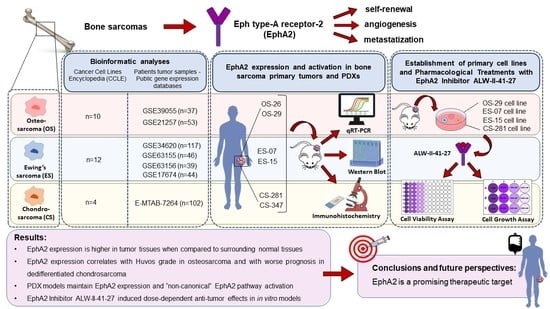
EphA2 Expression in Bone Sarcomas: Bioinformatic Analyses and Preclinical Characterization in Patient-Derived Models of Osteosarcoma, Ewing’s Sarcoma and Chondrosarcoma
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bioinformatics Analyses
2.2. Establishment of Patient-Derived Xenograft Models
2.3. RNA Extraction and Quantitative Real-Time PCR
2.4. Western Blot
2.5. Immunohistochemistry
2.6. Primary Cell Lines
2.7. Pharmacological Treatments with EphA Inhibitor ALW-II-41-27 and Cell Viability Assays
3. Results
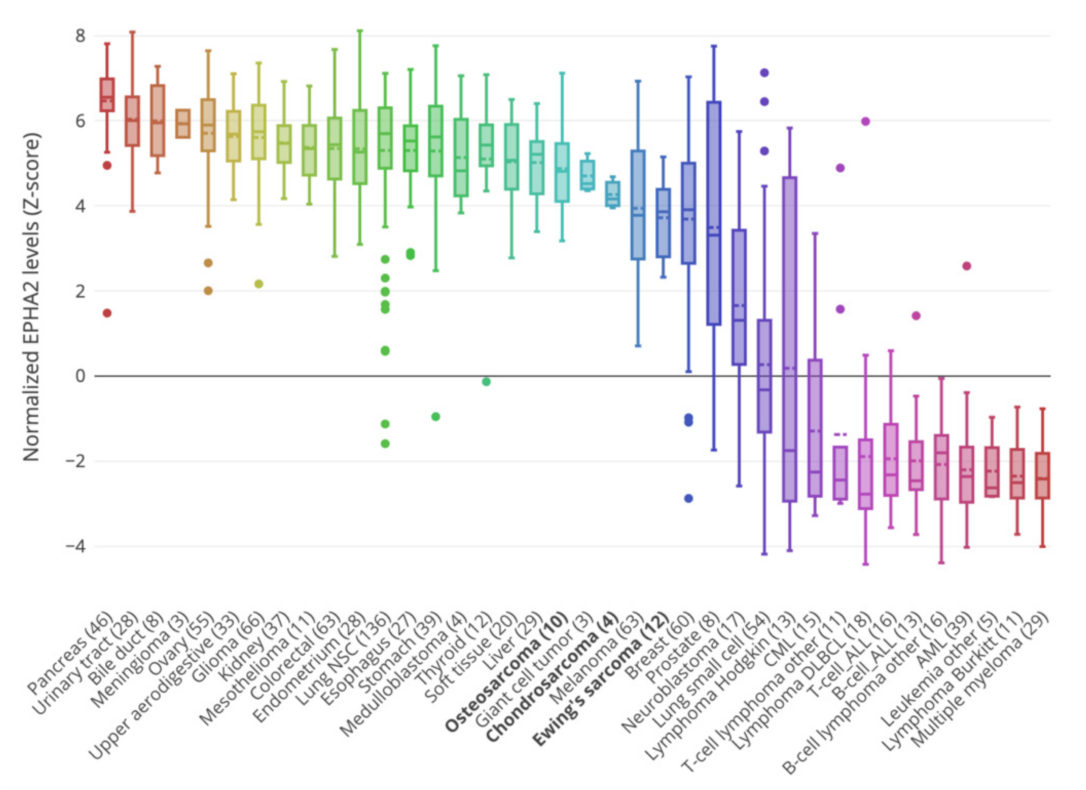
3.1. EphA2 Expression in Bone Sarcoma Cell Lines from the Cancer Cell lines Encyclopedia
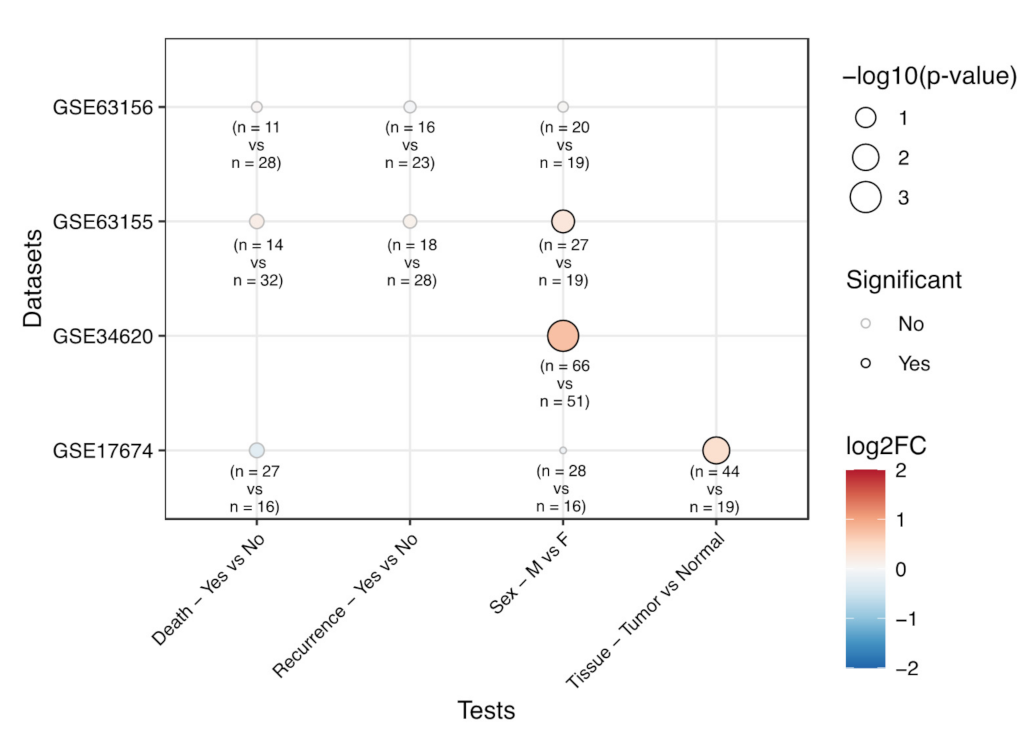
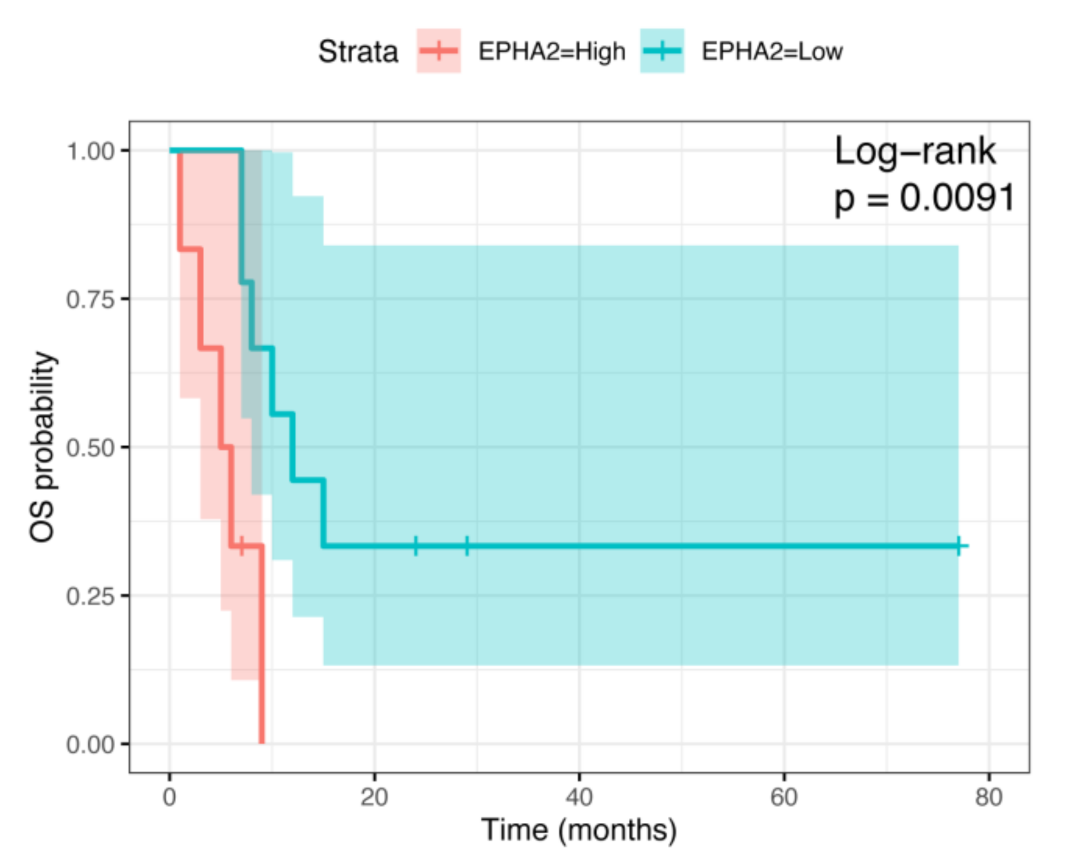
3.2. Correlation of EPHA2 Expression with Patients’ Characteristics and Clinical Outcome in Osteosarcoma
3.3. Correlation of EPHA2 Expression and Clinical Features in Ewing’s Sarcoma
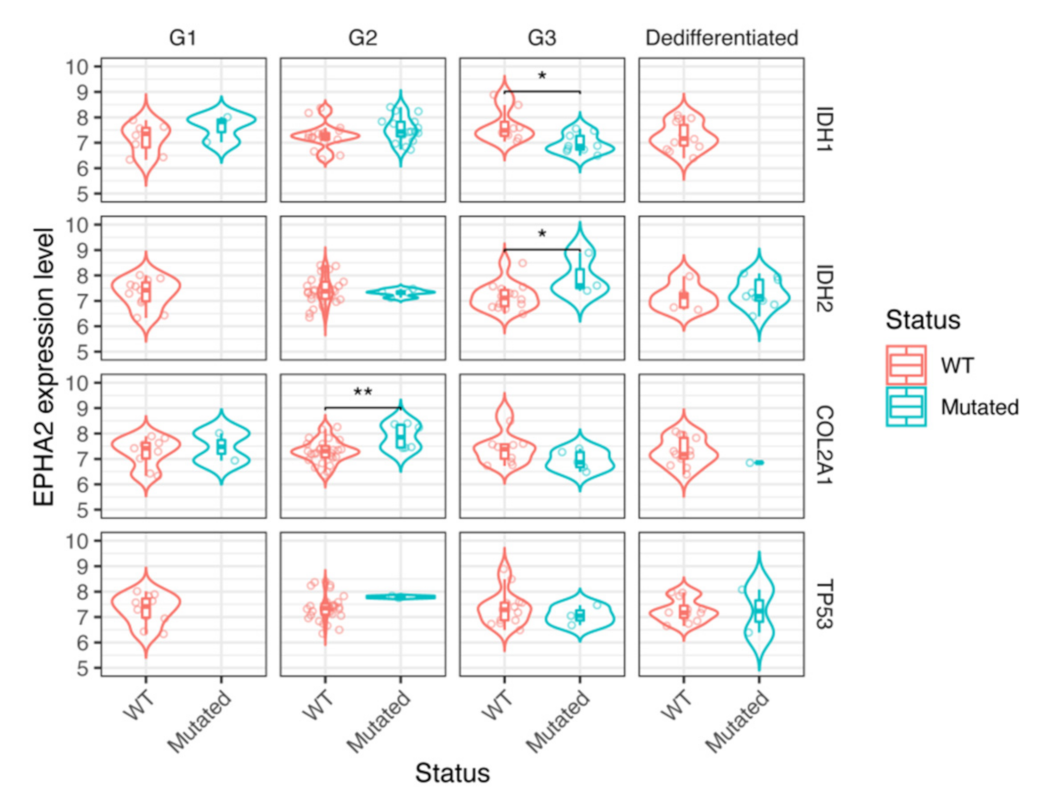
3.4. EPHA2 Expression in Chondrosarcoma and Correlation with Clinical and Molecular Features
3.5. EPHA2 Expression and Activation in Bone Sarcoma Patient-Derived Xenografts
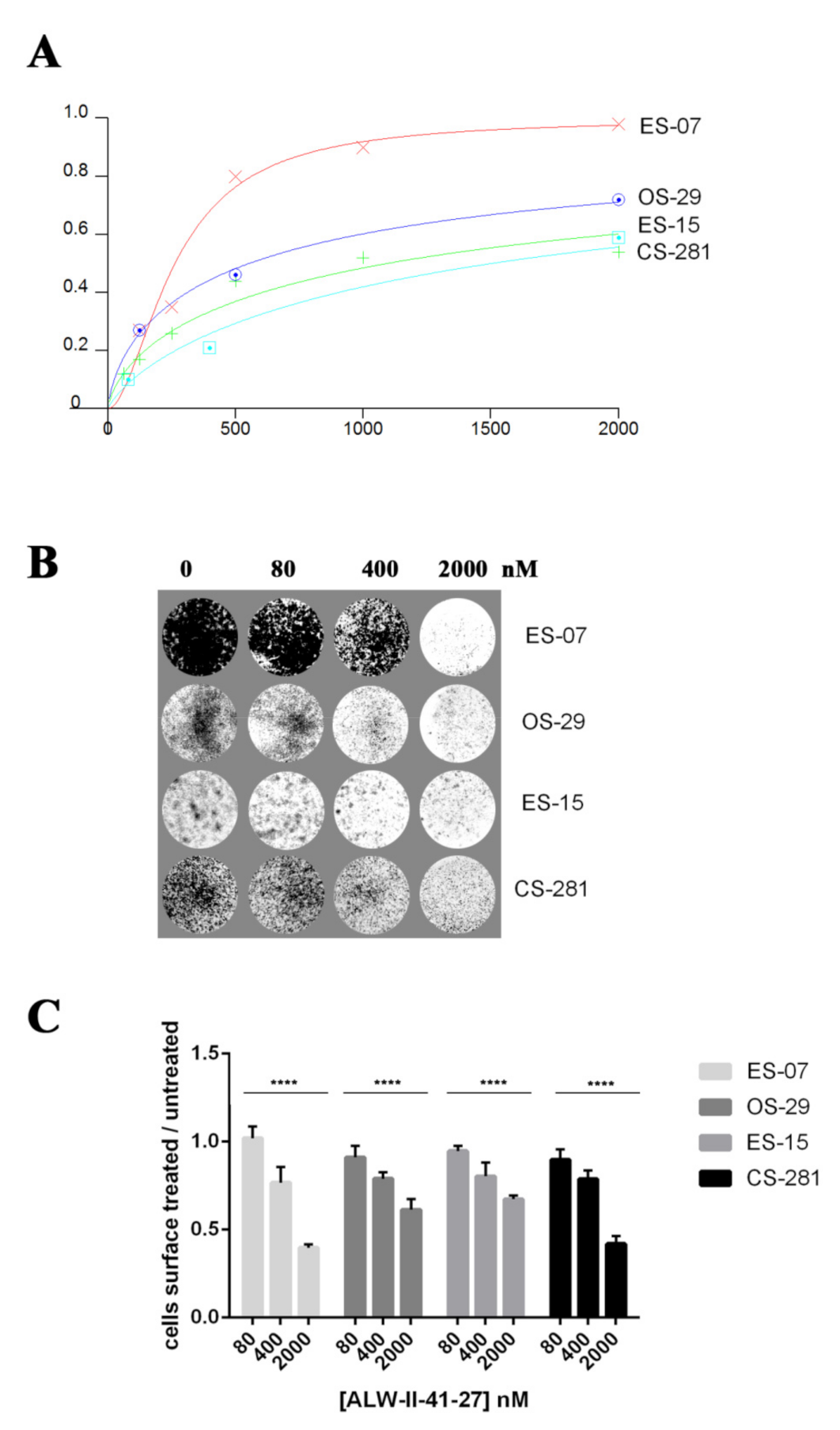
3.6. EphA2 Inhibitor ALW-II-41-27 Induced Dose-Dependent Anti-Tumor Effects in In Vitro Models
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Casali, P.G.; Bielack, S.; Abecassis, N.; Aro, H.T.; Bauer, S.; Biagini, R.; Bonvalot, S.; Boukovinas, I.; Bovee, J.V.M.G.; Brennan, B.; et al. Bone sarcomas: ESMO-PaedCan-EURACAN Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2018, 29, iv79–iv95. [Google Scholar] [CrossRef]
- Kager, L.; Pötschger, U.; Bielack, S. Review of mifamurtide in the treatment of patients with osteosarcoma. Ther. Clin. Risk Manag. 2010, 6, 279. [Google Scholar] [CrossRef] [Green Version]
- Duffaud, F.; Mir, O.; Boudou-Rouquette, P.; Piperno-Neumann, S.; Penel, N.; Bompas, E.; Delcambre, C.; Kalbacher, E.; Italiano, A.; Collard, O. Efficacy and safety of regorafenib in adult patients with metastatic osteosarcoma: A non-comparative, randomised, double-blind, placebo-controlled, phase 2 study. Lancet Oncol. 2019, 20, 120–133. [Google Scholar] [CrossRef]
