Abstract
Cholangiocarcinomas (CHOLs), hepatobiliary malignancies, are characterized by high genetic heterogeneity, a rich tumor microenvironment, therapeutic resistance, difficulty diagnosing, and poor prognoses. Current knowledge of genetic alterations and known molecular markers for CHOL is insufficient, necessitating the need for further evaluation of the genome and RNA expression data in order to identify potential therapeutic targets, clarify the roles of these targets in the tumor microenvironment, and explore novel therapeutic drugs against the identified targets. Consequently, in our attempt to explore novel genetic markers associated with the carcinogenesis of CHOL, five genes (SNX15, ATP2A1, PDCD10, BET1, and HMGA2), collectively termed CHOL-hub genes, were identified via integration of differentially expressed genes (DEGs) from relatively large numbers of samples from CHOL GEO datasets. We further explored the biological functions of the CHOL-hub genes and found significant enrichment in several biological process and pathways associated with stem cell angiogenesis, cell proliferation, and cancer development, while the interaction network revealed high genetic interactions with a number of onco-functional genes. In addition, we established associations between the CHOL-hub genes and tumor progression, metastasis, tumor immune and immunosuppressive cell infiltration, dysfunctional T-cell phenotypes, poor prognoses, and therapeutic resistance in CHOL. Thus, we proposed that targeting CHOL-hub genes could be an ideal therapeutic approach for treating CHOLs, and we explored the potential of HLC-018, a novel benzamide-linked small molecule, using molecular docking of ligand-receptor interactions. To our delight, HLC-018 was well accommodated with high binding affinities to binding pockets of CHOL-hub genes; more importantly, we found specific interactions of HLC-018 with the conserved sequence of the AT-hook DNA-binding motif of HMGA2. Altogether, our study provides insights into the immune-oncogenic phenotypes of CHOL and provides valuable information for our ongoing experimental validation.
1. Introduction
Cholangiocarcinomas (CHOLs) are hepatobiliary malignancies ranked as the 2nd most prevalent hepatic cancer next to hepatocellular carcinoma [1]. They are characterized by cholangiocyte phenotypic features and are anatomically classified into intrahepatic, perihilar, and distal CHOLs based on their respective locations in the periphery of second-order bile ducts, the right and/or left hepatic duct, and the common bile duct [2]. Percentage occurrences of the perihilar, distal, and intrahepatic types of CHOLs are 50%, 42%, and 8%, respectively [3]. These subtypes also differ in their etiology, epidemiology, pathogenesis, diagnosis, and treatment [4]. However, these subtypes are united by a late diagnosis, limited curative options, and poor prognoses [5]; they all have a median survival of 24 months after diagnosis [4].
Major risk factors for CHOLs include cirrhosis, hepatitis B virus (HBV), hepatitis C virus (HCV), liver flukes, toxins, excessive alcohol intake, and metabolic disorders such as diabetes, obesity, and nonalcoholic fatty liver disease [6,7]. These risk factors, however, greatly vary by geographic area. Although recent years have witnessed the emergence of novel treatment targets, medical therapy remains a compelling challenge in hepatobiliary malignancies [8,9,10]. The only curative treatment option for early-stage disease is surgery [11], and no conclusive evidence for the efficacy of chemotherapy has been documented [5]. The heterogenic complexity of CHOLs supported by the rich tumor microenvironment (TME) is a major contributor to high therapeutic failure [3]. Furthermore, dynamic regulatory mechanisms of interactions between the stromal and immune components of the TME in the progression of CHOL remain poorly understood.
Accumulating evidence has established that abnormal gene expression profiles are frontline etiologic factors in the carcinogenesis and progression of human cancers. However, considering the heterogeneity of CHOLs, current knowledge on genetic alterations and known molecular markers is insufficient, necessitating the need for further evaluation of genomic and RNA expression data of CHOLs to identify potential therapeutic targets, clarify the roles of those targets in modulating the immunophenotypes of tumors, and explore novel therapeutic drugs against the identified targets [12]. A myriad of large-scale human cancer genomics projects offer large quantities of genomic and clinical data of cancer patients to analyze prognostic-relevant differentially expressed genes (DEGs) and identify biomarkers of disease progression and therapeutic targets [13].
Herein, we integrated DEGs from relatively large CHOL datasets obtained from the Gene Expression Omnibus (GEO) to identify the most implicated genes (which we refer to as CHOL-hub genes) in the development of CHOLs. We further explored the biological functions of these DEGs and constructed a protein-protein interaction (PPI) network. In addition, we established associations between the CHOL-hub genes and tumor progression, metastasis, tumor immune and immunosuppressive cell infiltration, poor prognoses, and therapeutic resistance in CHOLs.
In our previous studies, a myriad of in-house-synthesized [14,15,16] multi-target small molecules were explored for preclinical efficacy in attenuating cell proliferation, therapeutic resistance, and aggressive phenotypes of various cancers [17,18,19,20,21,22,23,24]. Furthermore, these small molecules have demonstrated promising activities in the treatment of osteoarthritis [25,26]. In our ongoing efforts to obtain derivatives of great potency of these compounds, in the present study, we report a novel benzamide-linked small molecule, 6-(2,4-difluorophenyl)-3-(3-(trifluoromethyl)phenyl)-2H-benzo[e][1,3]oxazine-2,4(3H)-dione (HLC-018), and explored its potential as a therapeutic target against CHOL-hub genes through an in silico ligand-receptor interaction study. To our delight, HLC-018 was well accommodated with high binding affinities to the binding pockets of CHOL-hub genes, and more importantly, we found specific interactions of HLC-018 with the conserved sequence of the AT-hook DNA-binding motif of HMGA2. Altogether, our study provides insights into the immune-oncogenic phenotypes of CHOLs and provides valuable information for ongoing experimental validation.
2. Methods
2.1. Transcriptome Data Acquisition and DEG Identification
We mined the NCBI GEO, a public functional genomics data repository to obtain high-throughput sequencing and microarray-based gene expression profile data from five CHOL datasets (GSE132305, GSE31370, GSE38860, GSE45001, and GSE32225). The datasets consisted of gene expression profiles from CHOL patients vs. control cohorts. Detailed characteristics of the datasets are presented in Table 1. GEO2R tools were adopted to screen for DEGs from each dataset. The obtained DEGs were further screened to identify the most significant ones using the limma R package with a threshold of|log2multiple of change|of >1.5 and p < 0.05. DEGs from each dataset were superimposed to identify common intersecting DEGs, which were visualized using the Multiple List Comparator web tool.

Table 1.
Characteristic of the cholangiocarcinoma (CHOL) datasets used for differentially expressed gene identification.
2.2. Differential Expressions of CHOL-Hub Genes among Tumor Grades and with Nodal Metastasis
We analyzed differential expression levels of CHOL-hub genes between tumor and adjacent normal tissues of The Cancer Genome Atlas (TCGA) CHOL dataset using the Tumor Immune Estimation Resource (TIMER, vers. 2.0) [27]. We downloaded transcript expression levels of CHOL-hub genes in CHOL patients from TCGA cancer database via the UALCAN server [28] and compared expression levels of CHOL-hub genes among four types of tumor grades, including well-differentiated (low grade, grade 1), moderately differentiated (intermediate grade, grade 2), poorly differentiated (high grade, grade 3), and undifferentiated (high grade, grade 4). We also compared expression profiles of CHOL-hub genes between nodal metastasis statuses by comparing transcript expression levels of CHOL-hub genes between CHOL patients with no regional lymph node metastasis (N0) and patients with metastases in one to three axillary lymph nodes (N1). Differential expression was considered statistically significance at p < 0.05, <0.01, and <0.001.
2.3. Prognostic Analysis of CHOL-Hub Genes
Survival times (in days) and RNA expression data of CHOL-hub genes from tumor samples collected from CHOL patients in TCGA datasets were downloaded from the GDC portal using the Q-omics algorithm (wndows, vers. 0.9). Patients were split into high- and low-expression groups based on median expression levels of CHOL-hub genes. Overall survival (OS) and disease-free survival (DFS) of cohorts in each group were computed with a hazard ratio (HR), a 95% confidence interval (CI), and a log-rank test p value. In addition, we used the SurvExpress algorithm [29] to evaluate survival gene expression data of CHOL-hub genes using the CHOL-TCGA CHOL dataset, consisting of 35 CHOL patients. Survival times (in days) of the cohorts were censored and compared between cohorts with higher expression levels and those with lower expression levels of CHOL-hub genes. A risk index was computed, and hazard ratios (HRs) were estimated by fitting a CoxPH using the risk group as a covariate.
