Structural Insights into Ca2+ Permeation through Orai Channels
Abstract
:1. Introduction
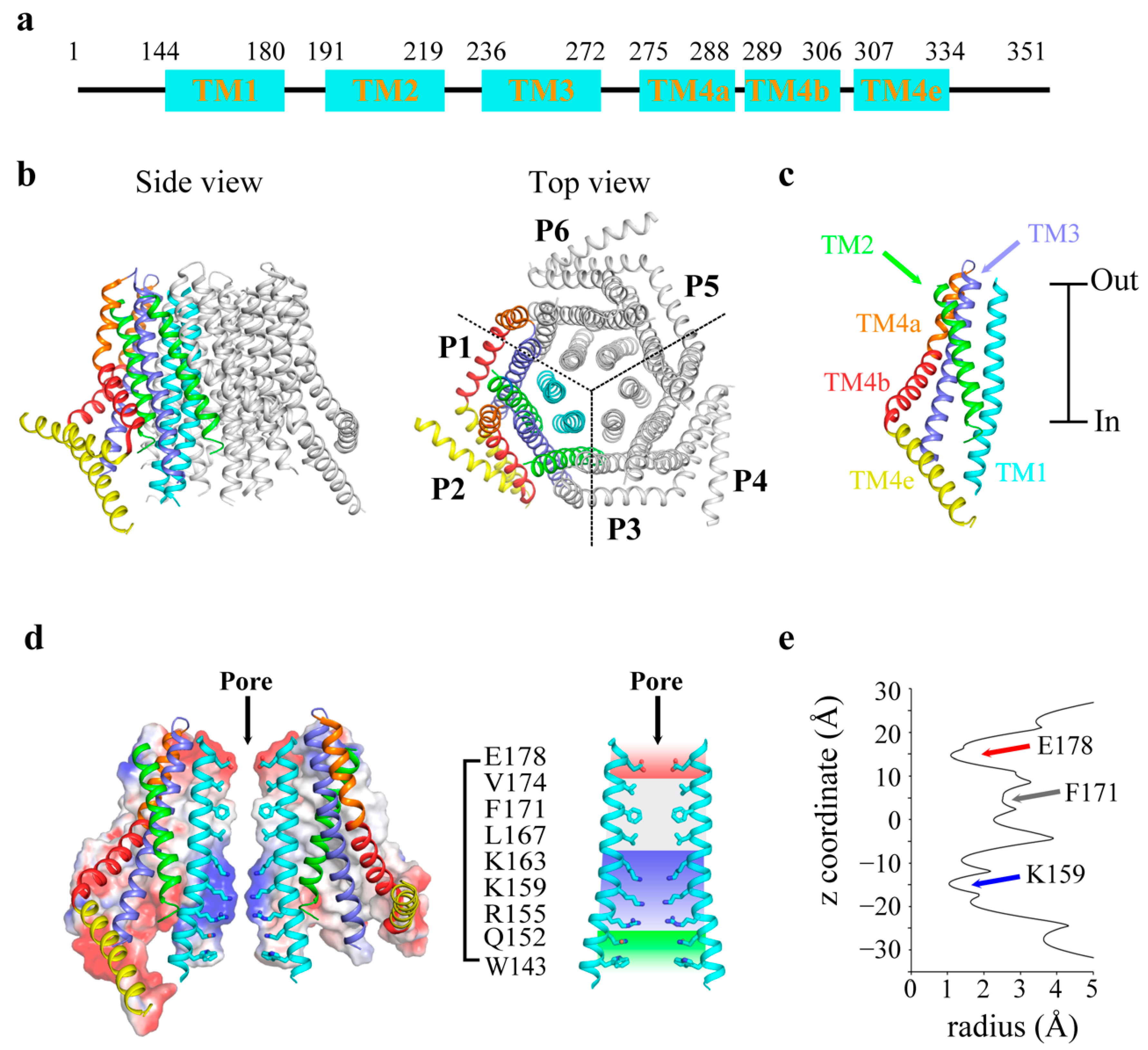
2. Overview of Orai Channels
3. Assembly of Orai Channels
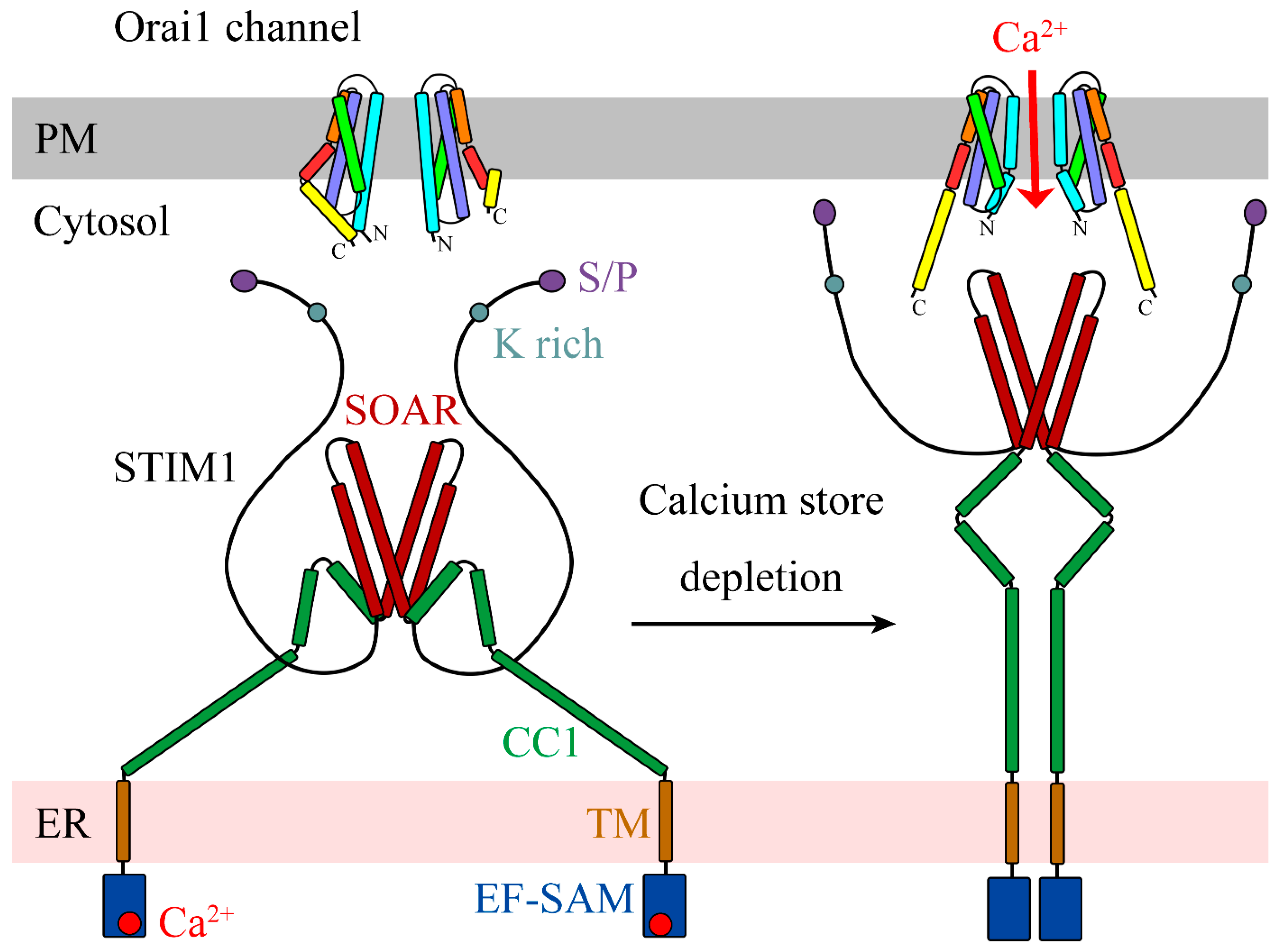
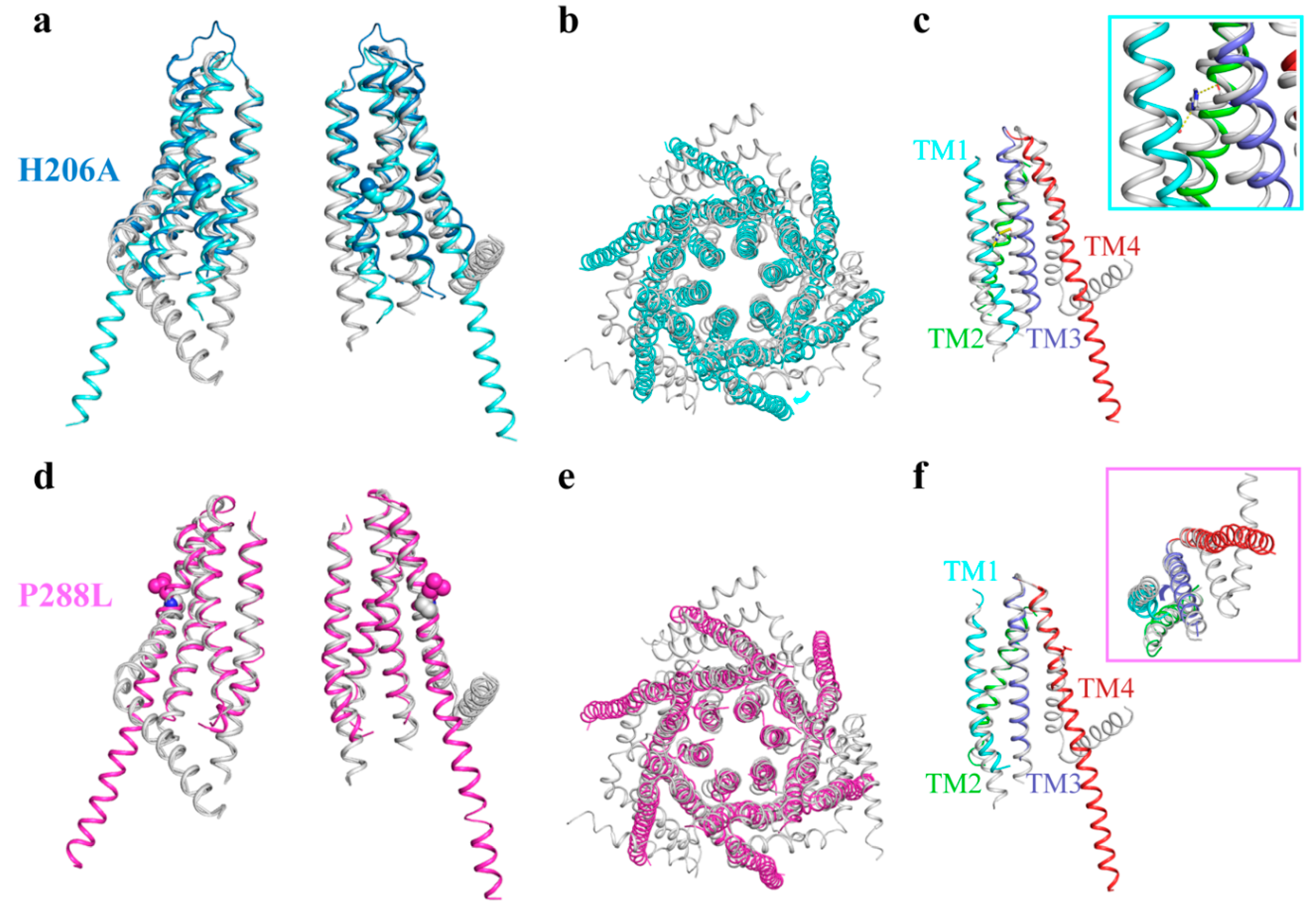
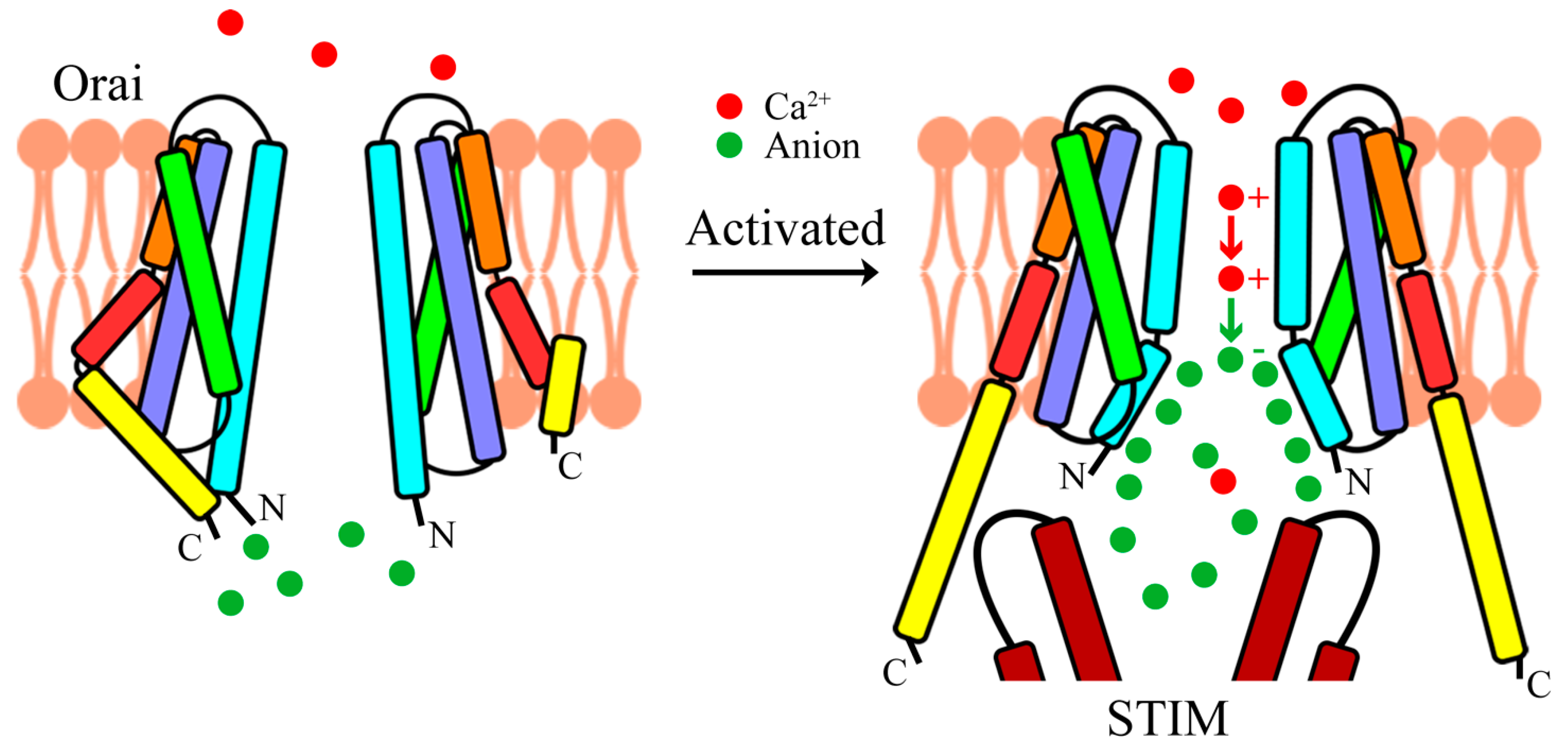
4. Activation of Orai Channels
5. Ca2+ Permeation
6. Perspective
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Clapham, D.E. Calcium signaling. Cell 2007, 131, 1047–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakowski, D.; Murray, F.; Parekh, A.B. Store-operated Ca2+ channels: Mechanism, function, pharmacology, and therapeutic targets. Annu. Rev. Pharmacol. Toxicol. 2021, 61, 629–654. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J.; Bootman, M.D.; Roderick, H.L. Calcium signalling: Dynamics, homeostasis and remodelling. Nat. Rev. Mol. Cell Biol. 2003, 4, 517–529. [Google Scholar] [CrossRef] [Green Version]
- Saheki, Y.; De Camilli, P. Endoplasmic reticulum-plasma membrane contact sites. Annu. Rev. Biochem. 2017, 86, 659–684. [Google Scholar] [CrossRef]
- Prakriya, M.; Lewis, R.S. Store-operated calcium channels. Physiol. Rev. 2015, 95, 1383–1436. [Google Scholar] [CrossRef] [Green Version]
- Parekh, A.B.; Putney, J.W. Store-operated calcium channels. Physiol. Rev. 2005, 85, 757–810. [Google Scholar] [CrossRef] [Green Version]
- Feske, S. CRAC channels and disease—From human CRAC channelopathies and animal models to novel drugs. Cell Calcium 2019, 80, 112–116. [Google Scholar] [CrossRef]
- Lacruz, R.S.; Feske, S. Diseases caused by mutations in ORAI1 and STIM1. Ann. N. Y. Acad. Sci. 2015, 1356, 45–79. [Google Scholar] [CrossRef] [Green Version]
- Fahrner, M.; Grabmayr, H.; Romanin, C. Mechanism of STIM activation. Curr. Opin. Physiol. 2020, 17, 74–79. [Google Scholar] [CrossRef]