- Grignani, G.; Palmerini, E.; Ferraresi, V.; D'Ambrosio, L.; Bertulli, R.; Asaftei, S.D.; Tamburini, A.; Pignochino, Y.; Sangiolo, D.; Marchesi, E. Sorafenib and everolimus for patients with unresectable high-grade osteosarcoma progressing after standard treatment: A non-randomised phase 2 clinical trial. Lancet Oncol. 2015, 16, 98–107. [Google Scholar] [CrossRef]
- Grignani, G.; Palmerini, E.; Dileo, P.; Asaftei, S.; D’Ambrosio, L.; Pignochino, Y.; Mercuri, M.; Picci, P.; Fagioli, F.; Casali, P. A phase II trial of sorafenib in relapsed and unresectable high-grade osteosarcoma after failure of standard multimodal therapy: An Italian Sarcoma Group study. Ann. Oncol. 2012, 23, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, J.; Federman, N.; Anderson, P.; Macy, M.; Davis, L.; Riedel, R.; Muscal, J.; Daw, N.; Ratan, R.; Toretsky, J. 1620O Phase I study of TK216, a novel anti-ETS agent for Ewing sarcoma. Ann. Oncol. 2020, 31, S972. [Google Scholar] [CrossRef]
- Waitkus, M.S.; Yan, H. Targeting Isocitrate Dehydrogenase Mutations in Cancer: Emerging Evidence and Diverging Strategies. Clin. Cancer Res. 2021, 27, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Tap, W.D.; Villalobos, V.M.; Cote, G.M.; Burris, H.; Janku, F.; Mir, O.; Beeram, M.; Wagner, A.J.; Jiang, L.; Wu, B. Phase I study of the mutant IDH1 inhibitor ivosidenib: Safety and clinical activity in patients with advanced chondrosarcoma. J. Clin. Oncol. 2020, 38, 1693. [Google Scholar] [CrossRef]
- Coulthard, M.G.; Duffy, S.; Down, M.; Evans, B.; Power, M.; Smith, F.; Stylianou, C.; Kleikamp, S.; Oates, A.; Lackmann, M.; et al. The role of the Eph-ephrin signalling system in the regulation of developmental patterning. Int. J. Dev. Biol. 2002, 46, 375–384. [Google Scholar] [PubMed]
- Fagotto, F.; Winklbauer, R.; Rohani, N. Ephrin-Eph signaling in embryonic tissue separation. Cell Adhes. Migr. 2014, 8, 308–326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.E.; Son, A.I.; Zhou, R. Roles of EphA2 in Development and Disease. Genes 2013, 4, 334–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Binda, E.; Visioli, A.; Giani, F.; Lamorte, G.; Copetti, M.; Pitter, K.L.; Huse, J.T.; Cajola, L.; Zanetti, N.; DiMeco, F.; et al. The EphA2 receptor drives self-renewal and tumorigenicity in stem-like tumor-propagating cells from human glioblastomas. Cancer Cell 2012, 22, 765–780. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Monclús, S.; López-Alemany, R.; Almacellas-Rabaiget, O.; Herrero-Martín, D.; Huertas-Martinez, J.; Lagares-Tena, L.; Alba-Pavón, P.; Hontecillas-Prieto, L.; Mora, J.; de Álava, E.; et al. EphA2 receptor is a key player in the metastatic onset of Ewing sarcoma. Int. J. Cancer 2018, 143, 1188–1201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barquilla, A.; Lamberto, I.; Noberini, R.; Heynen-Genel, S.; Brill, L.M.; Pasquale, E.B. Protein kinase A can block EphA2 receptor–mediated cell repulsion by increasing EphA2 S897 phosphorylation. Mol. Biol. Cell 2016, 27, 2757–2770. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Yamada, N.; Tanaka, T.; Hori, T.; Yokoyama, S.; Hayakawa, Y.; Yano, S.; Fukuoka, J.; Koizumi, K.; Saiki, I. Crucial roles of RSK in cell motility by catalysing serine phosphorylation of EphA2. Nat. Commun. 2015, 6, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Miao, H.; Li, D.-Q.; Mukherjee, A.; Guo, H.; Petty, A.; Cutter, J.; Basilion, J.P.; Sedor, J.; Wu, J.; Danielpour, D. EphA2 mediates ligand-dependent inhibition and ligand-independent promotion of cell migration and invasion via a reciprocal regulatory loop with Akt. Cancer Cell 2009, 16, 9–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, C.; Schmitz, G.; Meyer, S.; Bataille, F.; Hau, P.; Langmann, T.; Dietmaier, W.; Landthaler, M.; Vogt, T. Differential gene expression of Eph receptors and ephrins in benign human tissues and cancers. Clin. Chem. 2004, 50, 490–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wykosky, J.; Debinski, W. The EphA2 receptor and ephrinA1 ligand in solid tumors: Function and therapeutic targeting. Mol. Cancer Res. 2008, 6, 1795–1806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chiabotto, G.; Grignani, G.; Todorovic, M.; Martin, V.; Centomo, M.L.; Prola, E.; Giordano, G.; Merlini, A.; Miglio, U.; Berrino, E. Pazopanib and trametinib as a synergistic strategy against osteosarcoma: Preclinical activity and molecular insights. Cancers 2020, 12, 1519. [Google Scholar] [CrossRef]