2.4. Functional Enrichment and Interaction Network Analysis of CHOL-Hub Genes
The online gene set enrichment analysis (GSEA) server, Enrichr [30,31], was used to analyze functional enrichment profiles of CHOL-hub genes based on Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and gene ontology (GO) terms of biological processes. Enrichment terms were considered significant at p < 0.05, and visualization was achieved using the R package cluster Profiler [32]. The interaction analysis and network construction for gene-gene interactions (GGIs) of CHOL-hub genes were performed using GeneMANIA, a real-time multiple association network integration algorithm for predicting gene functions [33].
2.5. Analysis of CHOL-Hub Gene Associations with Drug Sensitivity
We downloaded the area under the dose-response curve (AUC) values for anticancer small molecules and CHOL-hub gene expression profiles in different cancer cell lines from the drug-cell response sensitivity repository data of the Genomics of Drug Sensitivity in Cancer (GDSC) and CTRP through the GSCALite server [34]. We then used Spearman correlation coefficients to analyze correlations between CHOL-hub gene expression levels and drug sensitivity (50% inhibitory concentration (IC50)) to 265 small molecules from the GDSC and CTRP databases. The top 30 most correlated compounds are presented in a bubble plot.
2.6. Analysis of CHOL-Hub Genes’ Associations with the TME and Immunophenotypes in CHOL
We used the ImmuCellAI (Immune Cell Abundance Identifier) algorithm to estimate the tumor infiltration of 24 immune cells including 18 T-cell subtypes and six other immune cells (natural killer (NK) cells, monocytes, macrophages, neutrophils, dendritic cells (DCs), and B cells) from RNA-sequencing (Seq) data of CHOL-hub genes. A correlation analysis was conducted using purity-corrected partial Spearman’s rho values and statistical significance based on p values of the Wilcoxon test and the false discovery rate (FDR). Data visualization was done using the GSCALite online server [34]. In addition, we used the QUERY module of the TIDE algorithm to evaluate the effect of CHOL-hub gene methylation on dysfunctional T-cell phenotypes [35].
2.7. Molecular Docking Studies of CHOL-Hub Genes with HLC-018
Crystal structures of CHOL-hub genes of SNX15 (PDB:6ECM), ATP2A1 (PDB:4H1W), PDCD10 (PDB:3AJM), BET1 (PDB:3EGX), and HMGA2 (PDB:3UXW) in PDB file format were obtained from the Protein Data Bank, while the three-dimensional (3D) structure of LCC18 in mol2 format was obtained using the Avogadro molecular builder and visualization tool vers. 1.XX [36]. These were converted to PDB files using the PyMOL Molecular Graphics System, vers. 1.2r3pre. All PDB files were subsequently converted to PDBQT files using AutoDock Vina (vers. 0.8, Scripps Research Institute, La Jolla, CA, USA) [37]. Each of the CHOL-hub gene targets was prepared for docking by removing water molecules and adding polar hydrogen atoms and charges [38,39,40]. Docking was conducted using AutoDock Vina as described in previous studies [38,41], and results were visualized using the PyMOL, Discovery studio visualizer vers. 19.1.0.18287 (BIOVIA, San Diego, CA, USA) [42] and protein-ligand interaction profiler web tool [43].
3. Results
3.1. SNX15, ATP2A1, PDCD10, BET1, and HMGA2 Are Biomarkers of Tumor Progression and Metastasis in CHOL Patients
A schematic flow chart summarizing the overall study design for the identification and evaluation of the pathological role of DEGs in cholangiocarcinoma is shown in Figure 1. With the aim of identifying biomarker hub genes associated with the etiology and progression of CHOLs, we integrated DEGs from five microarray datasets of CHOL cohorts vs. normal patients (Table 1, Figure 1) and identified SNX15, ATP2A1, PDCD10, BET1, and HMGA2 as the only overlapping DEGs across the five datasets (Figure 2A). Furthermore, we analyzed differential expression profiles of CHOL-hub genes between cancer and matched normal tissues across all TCGA tumors and found that CHOL cancer tissues expressed SNX15, ATP2A1, PDCD10, BET1, and HMGA2 at significantly (p < 0.001) higher levels than did adjacent normal tissues (Figure 2B). Gene expression comparisons revealed that expression levels of hub genes in CHOL patients occurred in a significant order of PDCD10 > BET1 > SNX15 > ATP2A1 > HMGA2 (Figure 2C). However, we compared expression profiles between tumor grades and found that SNX15, ATP2A1, PDCD10, BET1, and HMGA2 expressions were significantly (all p < 0.05) associated with tumor stages (Figure 2D). In addition, we found that CHOL patients with metastases in one to three axillary lymph nodes exhibited significantly (all p < 0.05) higher expression levels of SNX15, ATP2A1, PDCD10, BET1, and HMGA2 than patients with no regional lymph node metastasis (Figure 2E). Suggestively, our results indicated that SNX15, ATP2A1, PDCD10, BET1, and HMGA2 could serve as a biomarker signature of tumor progression and metastasis in CHOL patients.
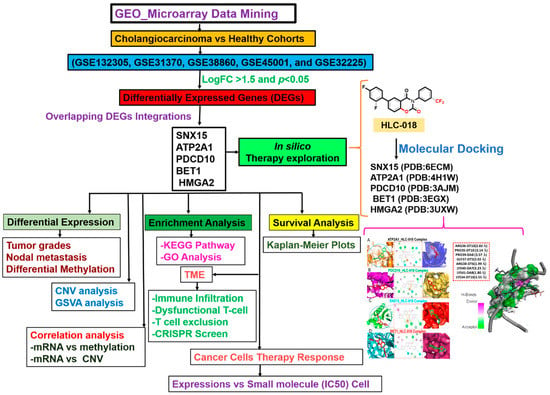
Figure 1.
Schematic flow chart of the study design for identifying and characterizing the pathological role of DEGs in cholangiocarcinoma and in silico exploration of HLC-018 as a potential therapeutic candidate.
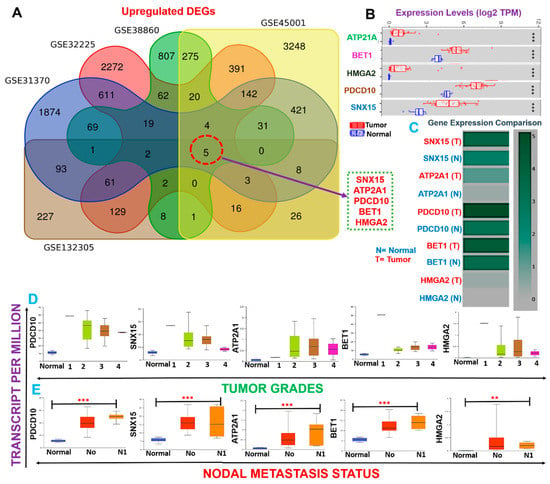
Figure 2.
Cholangiocarcinoma (CHOL)-hub genes are biomarkers of tumor progression and metastasis in CHOL patients. (A) Venn diagram showing the distribution of differentially expressed genes (DEGs) in each dataset and overlapping upregulated DEGs. (B) Boxplot showing differential mRNA expression levels of CHOL-hub genes between TCGA CHOL tumor samples and adjacent normal controls. (C) Heatmap of expression difference among CHOL-hub genes. (D) Box plot showing differential expression levels of hub genes among tumor grades (E) and nodal metastasis statuses in TCGA cohorts of CHOL. ** p < 0.01, *** p < 0.001.
3.2. SNX15, ATP2A1, PDCD10, BET1, and HMGA2 Are Associated with Higher Risks and Poor Prognoses of CHOL
Since CHOL-hub genes were found to be overexpressed and associated with tumor stages and metastasis (p < 0.05) in CHOL patients, we queried whether these genes could individually predict the survival of CHOL patients. Interestingly we found that out of the five hub genes, only HMGA2 (HR = 1.914, p = 0.048) demonstrated an independent significant association with a worse prognosis of CHOL patients (Figure 3A, Supplementary Table S1). Furthermore, we queried the collective survival effect of CHOL-hub genes and found that higher expression levels of CHOL-hub genes were significantly associated with higher risk and achieved significant worse prognoses (HR = 4.61, p = 0.044) of CHOL patients (Figure 3B). However, our multivariable and HR analysis revealed that the differential expression levels of the CHOL hub genes achieved no significant (p > 0.05) hazard risk among the tumor stages (Supplementary Table S1).
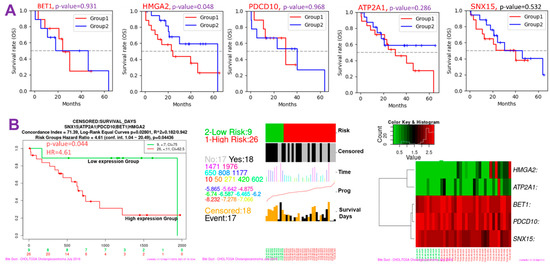
Figure 3.