- Vig, M.; Peinelt, C.; Beck, A.; Koomoa, D.L.; Rabah, D.; Koblan-Huberson, M.; Kraft, S.; Turner, H.; Fleig, A.; Penner, R.; et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science 2006, 312, 1220–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.L.; Yeromin, A.V.; Zhang, X.H.F.; Yu, Y.; Safrina, O.; Penna, A.; Roos, J.; Stauderman, K.A.; Cahalan, M.D. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc. Natl. Acad. Sci. USA 2006, 103, 9357–9362. [Google Scholar] [CrossRef] [Green Version]
- Feske, S.; Gwack, Y.; Prakriya, M.; Srikanth, S.; Puppel, S.H.; Tanasa, B.; Hogan, P.G.; Lewis, R.S.; Daly, M.; Rao, A. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature 2006, 441, 179–185. [Google Scholar] [CrossRef]
- Roos, J.; DiGregorio, P.J.; Yeromin, A.V.; Ohlsen, K.; Lioudyno, M.; Zhang, S.Y.; Safrina, O.; Kozak, J.A.; Wagner, S.L.; Cahalan, M.D.; et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J. Cell Biol. 2005, 169, 435–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liou, J.; Kim, M.L.; Heo, W.D.; Jones, J.T.; Myers, J.W.; Ferrell, J.E.; Meyer, T. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr. Biol. 2005, 15, 1235–1241. [Google Scholar] [CrossRef] [Green Version]
- Ma, H.T.; Patterson, R.L.; van Rossum, D.B.; Birnbaumer, L.; Mikoshiba, K.; Gill, D.L. Requirement of the inositol trisphosphate receptor for activation of store-operated Ca2+ channels. Science 2000, 287, 1647–1651. [Google Scholar] [CrossRef]
- Wang, Y.; Deng, X.; Mancarella, S.; Hendron, E.; Eguchi, S.; Soboloff, J.; Tang, X.D.; Gill, D.L. The calcium store sensor, STIM1, reciprocally controls Orai and Cav1.2 Channels. Science 2010, 330, 105–109. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.P.; Zeng, W.; Dorwart, M.R.; Choi, Y.-J.; Worley, P.F.; Muallem, S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat. Cell Biol. 2009, 11, 337–343. [Google Scholar] [CrossRef] [PubMed]