- Donovan, L.K.; Delaidelli, A.; Joseph, S.K.; Bielamowicz, K.; Fousek, K.; Holgado, B.L.; Manno, A.; Srikanthan, D.; Gad, A.Z.; Van Ommeren, R.; et al. Locoregional delivery of CAR T cells to the cerebrospinal fluid for treatment of metastatic medulloblastoma and ependymoma. Nat. Med. 2020, 26, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Shitara, K.; Satoh, T.; Iwasa, S.; Yamaguchi, K.; Muro, K.; Komatsu, Y.; Nishina, T.; Esaki, T.; Hasegawa, J.; Kakurai, Y.; et al. Safety, tolerability, pharmacokinetics, and pharmacodynamics of the afucosylated, humanized anti-EPHA2 antibody DS-8895a: A first-in-human phase I dose escalation and dose expansion study in patients with advanced solid tumors. J. Immunother. Cancer 2019, 7, 219. [Google Scholar] [CrossRef]
- Chow, K.K.; Naik, S.; Kakarla, S.; Brawley, V.S.; Shaffer, D.R.; Yi, Z.; Rainusso, N.; Wu, M.F.; Liu, H.; Kew, Y.; et al. T cells redirected to EphA2 for the immunotherapy of glioblastoma. Mol. Ther. 2013, 21, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Ba, T.; Ho, J.; Chen, D.; Cheng, Y.; Wang, L.; Xu, G.; Xu, L.; Zhou, Y.; Wei, Y.; et al. First-in-Human Trial of EphA2-Redirected CAR T-Cells in Patients With Recurrent Glioblastoma: A Preliminary Report of Three Cases at the Starting Dose. Front. Oncol. 2021, 11, 694941. [Google Scholar] [CrossRef]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicolle, R.; Ayadi, M.; Gomez-Brouchet, A.; Armenoult, L.; Banneau, G.; Elarouci, N.; Tallegas, M.; Decouvelaere, A.-V.; Aubert, S.; Rédini, F. Integrated molecular characterization of chondrosarcoma reveals critical determinants of disease progression. Nat. Commun. 2019, 10, 4622. [Google Scholar] [CrossRef]
- Xiao, T.; Xiao, Y.; Wang, W.; Tang, Y.Y.; Xiao, Z.; Su, M. Targeting EphA2 in cancer. J. Hematol. Oncol. 2020, 13, 114. [Google Scholar] [CrossRef] [PubMed]
- Barile, E.; Wang, S.; Das, S.K.; Noberini, R.; Dahl, R.; Stebbins, J.L.; Pasquale, E.B.; Fisher, P.B.; Pellecchia, M. Design, Synthesis and Bio-evaluation of an EphA2-based Targeted Delivery System. ChemMedChem 2014, 9, 1403. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurti, U.; Silverman, J.F. HER2 in breast cancer: A review and update. Adv. Anat. Pathol. 2014, 21, 100–107. [Google Scholar] [CrossRef]
- Ascierto, P.A.; Kirkwood, J.M.; Grob, J.-J.; Simeone, E.; Grimaldi, A.M.; Maio, M.; Palmieri, G.; Testori, A.; Marincola, F.M.; Mozzillo, N. The role of BRAF V600 mutation in melanoma. J. Transl. Med. 2012, 10, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, J.A.; Rubin, B.P. KIT mutations in GIST. Curr. Opin. Genet. Dev. 2007, 17, 3–7. [Google Scholar] [CrossRef]