Cholangiocarcinoma (CHOL)-hub genes are associated with higher risks and poorer prognoses of CHOL. (A) Kaplan–Meier survival plot (B) Kaplan–Meier plots and hazard plots of CHOL-hub genes in CHOL patients.
3.3. CHOL-Hub Genes Potentially Promote Tumor Progression by Crosstalking with Multiple Oncogenic Targets/Pathways
Deciphering genetic interactions is vital to understanding cellular and organismal responses to gene-level alterations in cancer. Under the hypothesis that functionally related genes tend to share common genetic interaction partners [44], we evaluated GGIs of CHOL-hub genes and observed high genetic interactions with a number of onco-functional genes, including STX5, STK25, SEC22B, STK24, SLN, GOSR1, GOSR2, HMGA1, NOTCH1, YKT6, NAPA, STK26, STX7, PDGFRB, E4F1, CCM2, ASF1A, UBN1, VTI1A, and HIRA (Figure 4A). Interestingly, the CHOL-hub genes exhibited coexpression interactions with the expression levels of the onco-functional genes (Supplementary Table S2). Additionally, we analyzed the KEGG and GO enrichment of hub genes and found significant enrichment in several biological processes and pathways associated with stem cell angiogenesis, cell proliferation, and cancer development (Figure 4B). Enriched KEGG pathways of hub genes were related to SNARE interactions in vesicular transport, cardiomyopathy, pancreatic secretion, microRNAs in cancer, transcriptional misregulation in cancer, thyroid hormone signaling pathway, calcium signaling pathway, cGMP-PKG signaling pathway, and cAMP signaling pathway. GO biological processes in which hub genes were enriched included regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis, regulation of stem cell proliferation, positive regulation of stress-activated protein kinase signaling cascade, wound healing, spreading of cells, regulation of cell migration involved in sprouting angiogenesis, mitotic G2/M transition checkpoint, and the epithelial-to-mesenchymal transition (EMT). Collectively, our results suggested that the CHOL-hub genes promote tumor progression by crosstalking with multiple oncogenic pathways and thus provides a rationale for targeting these genes as a therapeutic strategy to curb the activities of the multiple oncogenic pathways.
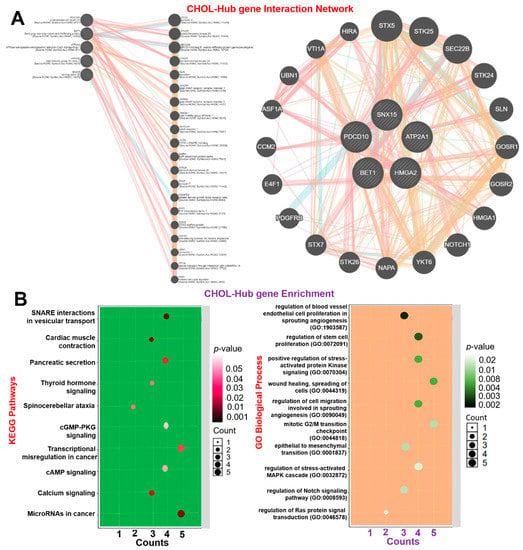
Figure 4.
Gene interaction and enrichment analysis of cholangiocarcinoma (CHOL)-hub genes and (A) gene-gene interaction (GGI) network. (B) Enrichment of KEGG pathways (left panel) and GO biological processes (right panel) of hub genes.
3.4. CHOL-Hub Genes Are Potential Drivers of Invasive Immunophenotypes of CHOL
Expression levels of HMGA2 and PDCD10 in CHOL were significantly correlated with tumor infiltration by seven and nine immune cell types, respectively. HMGA2 showed negative correlations (r = −0.30~−0.48, p < 0.05) with tumor infiltration by Th17 cells, monocytes, and macrophages and positive correlations (r = 0.33~−0.57, p < 0.05) with tumor infiltration of B cells, NKT cells, nTregs, and CD8-naive cells. PDCD10 expression was negatively correlated (r = −0.33~−0.43, p < 0.05) with tumor infiltration by MAIT, NK cells, and monocytes, and positively correlated (r = 0.29~−0.51, p < 0.05) with tumor infiltration by B cells, CD4_T cells, DCs, iTregs, nTregs, and Tr1s. Expression levels of ATP2A1 were negatively associated with monocytes and macrophages, and positively correlated with B-cell infiltration, while BET1 demonstrated positive correlations only with tumor infiltration by macrophages and Th1 cells in CHOL (Table 2).

Table 2.
Correlation of cholangiocarcinoma (CHOL)-hub gene mRNA expression levels and infiltration by tumor immune cells.
We queried the role of methylation and CNV of the CHOL-hub genes in mediating the invasive immunophenotypes of CHOL patients using the TCGA cohorts (Figure 5A–D). We found that HMGA2 and ATP2A1 are significantly (p < 0.05) hypermethylated, while SNX15 is hypomethylated in CHOL patients when compared with the normal cohort (Figure 5A). The differential methylation of CHOL-hub genes predicted dysfunctional T-cell phenotypes (Figure 5B,C) and a high survival risk of the cohorts (Figure 5D).
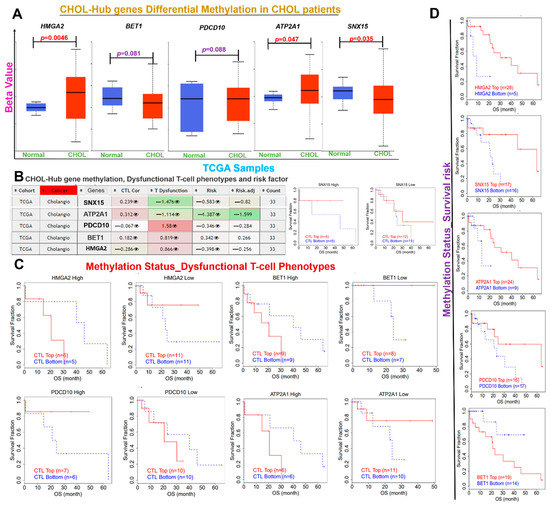
Figure 5.
Differential Methylation of CHOL-Hub gene is associated with Dysfunctional T-cell Phenotypes in CHOL patients. (A) Boxplot showing differential methylation levels of CHOL-hub genes between TCGA CHOL tumor samples and healthy. (B) Heatmap of CHOL-Hub gene methylation and its association with Dysfunctional T-cell phenotypes and risk factor of CHOL patients. (C) Kaplan–Meier plot showing the association between the methylation status of the CHOL-hub gene and dysfunctional T-cell phenotypes of CHOL patient (D) Kaplan–Meier plot showing the association between the methylation status of the CHOL-hub gene and survival risk of CHOL patient.
However, our exploration of correlations between mRNA expressions and the methylation or CNV of CHOL-hub genes in CHOL patients revealed no significant associations except for the positive correlation between mRNA expression and CNV of BET1 (Figure 6A). Survival analysis revealed that out of the five CHOL-hub genes, only HMGA2 exhibited CNV (amplification) and was significantly (p = 0.02) associated with a worse prognosis compared to patients with wide-type HMGA2 (Figure 6B). We queried the collective effect of the entire CHOL-hub gene set on tumor infiltrations of immune cells, and found that a higher GSVA score of CHOL-hub genes in CHOL tumor samples (Figure 6C) was positively correlated with higher tumor infiltrations of nTregs, iTregs, Th17 cells, central memory, MAIT, DCs, monocytes, macrophages, and Gamma-delta cells in CHOL patients, while negative corrections were observed between high GSVA scores of CHOL-hub genes and tumor infiltration by NKT, B-cells, CD4 and CD8, Tr1, and Tfh (Figure 6D). Collectively, our results suggested that the CHOL-hub genes are associated with tumor immune infiltrations and are potential drivers of dysfunctional T cell and invasive immunophenotypes of CHOL. We explored a single-cell RNA-sequencing dataset to evaluate which type of cells actually expressed the CHOL-hub genes. Our results revealed that SNX15, ATP2A1, PDCD10, BET1, HMGA2 are expressed by the T cells; however, only the BET1 and PDCD10 are expressed by the B cells (Supplementary Figure S1).
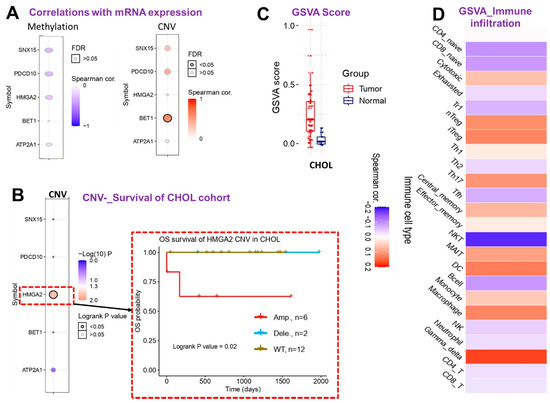
Figure 6.