- Park, C.Y.; Hoover, P.J.; Mullins, F.M.; Bachhawat, P.; Covington, E.D.; Raunser, S.; Walz, T.; Garcia, K.C.; Dolmetsch, R.E.; Lewis, R.S. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell 2009, 136, 876–890. [Google Scholar] [CrossRef] [Green Version]
- Muik, M.; Fahrner, M.; Derler, I.; Schindl, R.; Bergsmann, J.; Frischauf, I.; Groschner, K.; Romanin, C. A cytosolic homomerization and a modulatory domain within STIM1 C terminus determine coupling to ORAI1 channels. J. Biol. Chem. 2009, 284, 8421–8426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoth, M.; Penner, R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature 1992, 355, 353–356. [Google Scholar] [CrossRef] [PubMed]
- Prakriya, M.; Feske, S.; Gwack, Y.; Srikanth, S.; Rao, A.; Hogan, P.G. Orai1 is an essential pore subunit of the CRAC channel. Nature 2006, 443, 230–233. [Google Scholar] [CrossRef]
- Prakriya, M.; Lewis, R.S. Regulation of CRAC channel activity by recruitment of silent channels to a high open-probability gating mode. J. Gen. Physiol. 2006, 128, 373–386. [Google Scholar] [CrossRef] [PubMed]
- Prakriya, M.; Lewis, R.S. Separation and characterization of currents through store-operated CRAC channels and Mg2+-inhibited cation (MIC) channels. J. Gen. Physiol. 2002, 119, 487–507. [Google Scholar] [CrossRef] [Green Version]
- Prakriya, M.; Lewis, R.S. CRAC channels: Activation, permeation, and the search for a molecular identity. Cell Calcium 2003, 33, 311–321. [Google Scholar] [CrossRef]