- Napolitano, A.; Vincenzi, B. Secondary KIT mutations: The GIST of drug resistance and sensitivity. Br. J. Cancer 2019. [Google Scholar] [CrossRef] [Green Version]
- Da Cunha Santos, G.; Shepherd, F.A.; Tsao, M.S. EGFR mutations and lung cancer. Annu. Rev. Pathol. Mech. Dis. 2011, 6, 49–69. [Google Scholar] [CrossRef] [Green Version]
- Palmerini, E.; Lopez-Pousa, A.; Grignani, G.; Redondo, A.; Hindi, N.; Stacchiotti, S.; Sebio, A.; Lopez-Martin, J.A.; Valverde Morales, C.M.; Martinez-Trufero, J. IMMUNOSARC: A collaborative Spanish (GEIS) and Italian (ISG) sarcoma groups phase I/II trial of sunitinib and nivolumab in advanced soft tissue and bone sarcoma: Results from the phase II part, bone sarcoma cohort. J. Clin. Oncol. 2020. [Google Scholar] [CrossRef]
- Sainz-Jaspeado, M.; Huertas-Martinez, J.; Lagares-Tena, L.; Martin Liberal, J.; Mateo-Lozano, S.; de Alava, E.; de Torres, C.; Mora, J.; Muro, X.G.d.; Tirado, O.M. EphA2-induced angiogenesis in ewing sarcoma cells works through bFGF production and is dependent on caveolin-1. PLoS ONE 2013, 8, e71449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsche-Guenther, R.; Noske, A.; Ungethüm, U.; Kuban, R.J.; Schlag, P.M.; Tunn, P.U.; Karle, J.; Krenn, V.; Dietel, M.; Sers, C. De novo expression of EphA2 in osteosarcoma modulates activation of the mitogenic signalling pathway. Histopathology 2010, 57, 836–850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, J.; Xiao, D.; Li, G.; Ma, J.; Chen, P.; Yuan, W.; Hou, F.; Ge, J.; Zhong, M.; Tang, Y. EphA2 promotes epithelial–mesenchymal transition through the Wnt/β-catenin pathway in gastric cancer cells. Oncogene 2014, 33, 2737–2747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fattet, L.; Jung, H.-Y.; Matsumoto, M.W.; Aubol, B.E.; Kumar, A.; Adams, J.A.; Chen, A.C.; Sah, R.L.; Engler, A.J.; Pasquale, E.B. Matrix rigidity controls epithelial-mesenchymal plasticity and tumor metastasis via a mechanoresponsive EPHA2/LYN complex. Dev. Cell 2020, 54, 302–316.e307. [Google Scholar] [CrossRef] [PubMed]
- Parri, M.; Taddei, M.L.; Bianchini, F.; Calorini, L.; Chiarugi, P. EphA2 reexpression prompts invasion of melanoma cells shifting from mesenchymal to amoeboid-like motility style. Cancer Res. 2009, 69, 2072–2081. [Google Scholar] [CrossRef] [Green Version]