Cholangiocarcinoma (CHOL)-hub gene associations with tumor immune and immunosuppressive cell infiltration in CHOL patients. (A) Bubble plot showing correlations of the mRNA expression levels of CHOL-hub genes with methylation and copy number variation (CNV). (B) Heat map plot and Kaplan–Meier plot showing survival differences between CHOL patient with CNV of CHOL-hub genes and cohorts with wild-type genes. (C) Box plot showing GSVA scores of CHOL-hub genes between CHOL tumor and adjacent normal samples. (D) Heatmap showing correlations between GSVA scores of CHOL-hub genes and infiltration levels of various cells in CHOL patients.
3.5. The CHOL Hub Genes Are Associated with Worse Immune-Oncological Phenotypes and Therapy Outcomes
We evaluated the effects of CHOL-hub genes on anticancer drug sensitivity by analyzing associations between mRNA expression levels of CHOL-hub genes and IC50 values of anticancer small molecules against various cancer cell lines using the drug-cell response sensitivity repository data of the GDSC and CTRP. Interestingly, we found that out of the top 30 compounds for which data were explored from CTRP, HMGA2 demonstrated strong positive associations, while ATP2A1 demonstrated strong negative associations with IC50 values of all compounds, suggesting sharp contrasts in the roles of the former compared to the latter (Figure 7A). Similarly, IC50 values of most of the small molecules from the GDSC database showed positive correlations with mRNA expression levels of BET1 and HMGA2, while ATP2A1, in contrast to BET1 and HMGA2, demonstrated negative associations with IC50 values of the drugs (Figure 7B). In order to gain a generalized insight of the biomarker relevance and role of the hub genes, we compared the predictive value of the CHOL-Hub genes with 8 standardized biomarkers and accessed the gene prioritization of the CHOL-hub genes in various immune cohorts’ datasets, including the ICB therapy outcome, dysfunctional or exclusion T-cell phenotypes, and datasets of T-cell mediated tumor killing in CRISPR screens. Regarding biomarker relevance, out of the 8 standardized biomarkers of response outcome and overall survival, the CHOL-Hub gene set achieved higher counts of the predictive score (AUC > 0.5) than TMB, T-cell clonality, and B-cell clonality (Figure 7C). Our results suggest that high expression levels of the CHOL-Hub genes are associated with worse outcomes to PD1, CTL4A, and PDL1 immunotherapies in various ICB datasets (Figure 7D). In addition, high expression levels of the hub genes predicted dysfunctional T-cell phenotypes but demonstrate no significant association with T-cell exclusion phenotypic markers (MDSC, CAF, and M2_TAM). However, the overall prioritization of the genes in the various immune-related datasets occurs in the order of HMGA2 > SNX15 > ATP2A1 > BET1 > PDCD10. Collectively, these data suggest that the high expression levels of the CHOL-Hub gene are associated with worse immune-oncological phenotypes and treatment responses, hence serving as an attractive target for therapy exploration.
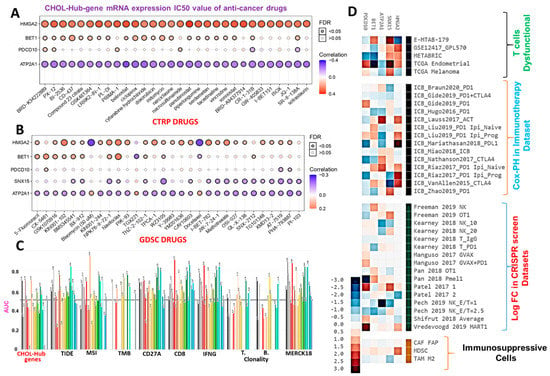
Figure 7.
Cholangiocarcinoma (CHOL)-hub genes exhibited different associations with the sensitivity of cancer cells to anticancer therapy. Bubble plot of correlations of mRNA expression levels of hub genes with (A) CTRP and (B) GDSC drug sensitivities. Colors from blue to red represent correlations between mRNA expressions and IC50 values of small-molecule drugs. A positive correlation means that the gene’s high expression was resistant to the drug and vice versa. The bubble size was positively correlated with the false discovery rate (FDR) significance. The black outline border indicates an FDR of <0.05. (C) Bar plot of the comparative biomarker relevance between the CHOL-Hub genes and standardized biomarkers. (D) Heat map depicting the association between the CHOL hub genes and outcomes of ICB therapy, T-cell dysfunctional and exclusion markers, and T cell-mediated tumor killing in CRISPR screens, CAFs; cancer-associated fibroblasts; MDSCs; myeloid-derived suppressor cells; M2-TAMs; M2 subtype of tumor-associated macrophages.
3.6. Discovery of a Novel Small Molecule, HLC-018, via Scaffold-Hopping of Bioactive Compounds
Scaffold-hopping of bioactive compounds is an important approach for novel drug design and development [45]. Biphenyl, flavones, and isoflavones are important natural product backbones and several bioactive compounds containing these backbones have been reported for a vast range of biological activities including anti-oxidative, anti-atherosclerosis, muscle relaxant, anti-microbial, anti-inflammatory, and anti-cancer effects [46,47]. Trifluoromethylphenyl is an important moiety that has been implicated in the activities of various drugs (Figure 8). A number of clinical drugs, e.g., nilotinib (a tyrosine kinase inhibitor with antineoplastic activity), fluoxetine (antidepressant, antiobsessional, anti-anxiety, and immunomodulating agent) and sorafenib (RAF/MEK/ERK inhibitor with anticancer activity) contain trifluoromethylphenyl as an important component responsible for their bioactivity. Niclosamide is a multipurpose compound with proven efficacy in the treatment of several diseases including oxidative stress, infection, metabolic disorders, inflammation and cancers [48,49]. In the present study, a scaffold-hopping (Figure 8) of these bioactive natural compounds (flavones and isoflavones), biphenyl, trifluoromethylphenyl, and niclosamide lead to the discovery of a novel multitarget small molecule; 6-(2,4-difluorophenyl)-3-(3-(trifluoromethyl)phenyl)-2H-benzo[e][1,3]oxazine-2,4(3H)-dione (HLC-018). Subsequently, we explored the potential of this compound as a therapeutic target against CHOL-hub genes through an in silico ligand-receptor interaction study.
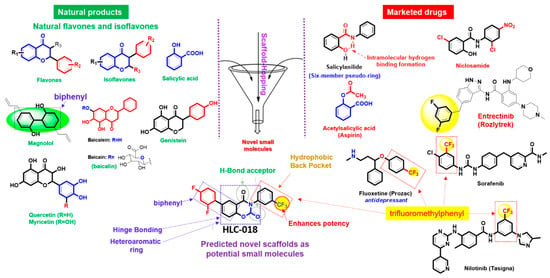
Figure 8.
Scaffold-hopping of bioactive natural products and marketed drugs for the discovery of HLC-018. Scaffold-hopping of bioactive natural compounds (flavones and isoflavones), biphenyl, trifluoromethylphenyl, and niclosamide lead to the discovery of a novel small molecule, 6-(2,4-difluorophenyl)-3-(3-(trifluoromethyl)phenyl)-2H-benzo[e][1,3]oxazine-2,4(3H)-dione (HLC-018).
3.7. Molecular Docking Reveals the Potential Druggability of CHOL-Hub Genes by HLC-018
Our molecular docking analysis revealed that HLC-018 exhibited high affinities for CHOL-hub genes with respective binding energies of −7.90, −10.40, −9.10, and −8.70 kcal/mol for SNX15, ATP2A1, PDCD10, and BET1 (Table 3). HLC-018 bound to the binding pocket of CHOL-hub genes by several conventional H-bonds, halogen bonds, and multiple π-interactions (Figure 9). Furthermore, several van der Waals forces were found around the HLC-018 backbone with the respective amino acid residues of CHOL-hub gene binding pockets. Furthermore, ligand-receptor complexes were stabilized by various hydrophobic contacts. Altogether, the receptor-ligand interaction profile suggested that HLC-018 has high potential to target the CHOL-hub gene, having higher affinity for PDCD10 and ATP2A1 than for SNX15 or BET1.

Table 3.
Docking profile of HLC-018 with cholangiocarcinoma (CHOL)-hub genes.
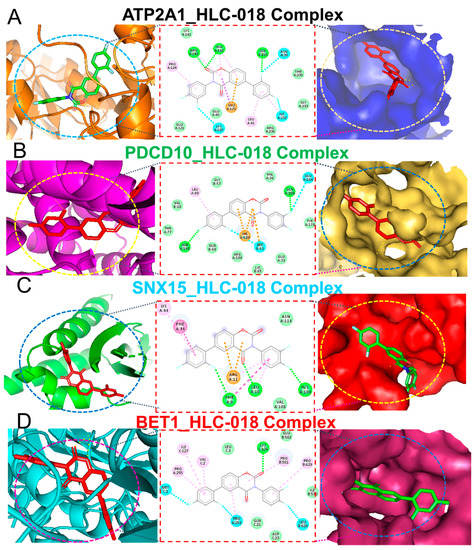
Figure 9.
