Sperm Cyst “Looping”: A Developmental Novelty Enabling Extreme Male Ornament Evolution
Abstract
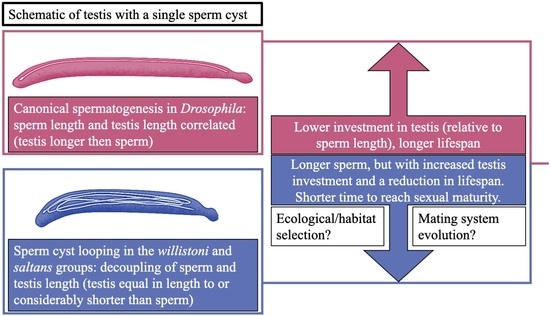
:1. Introduction
2. Materials and Methods
2.1. Experimental Organisms and Maintenance
2.2. Ultrastructure of Sperm Cysts and the Seminal Receptacle
2.3. Immunofluorescence Investigation of Sperm Cyst Development
2.4. Phylogeny
2.5. Data Acquisition
2.5.1. Sperm Cyst Looping (n = 5 Males per Species)
2.5.2. Sperm Length, Loop Length, and the Number of Loops per Cyst (n = 3–5 Males per Species)
2.5.3. Seminal Receptacle Length (n = 5 Females per Species)
2.5.4. Testis Length (n = 3–5 Males per Species)
2.5.5. Thorax Length and Gonadosomatic Index (GSI) (n = 5 Males per Species)
2.5.6. Male Post-Eclosion Maturation Time
2.5.7. Sex-Specific Lifespan (n = 80 per Sex per Species)
2.5.8. Number of Sperm Transferred per Copulation (n = 5 per Species) and Female Remating Interval (n = 25 per Species)
2.6. Statistical Analyses
3. Results
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Darwin, C. The Descent of Man, and Selection in Relation to Sex; Princeton University Press: Princeton, NJ, USA, 1981; ISBN 978-0-691-08278-3. [Google Scholar]
- Parker, G.A. How soon hath time… a history of two “seminal” publications. Cells 2021, 10, 287. [Google Scholar] [CrossRef]
- Parker, G.A. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 1970, 45, 525–567. [Google Scholar] [CrossRef]
- Eberhard, W.G. Female Control: Sexual Selection by Cryptic Female Choice; Monographs in Behavior and Ecology; Princeton University Press: Princeton, NJ, USA, 1996; ISBN 978-0-691-01085-4. [Google Scholar]
- Parker, G.A. Sexual selection and sexual conflict. Sex. Sel. Reprod. Compet. Insects 1979, 123, 166. [Google Scholar]
- Parker, G.A.; Baker, R.R.; Smith, V.G.F. The origin and evolution of gamete dimorphism and the male-female phenomenon. J. Theor. Biol. 1972, 36, 529–553. [Google Scholar] [CrossRef]
- Parker, G.A. Why are there so many tiny sperm? Sperm competition and the maintenance of two sexes. J. Theor. Biol. 1982, 96, 281–294. [Google Scholar] [CrossRef]
- Lessells, C.; Snook, R.; Hosken, D.J. The evolutionary origin and maintenance of sperm: Selection for a small, motile gamete mating type. In Sperm Biology: An Evolutionary Perspective; Birkhead, T., Hosken, D., Pitnick, S., Eds.; Academic Press: London, UK, 2009; pp. 43–67. ISBN 978-0-12-372568-4. [Google Scholar]
- Pitnick, S.; Hosken, D.J.; Birkhead, T.R. Sperm morphological diversity. In Sperm Biology: An Evolutionary Perspective; Birkhead, T., Hosken, D., Pitnick, S., Eds.; Academic Press: London, UK, 2009; pp. 69–149. ISBN 978-0-12-372568-4. [Google Scholar]
- Simmons, L.W. Sperm Competition and Its Evolutionary Consequences in the Insects; Princeton University Press: Princeton, NJ, USA, 2019; ISBN 978-0-69-120703-2. [Google Scholar]
- Fitzpatrick, J.L.; Lüpold, S. Sexual selection and the evolution of sperm quality. Mol. Hum. Reprod. 2014, 20, 1180–1189. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitnick, S.; Wolfner, M.F.; Suarez, S.S. Ejaculate–female and sperm–female interactions. In Sperm Biology: An Evolutionary Perspective; Birkhead, T., Hosken, D., Pitnick, S., Eds.; Academic Press: London, UK, 2009; pp. 247–304. ISBN 978-0-12-372568-4. [Google Scholar]
- Chapman, T. The soup in my fly: Evolution, form, and function of seminal fluid proteins. PLoS Biol. 2008, 6, e179. [Google Scholar] [CrossRef]