- Hogan, P.G.; Lewis, R.S.; Rao, A. Molecular basis of calcium signaling in lymphocytes: STIM and ORAI. Ann. Rev. Immunol. 2010, 28, 491–533. [Google Scholar] [CrossRef] [Green Version]
- Feske, S.; Wulff, H.; Skolnik, E.Y. Ion channels in innate and adaptive immunity. Ann. Rev. Immunol. 2015, 33, 291–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feske, S. Immunodeficiency due to defects in store-operated calcium entry. Ann. N. Y. Acad. Sci. 2011, 1238, 74–90. [Google Scholar] [CrossRef] [Green Version]
- Derler, I.; Fahrner, M.; Carugo, O.; Muik, M.; Bergsmann, J.; Schindl, R.; Frischauf, I.; Eshaghi, S.; Romanin, C. Increased hydrophobicity at the N-terminus/membrane interface impairs gating of the scid-related Orai1 mutant. J. Biol. Chem. 2009, 284, 15903–15915. [Google Scholar] [CrossRef] [Green Version]
- Bohm, J.; Laporte, J. Gain-of-function mutations in STIM1 and ORAI1 causing tubular aggregate myopathy and Stormorken syndrome. Cell Calcium 2018, 76, 1–9. [Google Scholar] [CrossRef]
- Sadaghiani, A.M.; Lee, S.M.; Odegaard, J.I.; Leveson-Gower, D.B.; McPherson, O.M.; Novick, P.; Kim, M.R.; Koehler, A.N.; Negrin, R.; Dolmetsch, R.E.; et al. Identification of Orai1 channel inhibitors by using minimal functional domains to screen small molecule microarrays. Chem. Biol. 2014, 21, 1278–1292. [Google Scholar] [CrossRef]
- Chen, G.; Panicker, S.; Lau, K.-Y.; Apparsundaram, S.; Patel, V.A.; Chen, S.-L.; Soto, R.; Jung, J.K.C.; Ravindran, P.; Okuhara, D.; et al. Characterization of a novel CRAC inhibitor that potently blocks human T cell activation and effector functions. Mol. Immunol. 2013, 54, 355–367. [Google Scholar] [CrossRef] [PubMed]
- Wen, L.; Voronina, S.; Javed, M.A.; Awais, M.; Szatmary, P.; Latawiec, D.; Chvanov, M.; Collier, D.; Huang, W.; Barrett, J.; et al. Inhibitors of ORAI1 prevent cytosolic calcium-associated injury of human pancreatic acinar cells and acute pancreatitis in 3 mouse models. Gastroenterology 2015, 149, 481–492. [Google Scholar] [CrossRef] [Green Version]