- Paul, C.D.; Mistriotis, P.; Konstantopoulos, K. Cancer cell motility: Lessons from migration in confined spaces. Nat. Rev. Cancer 2017, 17, 131–140. [Google Scholar] [CrossRef] [Green Version]
- Sabeh, F.; Shimizu-Hirota, R.; Weiss, S.J. Protease-dependent versus-independent cancer cell invasion programs: Three-dimensional amoeboid movement revisited. J. Cell Biol. 2009, 185, 11–19. [Google Scholar] [CrossRef] [Green Version]
- Condeelis, J.; Segall, J.E. Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 2003, 3, 921–930. [Google Scholar] [CrossRef] [PubMed]
- Jawad, M.U.; Cheung, M.C.; Min, E.S.; Schneiderbauer, M.M.; Koniaris, L.G.; Scully, S.P. Ewing sarcoma demonstrates racial disparities in incidence-related and sex-related differences in outcome: An analysis of 1631 cases from the SEER database, 1973-2005. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2009, 115, 3526–3536. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.; Wilhelm, M.; Conyers, R.; Herschtal, A.; Whelan, J.; Bielack, S.; Kager, L.; Kühne, T.; Sydes, M.; Gelderblom, H. Benefits and adverse events in younger versus older patients receiving neoadjuvant chemotherapy for osteosarcoma: Findings from a meta-analysis. J. Clin. Oncol. 2013, 31, 2303–2312. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.R.; Kanvinde, P.; King, C.; Pasquale, E.B.; Hristova, K. The EphA2 receptor is activated through induction of distinct, ligand-dependent oligomeric structures. Commun. Biol. 2018, 1, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, K.R.; Wang, S.; Hastings, A.K.; Youngblood, V.M.; Santapuram, P.R.; Chen, H.; Cates, J.M.; Colvin, D.C.; Ye, F.; Brantley-Sieders, D.M.; et al. Genetic and pharmacologic inhibition of EPHA2 promotes apoptosis in NSCLC. J. Clin. Investig. 2014, 124, 2037–2049. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amato, K.R.; Wang, S.; Tan, L.; Hastings, A.K.; Song, W.; Lovly, C.M.; Meador, C.B.; Ye, F.; Lu, P.; Balko, J.M.; et al. EPHA2 Blockade Overcomes Acquired Resistance to EGFR Kinase Inhibitors in Lung Cancer. Cancer Res. 2016, 76, 305–318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, W.; Hwang, Y.; Youngblood, V.M.; Cook, R.S.; Balko, J.M.; Chen, J.; Brantley-Sieders, D.M. Targeting EphA2 impairs cell cycle progression and growth of basal-like/triple-negative breast cancers. Oncogene 2017, 36, 5620–5630. [Google Scholar] [CrossRef] [Green Version]