- Avila, F.W.; Sirot, L.K.; LaFlamme, B.A.; Rubinstein, C.D.; Wolfner, M.F. Insect seminal fluid proteins: Identification and function. Annu. Rev. Entomol. 2011, 56, 21–40. [Google Scholar] [CrossRef] [Green Version]
- Lüpold, S.; Pitnick, S. Sperm form and function: What do we know about the role of sexual selection? Reproduction 2018, 155, R229–R243. [Google Scholar] [CrossRef]
- Kahrl, A.F.; Snook, R.R.; Fitzpatrick, J.L. Fertilization mode drives sperm length evolution across the animal tree of life. Nat. Ecol. Evol. 2021, 5, 1153–1164. [Google Scholar] [CrossRef]
- Higginson, D.M.; Miller, K.B.; Segraves, K.A.; Pitnick, S. Female reproductive tract form drives the evolution of complex sperm morphology. Proc. Natl. Acad. Sci. USA 2012, 109, 4538–4543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miller, G.T.; Pitnick, S. Sperm-female coevolution in Drosophila. Science 2002, 298, 1230–1233. [Google Scholar] [CrossRef] [PubMed]
- Pitnick, S.; Spicer, G.S.; Markow, T.A. How long is a giant sperm? Nature 1995, 375, 109. [Google Scholar] [CrossRef]
- Pitnick, S.; Marrow, T.; Spicer, G.S. Evolution of multiple kinds of female sperm-storage organs in Drosophila. Evolution 1999, 53, 1804–1822. [Google Scholar] [CrossRef]
- Lüpold, S.; Manier, M.K.; Puniamoorthy, N.; Schoff, C.; Starmer, W.T.; Luepold, S.H.B.; Belote, J.M.; Pitnick, S. How sexual selection can drive the evolution of costly sperm ornamentation. Nature 2016, 533, 535–538. [Google Scholar] [CrossRef] [Green Version]
- Pattarini, J.M.; Starmer, W.T.; Bjork, A.; Pitnick, S. Mechanisms underlying the sperm quality advantage in Drosophila melanogaster. Evolution 2006, 60, 2064–2080. [Google Scholar] [CrossRef]
- Lüpold, S.; Manier, M.K.; Berben, K.S.; Smith, K.J.; Daley, B.D.; Buckley, S.H.; Belote, J.M.; Pitnick, S. How multivariate ejaculate traits determine competitive fertilization success in Drosophila melanogaster. Curr. Biol. 2012, 22, 1667–1672. [Google Scholar] [CrossRef] [Green Version]
- Lüpold, S.; Reil, J.B.; Manier, M.K.; Zeender, V.; Belote, J.M.; Pitnick, S. How female × male and male × male interactions influence competitive fertilization in Drosophila melanogaster. Evol. Lett. 2020, 4, 416–429. [Google Scholar] [CrossRef]
- Pitnick, S.; Markow, T.A.; Spicer, G.S. Delayed male maturity is a cost of producing large sperm in Drosophila. Proc. Natl. Acad. Sci. USA 1995, 92, 10614–10618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lüpold, S.; Pitnick, S.; Berben, K.S.; Blengini, C.S.; Belote, J.M.; Manier, M.K. Female mediation of competitive fertilization success in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2013, 110, 10693–10698. [Google Scholar] [CrossRef] [Green Version]
- Fisher, R.A. The Genetical Theory of Natural Selection; The Clarendon Press: Oxford, UK, 1930; ISBN 978-1-176-62502-0. [Google Scholar]
- Kokko, H.; Brooks, R.; McNamara, J.M.; Houston, A.I. The sexual selection continuum. Proc. R. Soc. Lond. B 2002, 269, 1331–1340. [Google Scholar] [CrossRef]
- Henshaw, J.M.; Jones, A.G. Fisher’s lost model of runaway sexual selection. Evolution 2020, 74, 487–494. [Google Scholar] [CrossRef]
- Amitin, E.G.; Pitnick, S. Influence of developmental environment on male- and female-mediated sperm precedence in Drosophila melanogaster. J. Evol. Biol. 2007, 20, 381–391. [Google Scholar] [CrossRef]
- Miller, G.T.; Pitnick, S. Functional significance of seminal receptacle length in Drosophila melanogaster. J. Evol. Biol. 2003, 16, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Pitnick, S. Operational sex ratios and sperm limitation in populations of Drosophila pachea. Behav. Ecol. Sociobiol. 1993, 33, 383–391. [Google Scholar] [CrossRef]