- Waldron, R.T.; Chen, Y.; Pham, H.; Go, A.; Su, H.-Y.; Hu, C.; Wen, L.; Husain, S.Z.; Sugar, C.A.; Roos, J.; et al. The Orai Ca2+ channel inhibitor CM4620 targets both parenchymal and immune cells to reduce inflammation in experimental acute pancreatitis. J. Physiol. (Lond.) 2019, 597, 3085–3105. [Google Scholar] [CrossRef]
- Hou, X.; Pedi, L.; Diver, M.M.; Long, S.B. Crystal structure of the calcium release-activated calcium channel Orai. Science 2012, 338, 1308–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yeromin, A.V.; Zhang, S.L.; Jiang, W.; Yu, Y.; Safrina, O.; Cahalan, M.D. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature 2006, 443, 226–229. [Google Scholar] [CrossRef] [PubMed]
- McNally, B.A.; Somasundaram, A.; Yamashita, M.; Prakriya, M. Gated regulation of CRAC channel ion selectivity by STIM1. Nature 2012, 482, 241–245. [Google Scholar] [CrossRef]
- Yamashita, M.; Ing, C.E.; Yeung, P.S.; Maneshi, M.M.; Pomes, R.; Prakriya, M. The basic residues in the Orai1 channel inner pore promote opening of the outer hydrophobic gate. J. Gen. Physiol. 2020, 152, e201912397. [Google Scholar] [CrossRef]
- Hou, X.; Burstein, S.R.; Long, S.B. Structures reveal opening of the store-operated calcium channel Orai. eLife 2018, 7, e36758. [Google Scholar] [CrossRef]
- Liu, X.; Wu, G.; Yu, Y.; Chen, X.; Ji, R.; Lu, J.; Li, X.; Zhang, X.; Yang, X.; Shen, Y. Molecular understanding of calcium permeation through the open Orai channel. PLoS Biol. 2019, 17, e3000096. [Google Scholar] [CrossRef]