- Peng, Q.; Chen, L.; Wu, W.; Wang, J.; Zheng, X.; Chen, Z.; Jiang, Q.; Han, J.; Wei, L.; Wang, L.; et al. EPH receptor A2 governs a feedback loop that activates Wnt/β-catenin signaling in gastric cancer. Cell Death Dis. 2018, 9, 1146. [Google Scholar] [CrossRef] [Green Version]
- Martini, G.; Cardone, C.; Vitiello, P.P.; Belli, V.; Napolitano, S.; Troiani, T.; Ciardiello, D.; Della Corte, C.M.; Morgillo, F.; Matrone, N.; et al. EPHA2 Is a Predictive Biomarker of Resistance and a Potential Therapeutic Target for Improving Antiepidermal Growth Factor Receptor Therapy in Colorectal Cancer. Mol. Cancer Ther. 2019, 18, 845–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Hou, W.; Perera, A.; Bettler, C.; Beach, J.R.; Ding, X.; Li, J.; Denning, M.F.; Dhanarajan, A.; Cotler, S.J.; et al. Targeting EphA2 suppresses hepatocellular carcinoma initiation and progression by dual inhibition of JAK1/STAT3 and AKT signaling. Cell Rep. 2021, 34, 108765. [Google Scholar] [CrossRef] [PubMed]
- Swidergall, M.; Solis, N.V.; Lionakis, M.S.; Filler, S.G. EphA2 is an epithelial cell pattern recognition receptor for fungal β-glucans. Nat. MicroBiol. 2018, 3, 53–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheffield, N.C.; Pierron, G.; Klughammer, J.; Datlinger, P.; Schönegger, A.; Schuster, M.; Hadler, J.; Surdez, D.; Guillemot, D.; Lapouble, E. DNA methylation heterogeneity defines a disease spectrum in Ewing sarcoma. Nat. Med. 2017, 23, 386–395. [Google Scholar] [CrossRef] [PubMed]
- Franzetti, G.; Laud-Duval, K.; Van Der Ent, W.; Brisac, A.; Irondelle, M.; Aubert, S.; Dirksen, U.; Bouvier, C.; De Pinieux, G.; Snaar-Jagalska, E. Cell-to-cell heterogeneity of EWSR1-FLI1 activity determines proliferation/migration choices in Ewing sarcoma cells. Oncogene 2017, 36, 3505–3514. [Google Scholar] [CrossRef] [Green Version]


| PATIENT ID | Sex | Age | Anatomical Site | Histotype | Grade | Disease Stage |
|---|---|---|---|---|---|---|
| OS-026 | M | 67 | Upper limb | Osteoblastic osteosarcoma | G4 | Metastatic |
| OS-029 | M | 18 | Lower limb | Osteoblastic osteosarcoma | G4 | Metastatic |
| ES-07 | F | 19 | Lower limb | Ewing’s sarcoma | High grade | Localized |
| ES-15 | F | 65 | Lumbar spine | Ewing’s sarcoma | High grade | Localized |