- Pitnick, S.; Markow, T.A. Large-male advantages associated with costs of sperm production in Drosophila hydei, a species with giant sperm. Proc. Natl. Acad. Sci. USA 1994, 91, 9277–9281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pitnick, S.; Markow, T.A. Male gametic strategies: Sperm size, testes size, and the allocation of ejaculate among successive mates by the sperm-limited fly Drosophila pachea and its relatives. Am. Nat. 1994, 143, 785–819. [Google Scholar] [CrossRef]
- Pitnick, S. Investment in testes and the cost of making long sperm in Drosophila. Am. Nat. 1996, 148, 57–80. [Google Scholar] [CrossRef]
- Pitnick, S.; Miller, G.T.; Reagan, J.; Holland, B. Males’ evolutionary responses to experimental removal of sexual selection. Proc. R. Soc. Lond. B 2001, 268, 1071–1080. [Google Scholar] [CrossRef] [Green Version]
- Immler, S.; Pitnick, S.; Parker, G.A.; Durrant, K.L.; Lupold, S.; Calhim, S.; Birkhead, T.R. Resolving variation in the reproductive tradeoff between sperm size and number. Proc. Natl. Acad. Sci. USA 2011, 108, 5325–5330. [Google Scholar] [CrossRef] [Green Version]
- White-Cooper, H.; Doggett, K.; Ellis, R.E. The evolution of spermatogenesis. In Sperm Biology: An Evolutionary Perspective; Birkhead, T., Hosken, D., Pitnick, S., Eds.; Academic Press: London, UK, 2009; pp. 151–183. ISBN 978-0-12-372568-4. [Google Scholar]
- Fisher, H.S.; Jacobs-Palmer, E.; Lassance, J.-M.; Hoekstra, H.E. The genetic basis and fitness consequences of sperm midpiece size in deer mice. Nat. Commun. 2016, 7, 13652. [Google Scholar] [CrossRef] [Green Version]
- Gimond, C.; Vielle, A.; Silva-Soares, N.; Zdraljevic, S.; McGrath, P.T.; Andersen, E.C.; Braendle, C. Natural variation and genetic determinants of Caenorhabditis elegans sperm size. Genetics 2019, 213, 615–632. [Google Scholar] [CrossRef]
- Hime, G.; Brill, J.A.; Fuller, M. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 1996, 109, 2779–2788. [Google Scholar] [CrossRef]
- Casal, J.; González, C.; Ripoll, P. Spindles and centrosomes during male meiosis in Drosophila melanogaster. Eur. J. Cell Biol. 1990, 51, 38–44. [Google Scholar] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Grady, P.M.; DeSalle, R. Phylogeny of the genus Drosophila. Genetics 2018, 209, 1–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stamatakis, A. RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E.; Schliep, K. Ape 5.0: An Environment for modern phylogenetics and evolutionary analyses in R. Bioinformatics 2019, 35, 526–528. [Google Scholar] [CrossRef]
- Sanderson, M.J. Estimating absolute rates of molecular evolution and divergence times: A penalized likelihood approach. Mol. Biol. Evol. 2002, 19, 101–109. [Google Scholar] [CrossRef] [Green Version]
- Paradis, E. Molecular dating of phylogenies by likelihood methods: A comparison of models and a new information criterion. Mol. Phylogenet. Evol. 2013, 67, 436–444. [Google Scholar] [CrossRef] [Green Version]
- Russo, C.; Takezaki, N.; Nei, M. Molecular phylogeny and divergence times of drosophilid species. Mol. Biol. Evol. 1995, 12, 391–404. [Google Scholar] [CrossRef] [Green Version]
- Morales-Hojas, R.; Vieira, J. Phylogenetic patterns of geographical and ecological diversification in the subgenus Drosophila. PLoS ONE 2012, 7, e49552. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.yumpu.com/en/document/view/6853895/r-a-language-and-environment-for-statistical-computing (accessed on 31 May 2010).