- Zhou, Y.; Cai, X.; Loktionova, N.A.; Wang, X.; Nwokonko, R.M.; Wang, X.; Wang, Y.; Rothberg, B.S.; Trebak, M.; Gill, D.L. The STIM1-binding site nexus remotely controls Orai1 channel gating. Nat. Comm. 2016, 7, 13725. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tiffner, A.; Schober, R.; Hoglinger, C.; Bonhenry, D.; Pandey, S.; Lunz, V.; Sallinger, M.; Frischauf, I.; Fahrner, M.; Lindinger, S.; et al. CRAC channel opening is determined by a series of Orai1 gating checkpoints in the transmembrane and cytosolic regions. J. Biol. Chem. 2021, 296, 100224. [Google Scholar] [CrossRef] [PubMed]
- Hou, X.; Outhwaite, I.R.; Pedi, L.; Long, S.B. Cryo-EM structure of the calcium release-activated calcium channel Orai in an open conformation. eLife 2020, 9, e62772. [Google Scholar] [CrossRef]
- Rothberg, B.S.; Wang, Y.; Gill, D.L. Orai channel pore properties and gating by STIM: Implications from the Orai crystal structure. Sci. Signal. 2013, 6, pe9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, M.; Yeung, P.S.W.; Ing, C.E.; McNally, B.A.; Pomes, R.; Prakriya, M. STIM1 activates CRAC channels through rotation of the pore helix to open a hydrophobic gate. Nat. Comm. 2017, 8, 14512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palty, R.; Stanley, C.; Isacoff, E.Y. Critical role for Orai1 C-terminal domain and TM4 in CRAC channel gating. Cell Res. 2015, 25, 963–980. [Google Scholar] [CrossRef] [Green Version]
- Derler, I.; Plenk, P.; Fahrner, M.; Muik, M.; Jardin, I.; Schindl, R.; Gruber, H.J.; Groschner, K.; Romanin, C. The extended transmembrane Orai1 N-terminal (ETON) region combines binding interface and gate for Orai1 activation by STIM1. J. Biol. Chem. 2013, 288, 29025–29034. [Google Scholar] [CrossRef] [Green Version]