Ligand-receptor interaction profiles of HLC-018 in complex with (A) ATP2A1, (B) PDCD10, (C) SNX15, and (D) BET1. The left and middle panels respectively show two-(2D) and three-dimensional representations of ligand-receptor complexes, which reveal interacting amino acid residues and the type of interactions between the CHOL-hub genes and HLC-018. The right panels show the position of HLC-018 at the binding pocket flap of CHOL-hub genes.
3.8. HLC-018 Demonstrated Specific Interactions with the Conserved Sequence of the AT-Hook DNA-Binding Motif of HMGA2
High-mobility group A2 (HMGA2) is an AT-hook DNA-binding motif-containing protein, identifiable by a conserved core sequence of Pro-Arg-Gly-Arg-Pro [50]. Our molecular docking study revealed a unique interaction of HLC-018 with the unique Pro-Arg-Gly-Arg-Pro sequence of the AT-hook DNA-binding motif of HMGA2 (Figure 10). We found that HLC-018 bound with hydrogen bonds to the PRO35_DT10-ARG36_DT10-GLY37_DT5-ARG38_DT6-PRO39_DA8 residues of HMGA2 with a relatively close proximity range of 1.99~3.55 Å and a binding affinity of −8.8 kcal/mol. No hydrophobic interactions were found.
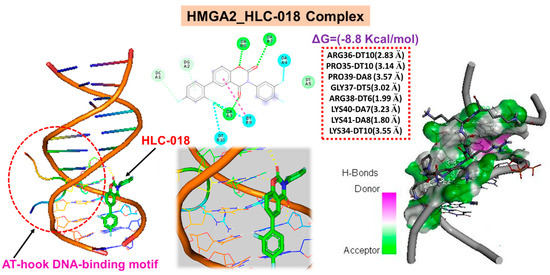
Figure 10.
Molecular docking of HLC-018 with the unique Pro-Arg-Gly-Arg-Pro sequence of the AT-hook DNA-binding motif of HMGA2.
4. Discussion
In recent years, great efforts have been put into studying the pathogenesis of CHOL, with much emphasis on the mechanisms of genetic and epigenetic alterations. However, our current knowledge of the genetic alterations and known molecular markers of CHOL is insufficient. Understanding the biology and pathogenesis of CHOL and its complex interactions with the TME could lead to better therapies and patient prognoses. The TME, a composite of various cells including stroma cells, infiltrating immune cells, and immunosuppressive cells, plays a crucial role in the initiation and progression of various human cancers [51].
In the present study, our attempt to explore novel genetic markers associated with the carcinogenesis of CHOL led to the identification of five genes (SNX15, ATP2A1, PDCD10, BET1, and HMGA2) as the most common deregulated overexpressed genes via integration of DEGs from relatively large-sample CHOL GEO datasets. We further explored the biological functions of CHOL-hub genes and found significant enrichment in several biological processes and pathways associated with stem cell angiogenesis, cell proliferation, and cancer development, while the interaction network revealed high genetic interactions with a number of onco-functional genes. In addition, we established associations between the CHOL-hub genes and tumor progression, metastasis, tumor immune and immunosuppressive cell infiltration, dysfunctional T-cell phenotypes, poor prognoses, and therapeutic resistance in CHOL. Therefore, it could plausibly be hypothesized that SNX15, ATP2A1, PDCD10, BET1, and HMGA2 are major factors in CHOL and potential drivers of invasive immunophenotypes of CHOL. Results of the present study therefore add to the existing knowledge of the oncogenic roles of these genes, promote our understanding of the biology and carcinogenesis of CHOL, and provide new targets for risk stratification, molecular diagnoses, therapy exploration, and follow-up of CHOL.
Suggestive evidence from previous studies revealed that aggressive phenotypes and prognoses of cancers are largely dependent on tumor histology, stages, and metastasis. Interestingly, our analysis revealed that SNX15, ATP2A1, PDCD10, BET1, and HMGA2 expressions were significantly associated with tumor stages and axillary lymph nodes metastasis of CHOL, and higher expression levels of CHOL-hub genes were significantly associated with higher risks and significantly worse prognoses (HR = 4.61, p = 0.044) of CHOL patients. In line with our observations, oncogenic roles of PDCD10 were reported in various cancers including breast [52], bladder [53], prostate [54], ovarian [55], and several other cancers. A previous study also identified ATP2A1 as an important driver of human papillomavirus (HPV)-associated oropharyngeal cancer [56]. The BET domain is regarded as a co-regulator of obesity, inflammation, and cancer [57]. Michalak et al. [58] identified BET1 as a malignancy-associated protein in human colorectal cancer, and inhibitors of BET1 were proposed for treating metastatic prostate cancer [59]. SNX15 is a regulator of intracellular protein trafficking consisting of endocytosis, endosomal sorting, and endosomal signaling and was associated with mammary adenocarcinoma metastases to the lungs [60].
HMGA2 is an AT-hook DNA-binding motif-containing protein, identifiable by a conserved core sequence of Pro-Arg-Gly-Arg-Pro [50]. It is a transcription factor that binds to the minor groove of the adenine-thymine (AT)-rich DNA sequence of several genes and alters the chromatin architecture with consequent modulation of their transcription [61]. As such, it influences various biological processes associated with cell proliferation and carcinogenesis. In agreement with our identification of HMGA2 as an important driver of CHOL carcinogenesis, overexpression of HMGA2 was reported in various cancers [62,63], and it was implicated in tumor metastasis, poor prognoses, and therapy failures in various cancer [64,65]. Thus, HMGA2 could serve as an important oncogenic molecule for exploration of targeted therapies.
Tumor microenvironment (TME), a composite of tumor cells, immune cells, stromal cells, cytokines, chemokines, and microvessels, has been proven to be crucial for the development of various tumors [39]. Although recent years have witnessed the emergence of novel treatment targets, medical therapy remains a compelling challenge in hepatobiliary malignancies [8,9,10]. The heterogenic complexity of CHOLs supported by the rich tumor microenvironment (TME) is a major contributor to high therapeutic failure [3]. Furthermore, dynamic regulatory mechanisms of interactions between the stromal and immune components of the TME in the progression of CHOL remain poorly understood. Results of the present study revealed that the CHOL-Hub genes are association with the infiltration levels of immune and immunosuppressive cells in cholangiocarcinoma. Tumor-infiltration of various immune and immunosuppressive cells vary with the host immune status and have latent prognostic value in various cancers. CAF, Treg, TAM, and MSDC are immunosuppressive cells that inhibit the function and abundance of cytotoxic lymphocytes leading to T-cell exclusion of the tumor and mediate invasive phenotypes [66]. Although, some tumors have abundant infiltration of immune cells, these immune cells are in a dysfunctional state and could not mediate any antitumor immune response, a condition known as T-cell allergy [41,51]. Interestingly, we found that high expression levels of the hub genes predicted dysfunctional T-cell phenotypes but demonstrate no significant association with T-cell exclusion phenotypic markers (MDSC, CAF, Treg, and TAM), suggesting that the CHOL hub genes are associated with invasive phenotypes of cholangiocarcinoma via dysfunctional T-cell phenotypes and not by T-cell exclusion mechanisms.
DNA methylation is an important epigenetic modulation in mammalian genomes, plays a crucial role in regulating gene expression, and may serve as a noninvasive biomarker for cancer diagnosis and prognosis [51,67]. It has been reported that DNA methylation induced genetic alterations in favor of carcinogenesis via the recruitment of the methyl moieties containing gene repressor proteins or by inhibiting the binding of the transcription factors to DNA [51,67]. Therefore, the differential methylation of CHOL-hub genes in cholangiocarcinoma when compared with the normal tissue strongly suggests that the CHOL-hub genes are involved in the epigenetic mechanism of the CHOL pathogenesis. In addition, the differential methylation of CHOL-hub genes predicted dysfunctional T-cell phenotypes and a high survival risk of the cohorts, suggesting that epigenetic modification of these genes does not only modulate the gene expression but also contributed to the immuno-invasive phenotypes and worse prognosis of cholangiocarcinoma.