- Orme, D.; Freckleton, R.; Thomas, G.; Petzoldt, T.; Fritz, S.; Isaac, N.; Pearse, W. The Caper package: Comparative analysis of phylogenetics and evolution in R. R Package Version 2013, 5, 1–36. [Google Scholar]
- Russo, C.A.; Mello, B.; Frazão, A.; Voloch, C.M. Phylogenetic analysis and a time tree for a large drosophilid data set (Diptera: Drosophilidae). Zool J. Linnean Soc. 2013, 169, 765–775. [Google Scholar] [CrossRef]
- Markow, T.A.; O’Grady, P.M. Drosophila: A Guide to Species Identification and Use; Elsevier: Amsterdam, The Netherlands, 2006; ISBN 978-0-12-473052-6. [Google Scholar]
- Nonidez, J.F. The internal phenomena of reproduction in Drosophila. Biol. Bull. 1920, 39, 207–230. [Google Scholar] [CrossRef] [Green Version]
- Parker, G.A.; Pizzari, T. Sperm competition and ejaculate economics. Biol. Rev. 2010, 85, 897–934. [Google Scholar] [CrossRef] [PubMed]
- Klingenberg, C.P. Heterochrony and allometry: The analysis of evolutionary change in ontogeny. Biol. Rev. Camb. Philos. Soc. 1998, 73, 79–123. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.K. Time’s arrow: Heterochrony and the evolution of development. Int. J. Dev. Biol. 2003, 47, 613–621. [Google Scholar] [CrossRef]
- Noguchi, T.; Koizumi, M.; Hayashi, S. Sustained elongation of sperm tail promoted by local remodeling of giant mitochondria in Drosophila. Curr. Biol. 2011, 21, 805–814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjork, A.; Pitnick, S. Intensity of sexual selection along the anisogamy–isogamy continuum. Nature 2006, 441, 742–745. [Google Scholar] [CrossRef]
- Pitnick, S.; Miller, G.T.; Schneider, K.; Markow, T.A. Ejaculate-Female coevolution in Drosophila mojavensis. Proc. R. Soc. Lond. B 2003, 270, 1507–1512. [Google Scholar] [CrossRef] [Green Version]
- Orr, T.J.; Brennan, P.L.R. Sperm storage: Distinguishing selective processes and evaluating criteria. Trends. Ecol. Evol. 2015, 30, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Roff, D.A. The Evolution of Life Histories: Theory and Analysis; Chapman & Hall: New York, NY, USA, 1992; ISBN 978-0-412-02381-1. [Google Scholar]
- Stearns, S.C. The Evolution of Life Histories; Oxford University Press: Oxford, UK, 1992; ISBN 978-0-19-857741-6. [Google Scholar]
- Charlesworth, B. Evolution in Age-Structured Populations, 2nd ed.; Cambridge University Press: Cambridge, UK, 1994; ISBN 978-0-521-45967-9. [Google Scholar]
- Lemaître, J.-F.; Ronget, V.; Tidière, M.; Allainé, D.; Berger, V.; Cohas, A.; Colchero, F.; Conde, D.A.; Garratt, M.; Liker, A.; et al. Sex differences in adult lifespan and aging rates of mortality across wild mammals. Proc. Natl. Acad. Sci. USA 2020, 117, 8546–8553. [Google Scholar] [CrossRef] [PubMed]
