Nuclear Death in Tetrahymena thermophila. FEBS J. 2018, 286, 913–929. [Google Scholar] [CrossRef] [PubMed]
- Osada, E.; Akematsu, T.; Asano, T.; Endoh, H. A Novel Mitochondrial Nuclease-Associated Protein: A Major Executor of the Programmed Nuclear Death in Tetrahymena thermophila. Biol. Cell 2014, 106, 97–109. [Google Scholar] [CrossRef]
- Akematsu, T.; Fukuda, Y.; Attiq, R.; Pearlman, E.R. Role of Class III Phosphatidylinositol 3-Kinase during Programmed Nuclear Death of Tetrahymena thermophila. Autophagy 2013, 10, 209–225. [Google Scholar] [CrossRef] [PubMed]
- Akematsu, T.; Endoh, H. Role of Apoptosis-Inducing Factor (AIF) in Programmed Nuclear Death During Conjugation in Tetrahymena thermophila. BMC Cell Biol. 2010, 11, 13. [Google Scholar] [CrossRef] [PubMed]
- Takahiko, A.; Pearlman, R.E.; Hiroshi, E.J.A. Gigantic Macroautophagy in Programmed Nuclear Death of Tetrahymena ther-mophila. Autophagy 2010, 6, 901–911. [Google Scholar]
- Liu, M.-L.; Yao, M.-C. Role of ATG8 and Autophagy in Programmed Nuclear Degradation in Tetrahymena thermophila. Eukaryot. Cell 2012, 11, 494–506. [Google Scholar] [CrossRef]
- Wenping, Z.; Tao, B.; Ya, L.; Jing, X.; Wei, W. Overexpression of Atg4.1 Promote Parental Macronucleus Programmed Deg-Radation in Tetrahymena thermophila. Chin. J. Cell. Biol. 2019, 41, 841–850. [Google Scholar]
- Qiao, J.; Jing, X.; Tao, B.; Wei, W.J.P.O. Micronucleus-Specific Histone H1 is Required for Micronuclear Chromosome Integrity in Tetrahymena Thermophila. PLoS ONE 2017, 12, e0187475. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Kuhn, M.; Simonovic, M.; Roth, A.; Minguez, P.; Doerks, T.; Stark, M.; Muller, J.; Bork, P.; et al. The STRING Database in 2011: Functional Interaction Networks of Proteins, Globally Integrated and Scored. Nucleic. Acids. Res. 2011, 39, D561–D568. [Google Scholar] [CrossRef]
- Franceschini, A.; Szklarczyk, D.; Frankild, S.; Kuhn, M.; Simonovic, M.; Roth, A.; Lin, J.; Minguez, P.; Bork, P.; von Mering, C.; et al. STRING v9.1: Protein-Protein Interaction Networks, with Increased Coverage and Integration. Nucleic Acids Res. 2013, 41, D808–D815. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 Years of the SMART Protein Domain Annotation Resource. Nucleic Acids Res. 2017, 46, D493–D496. [Google Scholar] [CrossRef]
- Larkin, M.; Blackshields, G.; Brown, N.; Chenna, R.; McGettigan, P.; McWilliam, H.; Valentin, F.; Wallace, I.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef]
- Clamp, M.; Cuff, J.; Searle, S.M.; Barton, G. The Jalview Java Alignment Editor. Bioinformatics 2004, 20, 426–427. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Cassidy-Hanley, D.; Bowen, J.; Lee, J.H.; Cole, E.; VerPlank, L.A.; Gaertig, J.; Gorovsky, M.A.; Bruns, P.J. Germline and Somatic Transformation of Mating Tetrahymena thermophila by Particle Bombardment. Genetics 1997, 146, 135–147. [Google Scholar] [CrossRef]
- Xu, J.; Tian, H.; Liu, X.; Wang, W.; Liang, A. Localization and Functional Analysis of HmgB3p, a Novel Protein Containing High-Mobility-Group-Box Domain from Tetrahymena thermophila. Gene 2013, 526, 87–95. [Google Scholar] [CrossRef]
- Howard-Till, R.A.; Lukaszewicz, A.; Novatchkova, M.; Loidl, J. A Single Cohesin Complex Performs Mitotic and Meiotic Functions in the Protist Tetrahymena. PLoS Genet. 2013, 9, e1003418. [Google Scholar] [CrossRef]
- Hanada, T.; Noda, N.; Satomi, Y.; Ichimura, Y.; Fujioka, Y.; Takao, T.; Inagaki, F.; Ohsumi, Y. The Atg12-Atg5 Conjugate Has a Novel E3-like Activity for Protein Lipidation in Autophagy. J. Biol. Chem. 2007, 282, 37298–37302. [Google Scholar] [CrossRef]