Deciphering of genetic interactions is vital to understanding cellular and organismal responses to gene-level alterations in cancer [44]. Results of our GGI analysis found that CHOL-hub genes exhibited high genetic interactions with a number of onco-functional genes, particularly members of the GCKIII protein kinase family, which comprises MST3/STK24, SOK1/STK25, and MST4/STK26, and the SNARE proteins (GOSR1 and GOSR2). These are recognized as functional genes implicated in cell proliferation and the developments of various cancers [68,69]. Coherently, our biological functional and pathway analysis of CHOL-hub genes achieved enrichment of SNARE interactions, microRNAs in cancer, transcriptional misregulation, regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis, regulation of stem cell proliferation, wound healing, spreading of cells, regulation of cell migration involved in sprouting angiogenesis, mitotic G2/M transition checkpoint, and the EMT. Altogether, our results suggested the CHOL-hub genes mediate the development and progression of CHOL via involvement in transcriptional misregulation, stem cell angiogenesis, cell proliferation, and cancer development.
Our in silico drug sensitivity analysis in various cancer cell lines suggested contrasting roles of CHOL-hub genes in potentiating therapeutic resistance. We found that HMGA2 was strongly positively correlated, while ATP2A1 was demonstrated to be strongly negatively correlated with IC50 values of all 30 CTRP small-molecule drugs, suggesting sharply contrasting roles of the former compared to the latter in modulating the sensitivity of cancer cell lines to therapy. Similarly, IC50 values of most of the small molecules from the GDSC database showed positive correlations with mRNA expression levels of BET1 and HMGA2, suggesting their associations with therapeutic resistance, while ATP2A1, in stark contrast to BET1 and HMGA2, demonstrated negative associations with IC50 values of the drugs. The potential mechanisms of the CHOL-hub genes’ effects on therapeutic responses require further investigation.
Nevertheless, we proposed that targeting CHOL-hub genes could become an ideal therapeutic approach for treating CHOL. To this end, we explored the potential of HLC-018, a novel small-molecule derivative using molecular docking of ligand-receptor complexes. Interestingly, our results revealed the potential druggability of CHOL-hub genes by HLC-018. To our delight, HLC-018 was well accommodated with high binding affinities to binding pockets of CHOL-hub genes. HLC-018 bound to the binding pockets of CHOL-hub genes by several conventional H-bonds, halogen bonds, and multiple π-interactions. Furthermore, several van der Waals forces were found around the HLC-018 backbone with respective amino acid residues of binding pockets of CHOL-hub genes. Furthermore, ligand-receptor complexes were stabilized by various hydrophobic contacts. Altogether, receptor-ligand interaction profiles suggested high potential of HLC-018 for targeting PDCD10 and ATP2A1 than for SNX15 or BET1. More, importantly, we found that HLC-018 bound to the AT-hook DNA-binding motif of HMGA2 specifically at the conserved sequence of PRO35_DT10-ARG36_DT10-GLY37_DT5-ARG38_DT6-PRO39_DA8 all due to hydrogen bonds and with a high affinity of −8.8 kcal/mol. Compared to the binding affinity of this conserved region to some clinical drugs reported in the literature, such as with aspirin (−7.85 kcal/mol) and sulindac sulfide (−7.68 kcal/mol) [62], HLC-018 demonstrated a higher potential for binding to the AT-hook DNA-binding motif of HMGA2 and consequently a higher potential for inhibiting the binding of the AT region of genes thereby affecting the transcriptional role of HMGA2. Altogether, our study provided insights into the immune-oncogenic phenotypes of CHOL and provided valuable information for ongoing experimental validation of targeted efficacy of HLC-018 in CHOL cell lines. Despite the advantages discussed above, the limitations of the present study merit discussion. This study represents an in silico and correlation analysis based on clinical data of cholangiocarcinoma patients and receptor-ligand interactions. Therefore, the reliability and clinical applicability of these findings required experimental validation. Furthermore, the full therapeutic potential of HLC-018 for targeting the CHOL-hub genes awaits experimental validation through in vitro and in vivo models.
5. Conclusions
In conclusion, the results of the present study suggest that the cholangiocarcinoma (CHOL)-hub genes (SNX15, ATP2A1, PDCD10, BET1, and HMGA2) can be used as biomarkers of tumor progression, metastasis, therapy outcome, and poor prognoses of CHOL. The CHOL-Hub genes potentially promote tumor progression by crosstalking with multiple oncogenic targets/pathways and potential drivers of invasive immunophenotypes of CHOL. To our delight, HLC-018, a novel benzamide-linked small molecule, demonstrated a high potential for targeting these genes and is currently under detailed experimental validation in our lab.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/cells10112873/s1, Table S1: Multivariable and HR analysis of the CHOL hub genes in cholangiocarcinoma. Table S2: Coexpression interactions of CHOL-hub genes with the expression levels of the onco-functional genes. Figure S1: Heatmap of the expression levels of the CHOL-Hub genes based on single-cell RNA-sequencing dataset.
Author Contributions
B.L. carried out the study and wrote the manuscript; S.-L.T. and F.-C.L. participated in data collection and analyses; H.-S.H. synthesized and provided the drug; Y.-C.K., H.-Y.L., A.T.H.W., and H.-S.H. designed and oversaw the study. All authors have read and agreed to the published version of the manuscript.
Funding
This study was funded by the Ministry of Science and Technology (MOST110-2314-B-038-120 to H.-S.H. and MOST110-2314-B-038 -115 to H.-Y.L.) and the TMU Research Center of Cancer Translational Medicine from The Featured Areas Research Center Program within the framework of the Higher Education Sprout Project by the Ministry of Education (MOE) in Taiwan (DP2-107-20000).
Institutional Review Board Statement
Not Applicable.
Informed Consent Statement
Not Applicable.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Welzel, T.M.; McGlynn, K.A.; Hsing, A.W.; O’Brien, T.R.; Pfeiffer, R.M. Impact of classification of hilar cholangiocarcinomas (Klatskin tumors) on the incidence of intra-and extrahepatic cholangiocarcinoma in the United States. J. Natl. Cancer Inst. 2006, 98, 873–875. [Google Scholar] [CrossRef] [PubMed]
- Blechacz, B.; Komuta, M.; Roskams, T.; Gores, G.J. Clinical diagnosis and staging of cholangiocarcinoma. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 512–522. [Google Scholar] [CrossRef] [PubMed]
- DeOliveira, M.L.; Cunningham, S.C.; Cameron, J.L.; Kamangar, F.; Winter, J.M.; Lillemoe, K.D.; Choti, M.A.; Yeo, C.J.; Schulick, R.D. Cholangiocarcinoma: Thirty-one-year experience with 564 patients at a single institution. Ann. Surg. 2007, 245, 755. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, S.; Gores, G.J. Pathogenesis, Diagnosis, and Management of Cholangiocarcinoma. Gastroenterology 2013, 145, 1215–1229. [Google Scholar] [CrossRef]
- Razumilava, N.; Gores, G.J. Cholangiocarcinoma. Lancet 2014, 383, 2168–2179. [Google Scholar] [CrossRef]