- Paoli, F.; Roversi, P.F.; Mercati, D.; Marziali, L.; Cocco, A.; Dallai, R. The ultrastructure of spermiogenesis in four species of Coccoidea (Insecta, Homoptera). Zool. Anz. 2015, 258, 69–81. [Google Scholar] [CrossRef]
- Schumacher, J.; Hooton, D. Population genetics of two neotropical Drosophila saltans group species. Drosoph. Info. Serv. 2010, 93, 69. [Google Scholar]
- Bicudo, H. Reproductive isolation in Drosophila prosaltans (Saltans Group). Braz. J. Genet. 1978, 1, 11–27. [Google Scholar]
- O’Grady, P.M.; Clark, J.B.; Kidwell, M.G. Phylogeny of the Drosophila saltans species group based on combined analysis of nuclear and mitochondrial DNA sequences. Mol. Biol. Evol. 1998, 15, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Döge, J.S.; Gottschalk, M.S.; De Toni, D.C.; Bizzo, L.; Oliveira, S.C.; Valente, V.L.; Hofmann, P.R. New records of six species of subgenus Sophophora (Drosophila, Drosophilidae) collected in Brazil. Zootaxa 2004, 675, 1–6. [Google Scholar] [CrossRef]
- Tidon, R. Relationships between drosophilids (Diptera, Drosophilidae) and the environment in two contrasting tropical vegetations. Biol. J. Linn. Soc. Lond. 2006, 87, 233–247. [Google Scholar] [CrossRef] [Green Version]
- Emlen, S.; Oring, L. Ecology, sexual selection, and the evolution of mating systems. Science 1977, 197, 215–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valadão, H.; Proença, C.E.B.; Kuhlmann, M.P.; Harris, S.A.; Tidon, R. Fruit-breeding drosophilids (Diptera) in the Neotropics: Playing the field and specialising in generalism? Ecol. Entomol. 2019, 44, 721–737. [Google Scholar] [CrossRef] [Green Version]
- Da Mata, R.A.; Valadão, H.; Tidon, R. Spatial and temporal dynamics of drosophilid larval assemblages associated to fruits. Rev. Bras. Entomol. 2015, 59, 50–57. [Google Scholar] [CrossRef] [Green Version]
- Roque, F.; Hay, J.D.V.; Tidon, R. Breeding sites of drosophilids (Diptera) in the Brazilian Savanna. I. Fallen Fruits of Emmotum nitens (Icacinaceae), Hancornia speciosa (Apocynaceae) and Anacardium humile (Anacardiaceae). Rev. Bras. Entomol. 2009, 53, 308–313. [Google Scholar] [CrossRef]
- Pipkin, S.B. The influence of adult and larval food habits on population size of Neotropical ground-feeding Drosophila. Am. Midl. Nat. 1965, 74, 1–27. [Google Scholar] [CrossRef]
- Patterson, J.T.; Stone, W.S. Evolution in the Genus Drosophila; The Macmillan Company: New York, NY, USA, 1954; Volume 38, p. 315. [Google Scholar] [CrossRef]
- Markow, T.A.; O’Grady, P.M. Evolutionary genetics of reproductive behavior in Drosophila: Connecting the dots. Annu. Rev. Genet. 2005, 39, 263–291. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Keller, L.; Reeve, H. Why do females mate with multiple males? The Sexually Selected Sperm Hypothesis. In Advances in The Study of Behavior; Academic Press: San Deigo, CA, USA, 1995; Volume 24, pp. 291–315. [Google Scholar] [CrossRef]