- Arakawa, S.; Honda, S.; Yamaguchi, H.; Shimizu, S. Molecular Mechanisms and Physiological Roles of Atg5/Atg7-Independent Alternative Autophagy. Proc. Jpn. Acad. Ser. B 2017, 93, 378–385. [Google Scholar] [CrossRef]
- Soubannier, V.; McLelland, G.-L.; Zunino, R.; Braschi, E.; Rippstein, P.; Fon, E.A.; McBride, H.M. A Vesicular Transport Pathway Shuttles Cargo from Mitochondria to Lysosomes. Curr. Biol. 2012, 22, 135–141. [Google Scholar] [CrossRef]
- Soubannier, V.; Rippstein, P.; Kaufman, B.A.; Shoubridge, E.A.; McBride, H.M. Reconstitution of Mitochondria Derived Vesicle Formation Demonstrates Selective Enrichment of Oxidized Cargo. PLoS ONE 2012, 7, e52830. [Google Scholar] [CrossRef]
- Lemasters, J.J. Variants of Mitochondrial Autophagy: Types 1 and 2 Mitophagy and Micromitophagy (Type 3). Redox Biol. 2014, 2, 749–754. [Google Scholar] [CrossRef]
- McLelland, G.L.; Soubannier, V.; Chen, C.X.; McBride, H.M.; Fon, E.A. Parkin and PINK1 function in a vesicular trafficking pathway regulating mitochondrial quality control. Embo. J. 2014, 33, 282–295. [Google Scholar] [CrossRef]
- Tsuboyama, K.; Koyama-Honda, I.; Sakamaki, Y.; Koike, M.; Morishita, H.; Mizushima, N. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science 2016, 354, 1036–1041. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Padman, B.S.; Usher, J.; Oorschot, V.; Ramm, G.; Lazarou, M. Atg8 family LC3/GABARAP Proteins are Crucial for Autophagosome-Lysosome Fusion but not Autophagosome Formation during PINK1/Parkin Mitophagy and Starvation. J. Cell. Biol. 2016, 215, 857–874. [Google Scholar] [CrossRef]
- Wang, H.; Sun, H.Q.; Zhu, X.; Zhang, L.; Albanesi, J.; Levine, B.; Yin, H. GABARAPs regulate PI4P-dependent autophago-some:lysosome fusion. Proc. Natl. Acad. Sci. USA 2015, 112, 7015–7020. [Google Scholar] [CrossRef]
- Manil-Ségalen, M.; Lefebvre, C.; Jenzer, C.; Trichet, M.; Boulogne, C.; Satiat-Jeunemaitre, B.; Legouis, R. The C. elegans LC3 acts ownstream of GABARAP to Degrade Autophagosomes by Interacting with the HOPS subunit VPS39. Dev. Cell 2014, 28, 43–55. [Google Scholar] [CrossRef]
- Fernández, Á.F.; López-Otín, C. The Functional and Pathologic Relevance of Autophagy Proteases. J. Clin. Investig. 2015, 125, 33–41. [Google Scholar] [CrossRef]
- Yu, Z.-Q.; Ni, T.; Hong, B.; Wang, H.-Y.; Jiang, F.-J.; Zou, S.; Chen, Y.; Zheng, X.-L.; Klionsky, D.J.; Liang, Y. Dual Roles of Atg8− PE Deconjugation by Atg4 in Autophagy. Autophagy 2012, 8, 883–892. [Google Scholar] [CrossRef]
- Lőrincz, P.; Juhász, G. Autophagosome-Lysosome Fusion. J. Mol. Biol. 2020, 432, 2462–2482. [Google Scholar] [CrossRef]
- Nakamura, S.; Yoshimori, T. New Insights into Autophagosome-Lysosome Fusion. J. Cell. Sci. 2017, 130, 1209–1216. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bo, T.; Kang, Y.; Liu, Y.; Xu, J.; Wang, W. Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction. Cells 2021, 10, 3071. https://doi.org/10.3390/cells10113071
Bo T, Kang Y, Liu Y, Xu J, Wang W. Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction. Cells. 2021; 10(11):3071. https://doi.org/10.3390/cells10113071
Chicago/Turabian StyleBo, Tao, Yu Kang, Ya Liu, Jing Xu, and Wei Wang. 2021. "Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction" Cells 10, no. 11: 3071. https://doi.org/10.3390/cells10113071
APA StyleBo, T., Kang, Y., Liu, Y., Xu, J., & Wang, W. (2021). Atg5 Regulates Selective Autophagy of the Parental Macronucleus during Tetrahymena Sexual Reproduction. Cells, 10(11), 3071. https://doi.org/10.3390/cells10113071