| CS-281 | F | 58 | Hip | Chondrosarcoma, conventional | G3 | Locally advanced |
| CS-347 | M | 56 | Upper limb | Chondrosarcoma, conventional | G2 | Locally advanced |
| Dataset | Category | Test | EphA2 log 2 FC | p-Value |
|---|---|---|---|---|
| GSE39055 | Death | Yes vs. No | 0.24 | 0.596 |
| Sex | Male vs. Female | -0.32 | 0.515 | |
| Recurrence | Yes vs. No | -0.01 | 0.979 | |
| GSE21257 | Sex | Male vs. Female | 0.64 | 0.019 |
| Huvos Grade | 2 vs. 1 | 0.24 | 0.474 | |
| Huvos Grade | 3 vs. 1 | 0.73 | 0.030 | |
| Huvos Grade | 3 vs. 2 | 0.50 | 0.081 | |
| Huvos Grade | 4 vs. 1 | 0.11 | 0.835 | |
| Huvos Grade | 3-4 vs. 2-1 | 0.43 | 0.088 | |
| TARGET-OS | Sex | Male vs. Female | 0.06 | 0.628 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giordano, G.; Merlini, A.; Ferrero, G.; Mesiano, G.; Fiorino, E.; Brusco, S.; Centomo, M.L.; Leuci, V.; D’Ambrosio, L.; Aglietta, M.; et al. EphA2 Expression in Bone Sarcomas: Bioinformatic Analyses and Preclinical Characterization in Patient-Derived Models of Osteosarcoma, Ewing’s Sarcoma and Chondrosarcoma. Cells 2021, 10, 2893. https://doi.org/10.3390/cells10112893
Giordano G, Merlini A, Ferrero G, Mesiano G, Fiorino E, Brusco S, Centomo ML, Leuci V, D’Ambrosio L, Aglietta M, et al. EphA2 Expression in Bone Sarcomas: Bioinformatic Analyses and Preclinical Characterization in Patient-Derived Models of Osteosarcoma, Ewing’s Sarcoma and Chondrosarcoma. Cells. 2021; 10(11):2893. https://doi.org/10.3390/cells10112893
Chicago/Turabian StyleGiordano, Giorgia, Alessandra Merlini, Giulio Ferrero, Giulia Mesiano, Erika Fiorino, Silvia Brusco, Maria Laura Centomo, Valeria Leuci, Lorenzo D’Ambrosio, Massimo Aglietta, and et al. 2021. "EphA2 Expression in Bone Sarcomas: Bioinformatic Analyses and Preclinical Characterization in Patient-Derived Models of Osteosarcoma, Ewing’s Sarcoma and Chondrosarcoma" Cells 10, no. 11: 2893. https://doi.org/10.3390/cells10112893
APA StyleGiordano, G., Merlini, A., Ferrero, G., Mesiano, G., Fiorino, E., Brusco, S., Centomo, M. L., Leuci, V., D’Ambrosio, L., Aglietta, M., Sangiolo, D., Grignani, G., & Pignochino, Y. (2021). EphA2 Expression in Bone Sarcomas: Bioinformatic Analyses and Preclinical Characterization in Patient-Derived Models of Osteosarcoma, Ewing’s Sarcoma and Chondrosarcoma. Cells, 10(11), 2893. https://doi.org/10.3390/cells10112893