| Species | SR Length (mm) | Sperm Length (mm) | Testis Length (mm) | Loop no. | GSI (%) | Maturity (Days) | RI50 (Days) * | Sperm per Copulation | Female Median Lifespan (Days) | Male Median Lifespan (Days) |
|---|---|---|---|---|---|---|---|---|---|---|
| D equinoxialis | 12.60 ± 0.05 | 4.70 ± 0.42 | 1.57 ± 0.02 | 8 | 9.35 | 0.5 | 14 | 128 ± 6 | 39 | 32 |
| D nebulosa | 11.45 ± 0.27 | 6.05 ± 0.42 | 1.54 ± 0.01 | 7 | 6.20 | 0.5 | 21 | 118 ± 9 | 42 | 28 |
| D paulistorum | 10.32 ± 0.29 | 8.81 ± 0.53 | 1.68 ± 0.01 | 9 | 9.23 | 0.5 | 21 | 141 ± 10 | 42 | 32 |
| D sucinea | 9.78 ± 0.10 | 5.50 ± 0.29 | 1.77 ± 0.01 | 8 | 8.09 | 0.5 | 21 | 87 ± 9 | 35 | 32 |
| D tropicalis | 2.87 ± 0.04 | 1.56 ± 0.20 | 1.57 ± 0.01 | 4 | 5.66 | 0.5 | 21 | 100 ± 10 | 51 | 32 |
| D willistoni | 8.87 ± 0.12 | 6.87 ± 0.32 | 1.75 ± 0.02 | 12 | 9.12 | 0.5 | 14 | 114 ± 9 | 28 | 21 |
| D insularis | 6.80 ± 0.27 | 5.96 ± 0.39 | 1.86 ± 0.06 | 4 | 10.23 | 1.5 | 21 | 97 ± 10 | 42 | 39 |
| D saltans | 13.35 ± 0.17 | 8.96 ± 0.45 | 4.10 ± 0.08 | 7 | 5.83 | 3 | 21 | 73 ± 8 | 59 | 51 |
| D austrosaltans | 13.66 ± 0.22 | 9.96 ± 0.33 | 3.91 ± 0.09 | 9 | 4.26 | 5 | 7 | 60 ± 8 | 39 | 38 |
| D prosaltans | 12.24 ± 0.05 | 8.24 ± 0.28 | 3.90 ± 0.05 | 8 | 7.02 | 5 | 10 | 49 ± 8 | 51 | 44 |
| D sturtevanti | 35.60 ± 1.12 | 19.80 ± 0.45 | 8.24 ± 0.09 | 3 | 7.45 | 9 | 2 | 78 ± 8 | 36 | 33 |
| D emarginata | 29.43 ± 2.05 | 10.60 ± 0.72 | 6.69 ± 0.02 | NA | 6.12 | 7 | NA | NA | NA | NA |
| D americana | 10.54 ± 0.53 | 5.22 ± 0.02 | 7.02 ± 0.34 | NA | 6.50 | 6 | 0.5 | NA | 77 | 66 |
| D ananassae | 4.55 ± 0.23 | 2.16 ± 0.02 | 2.61 ± 0.11 | NA | 1.86 | 4 | 21 | NA | 43 | 40 |
| D biarmipes | 3.08 ± 0.06 | 1.91 ± 0.03 | 2.41 ± 0.05 | NA | 1.47 | 2 | M | NA | 42 | 53 |
| D bipectinata | 4.28 ± 0.07 | 1.75 ± 0.02 | 2.14 ± 0.07 | NA | 2.52 | 2 | 14 | 126 ± 8 | 36 | 36 |
| D borealis | 13.78 ± 0.17 | 7.54 ± 0.05 | 8.05 ± 0.31 | NA | 8.96 | 9 | 1 | NA | 49 | 45 |
| D immigrans | 3.78 ± 0.05 | 1.70 ± 0.11 | 2.62 ± 0.19 | NA | 5.28 | 3 | M | NA | 51 | 54 |
| D lummei | 11.12 ± 0.42 | 7.79 ± 0.02 | 8.34 ± 0.13 | NA | 6.23 | 7 | 7 | NA | 42 | 42 |
| D melanogaster | 2.08 ± 0.16 | 1.85 ± 0.01 | 1.91 ± 0.01 | NA | 5.00 | 2 | 3 | 1414 ± 32 | 45 | 41 |
| D montana | 5.14 ± 0.30 | 3.34 ± 0.02 | 4.60 ± 0.08 | NA | 7.87 | 8 | 1 | NA | 72 | 68 |
| D serrata | 4.44 ± 0.21 | 3.44 ± 0.09 | 4.14 ± 0.26 | NA | 5.11 | 5 | 14 | NA | 67 | 63 |
| D siamana | 7.65 ± 0.07 | 4.42 ± 0.04 | 5.19 ± 0.13 | NA | 2.31 | 4 | 21 | NA | 58 | 54 |