- Penna, A.; Demuro, A.; Yeromin, A.V.; Zhang, S.L.; Safrina, O.; Parker, I.; Cahalan, M.D. The CRAC channel consists of a tetramer formed by Stim-induced dimerization of Orai dimers. Nature 2008, 456, 116–120. [Google Scholar] [CrossRef]
- Ji, W.; Xu, P.; Li, Z.; Lu, J.; Liu, L.; Zhan, Y.; Chen, Y.; Hille, B.; Xu, T.; Chen, L. Functional stoichiometry of the unitary calcium-release-activated calcium channel. Proc. Natl. Acad. Sci. USA 2008, 105, 13668–13673. [Google Scholar] [CrossRef] [Green Version]
- Mignen, O.; Thompson, J.L.; Shuttleworth, T.J. The molecular architecture of the arachidonate-regulated Ca2+-selective ARC channel is a pentameric assembly of Orai1 and Orai3 subunits. J. Physiol. 2009, 587, 4181–4197. [Google Scholar] [CrossRef]
- Cai, X.; Zhou, Y.; Nwokonko, R.M.; Loktionova, N.A.; Wang, X.; Xin, P.; Trebak, M.; Wang, Y.; Gill, D.L. The Orai1 store-operated calcium channel functions as a hexamer. J. Biol. Chem. 2016, 291, 25764–25775. [Google Scholar] [CrossRef] [Green Version]
- Yen, M.; Lokteva, L.A.; Lewis, R.S. Functional analysis of Orai1 concatemers supports a hexameric stoichiometry for the CRAC channel. Biophys. J. 2016, 111, 1897–1907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Yang, X.; Shen, Y. Structural Insights into Ca2+ Permeation through Orai Channels. Cells 2021, 10, 3062. https://doi.org/10.3390/cells10113062
Li Y, Yang X, Shen Y. Structural Insights into Ca2+ Permeation through Orai Channels. Cells. 2021; 10(11):3062. https://doi.org/10.3390/cells10113062
Chicago/Turabian StyleLi, Yang, Xue Yang, and Yuequan Shen. 2021. "Structural Insights into Ca2+ Permeation through Orai Channels" Cells 10, no. 11: 3062. https://doi.org/10.3390/cells10113062
APA StyleLi, Y., Yang, X., & Shen, Y. (2021). Structural Insights into Ca2+ Permeation through Orai Channels. Cells, 10(11), 3062. https://doi.org/10.3390/cells10113062