- Petrick, J.L.; Yang, B.; Altekruse, S.F.; Van Dyke, A.L.; Koshiol, J.; Graubard, B.I.; McGlynn, K.A. Risk factors for intrahepatic and extrahepatic cholangiocarcinoma in the United States: A population-based study in SEER-Medicare. PLoS ONE 2017, 12, e0186643. [Google Scholar] [CrossRef]
- Welzel, T.M.; Mellemkjaer, L.; Gloria, G.; Sakoda, L.C.; Hsing, A.W.; Ghormli, L.E.; Olsen, J.H.; McGlynn, K.A. Risk factors for intrahepatic cholangiocarcinoma in a low-risk population: A nationwide case-control study. Int. J. Cancer 2007, 120, 638–641. [Google Scholar] [CrossRef]
- Ricci, A.D.; Rizzo, A.; Brandi, G. Immunotherapy in Biliary Tract Cancer: Worthy of a Second Look. Cancer Control 2020, 27, 1–3. [Google Scholar] [CrossRef]
- Rizvi, S.; Khan, S.A.; Hallemeier, C.L.; Kelley, R.K.; Gores, G.J. Cholangiocarcinoma-evolving concepts and therapeutic strategies. Nat. Rev. Clin. Oncol. 2018, 15, 95–111. [Google Scholar] [CrossRef]
- Rizzo, A.; Ricci, A.D.; Brandi, G. Recent advances of immunotherapy for biliary tract cancer. Expert Rev. Gastroenterol. Hepatol. 2021, 15, 527–536. [Google Scholar] [CrossRef]
- Blechacz, B.; Gores, G.J. Tumors of the bile ducts, gallbladder, and ampulla. In Sleisenger and Fordtran’s Gastrointestinal and Liver Disease; Elsevier: Amsterdam, The Netherlands, 2010; pp. 1171–1184. [Google Scholar]
- Ebata, N.; Fujita, M.; Sasagawa, S.; Maejima, K.; Okawa, Y.; Hatanaka, Y.; Mitsuhashi, T.; Oosawa-Tatsuguchi, A.; Tanaka, H.; Miyano, S.; et al. Molecular Classification and Tumor Microenvironment Characterization of Gallbladder Cancer by Comprehensive Genomic and Transcriptomic Analysis. Cancers 2021, 13, 733. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, H.; Li, H. Identification of Gene Expression Pattern Related to Breast Cancer Survival Using Integrated TCGA Datasets and Genomic Tools. Biomed. Res. Int. 2015, 2015, 878546. [Google Scholar] [CrossRef]
- Lee, C.-C.; Liu, F.-L.; Chen, C.-L.; Chen, T.-C.; Liu, F.-C.; Ali, A.A.A.; Chang, D.-M.; Huang, H.-S. Novel inhibitors of RANKL-induced osteoclastogenesis: Design, synthesis, and biological evaluation of 6-(2, 4-difluorophenyl)-3-phenyl-2H-benzo [e][1,3] oxazine-2, 4 (3H)-diones. Bioorgan. Med. Chem. 2015, 23, 4522–4532. [Google Scholar] [CrossRef]
- Lee, C.-C.; Liu, F.-L.; Chen, C.-L.; Chen, T.-C.; Chang, D.-M.; Huang, H.-S. Discovery of 5-(2′,4′-difluorophenyl)-salicylanilides as new inhibitors of receptor activator of NF-κB ligand (RANKL)-induced osteoclastogenesis. Eur. J. Med. Chem. 2015, 98, 115–126. [Google Scholar] [CrossRef]
- Chen, T.C.; Wu, C.L.; Lee, C.C.; Chen, C.L.; Yu, D.S.; Huang, H.S. Structure-based hybridization, synthesis and biological evaluation of novel tetracyclic heterocyclic azathioxanthone analogues as potential antitumor agents. Eur. J. Med. Chem. 2015, 103, 615–627. [Google Scholar] [CrossRef]
- Chen, T.-C.; Yu, D.-S.; Chen, S.-J.; Chen, C.-L.; Lee, C.-C.; Hsieh, Y.-Y.; Chang, L.-C.; Guh, J.-H.; Lin, J.-J.; Huang, H.-S. Design, synthesis and biological evaluation of tetracyclic azafluorenone derivatives with topoisomerase I inhibitory properties as potential anticancer agents. Arab. J. Chem. 2019, 12, 4348–4364. [Google Scholar] [CrossRef]
- Ali, A.A.A.; Lee, Y.-R.; Wu, A.T.H.; Yadav, V.K.; Yu, D.-S.; Huang, H.-S. Structure-based strategies for synthesis, lead optimization and biological evaluation of N-substituted anthra[1,2-c][1,2,5]thiadiazole-6,11-dione derivatives as potential multi-target anticancer agents. Arab. J. Chem. 2021, 14, 102884. [Google Scholar] [CrossRef]
- Huang, H.S.; Chen, T.C.; Chen, R.H.; Huang, K.F.; Huang, F.C.; Jhan, J.R.; Chen, C.L.; Lee, C.C.; Lo, Y.; Lin, J.J. Synthesis, cytotoxicity and human telomerase inhibition activities of a series of 1,2-heteroannelated anthraquinones and anthra[1,2-d]imidazole-6,11-dione homologues. Bioorgan. Med. Chem. 2009, 17, 7418–7428. [Google Scholar] [CrossRef]
- Huang, H.-S.; Chiu, H.-F.; Lee, A.-L.; Guo, C.-L.; Yuan, C.-L. Synthesis and structure–activity correlations of the cytotoxic bifunctional 1,4-diamidoanthraquinone derivatives. Bioorg. Med. Chem. 2004, 12, 6163–6170. [Google Scholar] [CrossRef]
- Lee, J.C.; Wu, A.T.H.; Chen, J.H.; Huang, W.Y.; Lawal, B.; Mokgautsi, N.; Huang, H.S.; Ho, C.L. HNC0014, a Multi-Targeted Small-Molecule, Inhibits Head and Neck Squamous Cell Carcinoma by Suppressing c-Met/STAT3/CD44/PD-L1 Oncoimmune Signature and Eliciting Antitumor Immune Responses. Cancers 2020, 12, 3759. [Google Scholar] [CrossRef]
- Yadav, V.K.; Huang, Y.-J.; George, T.A.; Wei, P.-L.; Sumitra, M.R.; Ho, C.-L.; Chang, T.-H.; Wu, A.T.H.; Huang, H.-S. Preclinical Evaluation of the Novel Small-Molecule MSI-N1014 for Treating Drug-Resistant Colon Cancer via the LGR5/β-catenin/miR-142-3p Network and Reducing Cancer-Associated Fibroblast Transformation. Cancers 2020, 12, 1590. [Google Scholar] [CrossRef]
- Lawal, B.; Kuo, Y.-C.; Wu, A.T.H.; Huang, H.-S. BC-N102 suppress breast cancer tumorigenesis by interfering with cell cycle regulatory proteins and hormonal signaling, and induction of time-course arrest of cell cycle at G1/G0 phase. Int. J. Biol. Sci. 2021, 17, 3224–3238. [Google Scholar] [CrossRef]
- Wu, D.W.; Chen, T.C.; Huang, H.S.; Lee, H. TC-N19, a novel dual inhibitor of EGFR and cMET, efficiently overcomes EGFR-TKI resistance in non-small-cell lung cancer cells. Cell Death Dis 2016, 7, e2290. [Google Scholar] [CrossRef][Green Version]
- Liu, F.C.; Lu, J.W.; Chien, C.Y.; Huang, H.S.; Lee, C.C.; Lien, S.B.; Lin, L.C.; Chen, L.W.; Ho, Y.J.; Shen, M.C.; et al. Arthroprotective Effects of Cf-02 Sharing Structural Similarity with Quercetin. Int. J. Mol. Sci. 2018, 19, 1453. [Google Scholar] [CrossRef]
- Cheng, C.-P.; Huang, H.-S.; Hsu, Y.-C.; Sheu, M.-J.; Chang, D.-M. A Benzamide-linked Small Molecule NDMC101 Inhibits NFATc1 and NF-κB Activity: A Potential Osteoclastogenesis Inhibitor for Experimental Arthritis. J. Clin. Immunol. 2012, 32, 762–777. [Google Scholar] [CrossRef]
- Li, T.; Fan, J.; Wang, B.; Traugh, N.; Chen, Q.; Liu, J.S.; Li, B.; Liu, X.S. TIMER: A Web Server for Comprehensive Analysis of Tumor-Infiltrating Immune Cells. Cancer Res. 2017, 77, e108–e110. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martínez-Ledesma, E.; Martínez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Peña, J.G.; Treviño, V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- Chen, E.Y.; Tan, C.M.; Kou, Y.; Duan, Q.; Wang, Z.; Meirelles, G.V.; Clark, N.R.; Ma’ayan, A. Enrichr: Interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinform. 2013, 14, 128. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R package for comparing biological themes among gene clusters. Omics: A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Mostafavi, S.; Ray, D.; Warde-Farley, D.; Grouios, C.; Morris, Q. GeneMANIA: A real-time multiple association network integration algorithm for predicting gene function. Genome Biol. 2008, 9, S4. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-J.; Hu, F.-F.; Xia, M.-X.; Han, L.; Zhang, Q.; Guo, A.-Y. GSCALite: A web server for gene set cancer analysis. Bioinformatics 2018, 34, 3771–3772. [Google Scholar] [CrossRef]
- Fu, J.; Li, K.; Zhang, W.; Wan, C.; Zhang, J.; Jiang, P.; Liu, X.S. Large-scale public data reuse to model immunotherapy response and resistance. Genome Med. 2020, 12, 21. [Google Scholar] [CrossRef]