| D simulans | 1.46 ± 0.04 | 1.10 ± 0.01 | 1.66 ± 0.03 | NA | 4.98 | 1 | 4 | 1340 ± 35 | 27 | 34 |
| D sulfurigaster | 8.26 ± 0.08 | 4.51 ± 0.60 | 5.14 ± 0.09 | NA | 4.36 | 5 | 14 | NA | 44 | 47 |
| D suzukii | 3.27 ± 0.23 | 1.67 ± 0.04 | 2.83 ± 0.04 | NA | 2.90 | 3 | 7 | NA | 35 | 25 |
| D virilis | 8.21 ± 0.26 | 5.70 ± 0.16 | 6.94 ± 0.30 | NA | 8.50 | 6 | 1 | NA | 73 | 73 |
| D yakuba | 1.77 ± 0.03 | 1.75 ± 0.01 | 2.31 ± 0.02 | NA | 3.47 | 3 | M | 1068 ± 56 | 59 | 59 |
| (A) Testis Mass | |||
|---|---|---|---|
| Variable | β (Slope) | SE β | Partial R2 |
| Body mass | 0.09 * | 0.03 | 0.60 |
| Sperm length | 0.008 | 0.006 | 0.44 |
| Sperm length x looping | 0.76 | ||
| Absent | 0.07 | 0.01 | |
| Partial | 0.09 | 0.02 | |
| Complete | 0.11 * | 0.02 | |
| Adjusted R-squared: 0.50; F5,21 = 6.09; p = 0.001, λ = 1(0.93,1) | |||
| (B) Female Median Lifespan | |||
| Variable | β (slope) | SE β | Partial R2 |
| Body mass | 0.53 | 0.42 | 0.009 |
| SR length | 0.01 | 0.03 | 0.02 |
| SR length x SR looping | 0.05 | ||
| Absent | −0.02 | 0.06 | |
| Present | −0.04 | 0.07 | |
| Adjusted R-squared: 0.08; F4,22 = 1.61; p = 0.21, λ = 0(0,0.74) | |||
| (C) Male Median Lifespan | |||
| Variable | β (slope) | SE β | Partial R2 |
| Body mass | 1.82 * | 0.25 | 0.34 |
| Sperm length | 0.07 | 0.06 | 0.07 |
| Sperm length x looping | 0.75 | ||
| Absent | −0.23 | 0.14 | |
| Partial | −0.29 | 0.20 | |
| Complete | −0.45 * | 0.22 | |
| Adjusted R-squared: 0.34; F5,21 = 3.68; p = 0.015, λ = 0(0,0.43) | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Syed, Z.A.; Dallai, R.; Nasirzadeh, N.; Brill, J.A.; O’Grady, P.M.; Cong, S.; Leef, E.M.; Rice, S.; Asif, A.; Nguyen, S.; et al. Sperm Cyst “Looping”: A Developmental Novelty Enabling Extreme Male Ornament Evolution. Cells 2021, 10, 2762. https://doi.org/10.3390/cells10102762
Syed ZA, Dallai R, Nasirzadeh N, Brill JA, O’Grady PM, Cong S, Leef EM, Rice S, Asif A, Nguyen S, et al. Sperm Cyst “Looping”: A Developmental Novelty Enabling Extreme Male Ornament Evolution. Cells. 2021; 10(10):2762. https://doi.org/10.3390/cells10102762
Chicago/Turabian StyleSyed, Zeeshan A., Romano Dallai, Negar Nasirzadeh, Julie A. Brill, Patrick M. O’Grady, Siyuan Cong, Ethan M. Leef, Sarah Rice, Amaar Asif, Stephanie Nguyen, and et al. 2021. "Sperm Cyst “Looping”: A Developmental Novelty Enabling Extreme Male Ornament Evolution" Cells 10, no. 10: 2762. https://doi.org/10.3390/cells10102762
APA StyleSyed, Z. A., Dallai, R., Nasirzadeh, N., Brill, J. A., O’Grady, P. M., Cong, S., Leef, E. M., Rice, S., Asif, A., Nguyen, S., Hansen, M. M., Dorus, S., & Pitnick, S. (2021). Sperm Cyst “Looping”: A Developmental Novelty Enabling Extreme Male Ornament Evolution. Cells, 10(10), 2762. https://doi.org/10.3390/cells10102762