- Marcus D Hanwell, D.E.C.; Lonie, D.C.; Vandermeersch, T.; Zurek, E.; Hutchison, G.R. Avogadro: An advanced semantic chemical editor, visualization, and analysis platform. J. Cheminform. 2012, 4, 17. [Google Scholar] [CrossRef]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef]
- Lawal, B.; Liu, Y.-L.; Mokgautsi, N.; Khedkar, H.; Sumitra, M.R.; Wu, A.T.H.; Huang, H.-S. Pharmacoinformatics and Preclinical Studies of NSC765690 and NSC765599, Potential STAT3/CDK2/4/6 Inhibitors with Antitumor Activities against NCI60 Human Tumor Cell Lines. Biomedicines 2021, 9, 92. [Google Scholar] [CrossRef]
- Wu, S.-Y.; Lin, K.-C.; Lawal, B.; Wu, A.T.H.; Wu, C.-Z. MXD3 as an onco-immunological biomarker encompassing the tumor microenvironment, disease staging, prognoses, and therapeutic responses in multiple cancer types. Comput. Struct. Biotechnol. J. 2021, 19, 4970–4983. [Google Scholar] [CrossRef]
- Wu, A.T.H.; Lawal, B.; Wei, L.; Wen, Y.-T.; Tzeng, D.T.W.; Lo, W.-C. Multiomics Identification of Potential Targets for Alzheimer Disease and Antrocin as a Therapeutic Candidate. Pharmaceutics 2021, 13, 1555. [Google Scholar] [CrossRef]
- Lawal, B.; Lee, C.-Y.; Mokgautsi, N.; Sumitra, M.R.; Khedkar, H.; Wu, A.T.H.; Huang, H.-S. mTOR/EGFR/iNOS/MAP2K1/FGFR/TGFB1 Are Druggable Candidates for N-(2,4-Difluorophenyl)-2′,4′-Difluoro-4-Hydroxybiphenyl-3-Carboxamide (NSC765598), With Consequent Anticancer Implications. Front. Oncol. 2021, 11, 932. [Google Scholar] [CrossRef]
- Visualizer, D.S. BIOVIA, Dassault Systèmes, BIOVIA Workbook, Release 2020; BIOVIA Pipeline Pilot: San Diego, CA, USA, 2020. [Google Scholar]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef] [PubMed]
- Young, J.H.; Marcotte, E.M. Predictability of Genetic Interactions from Functional Gene Modules. G3 Genes|Genomes|Genet. 2017, 7, 617–624. [Google Scholar] [CrossRef]
- Jasial, S.; Hu, Y.; Bajorath, J.r. Assessing the growth of bioactive compounds and scaffolds over time: Implications for lead discovery and scaffold hopping. J. Chem. Inf. Model. 2016, 56, 300–307. [Google Scholar] [CrossRef] [PubMed]
- SAMPLES, I.B. Serial Review: Flavonoids and Isoflavones (Photoestrogens): Absorption, Metabolism, and Bioactivity. Free Radic. Biol. Med. 2004, 37, 1324–1350. [Google Scholar]
- Erlund, I. Review of the flavonoids quercetin, hesperetin, and naringenin. Dietary sources, bioactivities, bioavailability, and epidemiology. Nutr. Res. 2004, 24, 851–874. [Google Scholar] [CrossRef]
- Li, Y.; Li, P.-K.; Roberts, M.J.; Arend, R.C.; Samant, R.S.; Buchsbaum, D.J. Multi-targeted therapy of cancer by niclosamide: A new application for an old drug. Cancer Lett. 2014, 349, 8–14. [Google Scholar] [CrossRef]
- Kadri, H.; Lambourne, O.A.; Mehellou, Y. Niclosamide, a drug with many (re) purposes. ChemMedChem 2018, 13, 1088. [Google Scholar] [CrossRef]
- Reeves, R.; Beckerbauer, L. HMGI/Y proteins: Flexible regulators of transcription and chromatin structure. Biochim. Et Biophys. Acta (BBA) Gene Struct. Expr. 2001, 1519, 13–29. [Google Scholar] [CrossRef]
- Lawal, B.; Lin, L.-C.; Lee, J.-C.; Chen, J.-H.; Bekaii-Saab, T.S.; Wu, A.T.H.; Ho, C.-L. Multi-Omics Data Analysis of Gene Expressions and Alterations, Cancer-Associated Fibroblast and Immune Infiltrations, Reveals the Onco-Immune Prognostic Relevance of STAT3/CDK2/4/6 in Human Malignancies. Cancers 2021, 13, 954. [Google Scholar] [CrossRef]
- Tan, P.; Ye, Y.; He, L.; Xie, J.; Jing, J.; Ma, G.; Pan, H.; Han, L.; Han, W.; Zhou, Y. TRIM59 promotes breast cancer motility by suppressing p62-selective autophagic degradation of PDCD10. PLoS Biol. 2018, 16, e3000051. [Google Scholar] [CrossRef]
- Wu, K.; Mu, X.Y.; Jiang, J.T.; Tan, M.Y.; Wang, R.J.; Zhou, W.J.; Wang, X.; He, Y.Y.; Li, M.Q.; Liu, Z.H. miRNA-26a-5p and miR-26b-5p inhibit the proliferation of bladder cancer cells by regulating PDCD10. Oncol. Rep. 2018, 40, 3523–3532. [Google Scholar] [CrossRef]
- Fu, X.; Zhang, W.; Su, Y.; Lu, L.; Wang, D.; Wang, H. MicroRNA-103 suppresses tumor cell proliferation by targeting PDCD10 in prostate cancer. Prostate 2016, 76, 543–551. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Lei, H.; Zhang, S.; Peng, Y.; Fu, C.; Shu, G.; Yin, G. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics 2020, 10, 5895. [Google Scholar] [CrossRef]
- Misawa, K.; Imai, A.; Matsui, H.; Kanai, A.; Misawa, Y.; Mochizuki, D.; Mima, M.; Yamada, S.; Kurokawa, T.; Nakagawa, T.; et al. Identification of novel methylation markers in HPV-associated oropharyngeal cancer: Genome-wide discovery, tissue verification and validation testing in ctDNA. Oncogene 2020, 39, 4741–4755. [Google Scholar] [CrossRef]
- Belkina, A.C.; Denis, G.V. BET domain co-regulators in obesity, inflammation and cancer. Nat. Rev. Cancer 2012, 12, 465–477. [Google Scholar] [CrossRef]
- Michalak, M.; Warnken, U.; Schnölzer, M.; Gabius, H.-J.; Kopitz, J. Detection of malignancy-associated phosphoproteome changes in human colorectal cancer induced by cell surface binding of growth-inhibitory galectin-4. IUBMB Life 2019, 71, 364–375. [Google Scholar] [CrossRef]
- Markowski, M.C.; De Marzo, A.M.; Antonarakis, E.S. BET inhibitors in metastatic prostate cancer: Therapeutic implications and rational drug combinations. Expert Opin. Investig. Drugs 2017, 26, 1391–1397. [Google Scholar] [CrossRef]
- Król, M.; Polańska, J.; Pawłowski, K.M.; Turowski, P.; Skierski, J.; Majewska, A.; Ugorski, M.; Morty, R.E.; Motyl, T. Transcriptomic signature of cell lines isolated from canine mammary adenocarcinoma metastases to lungs. J. Appl. Genet. 2010, 51, 37–50. [Google Scholar] [CrossRef]
- Zhang, S.; Mo, Q.; Wang, X. Oncological role of HMGA2 (Review). Int. J. Oncol. 2019, 55, 775–788. [Google Scholar] [CrossRef]
- Ekanem, T.I.; Tsai, W.L.; Lin, Y.H.; Tan, W.Q.; Chang, H.Y.; Huang, T.C.; Chen, H.Y.; Lee, K.H. Identification of the Effects of Aspirin and Sulindac Sulfide on the Inhibition of HMGA2-Mediated Oncogenic Capacities in Colorectal Cancer. Molecules 2020, 25, 3826. [Google Scholar] [CrossRef]
- Lee, Y.S.; Dutta, A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007, 21, 1025–1030. [Google Scholar] [CrossRef]
- Califano, D.; Pignata, S.; Losito, N.S.; Ottaiano, A.; Greggi, S.; De Simone, V.; Cecere, S.; Aiello, C.; Esposito, F.; Fusco, A. High HMGA2 expression and high body mass index negatively affect the prognosis of patients with ovarian cancer. J. Cell Physiol. 2014, 229, 53–59. [Google Scholar] [CrossRef]
- Morishita, A.; Zaidi, M.R.; Mitoro, A.; Sankarasharma, D.; Szabolcs, M.; Okada, Y.; D’Armiento, J.; Chada, K. HMGA2 is a driver of tumor metastasis. Cancer Res. 2013, 73, 4289–4299. [Google Scholar] [CrossRef]
- Chen, J.-H.; Wu, A.T.H.; Lawal, B.; Tzeng, D.T.W.; Lee, J.-C.; Ho, C.-L.; Chao, T.-Y. Identification of Cancer Hub Gene Signatures Associated with Immune-Suppressive Tumor Microenvironment and Ovatodiolide as a Potential Cancer Immunotherapeutic Agent. Cancers 2021, 13, 3847. [Google Scholar] [CrossRef]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Zalvide, J.; Almengló, C.; Vázquez, S.; García-Colomer, M.; Sartages, M.; Pombo, C.M. Measuring the Kinase Activity of GCKIII Proteins In Vitro. Methods Mol. Biol. 2020, 2152, 437–443. [Google Scholar] [CrossRef]
- Meng, J.; Wang, J. Role of SNARE proteins in tumourigenesis and their potential as targets for novel anti-cancer therapeutics. Biochim. Et Biophys. Acta (BBA)-Rev. Cancer 2015, 1856, 1–12. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).