Origin and Isoform Specific Functions of Exchange Proteins Directly Activated by cAMP: A Phylogenetic Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Protein Sequence Mining and Alignment
2.2. Phylogenetic Tree Construction
2.3. Ancestral Sequence Reconstruction
2.4. Amino Acid Composition of EPAC Isoform Specific Sequence Motifs
3. Results
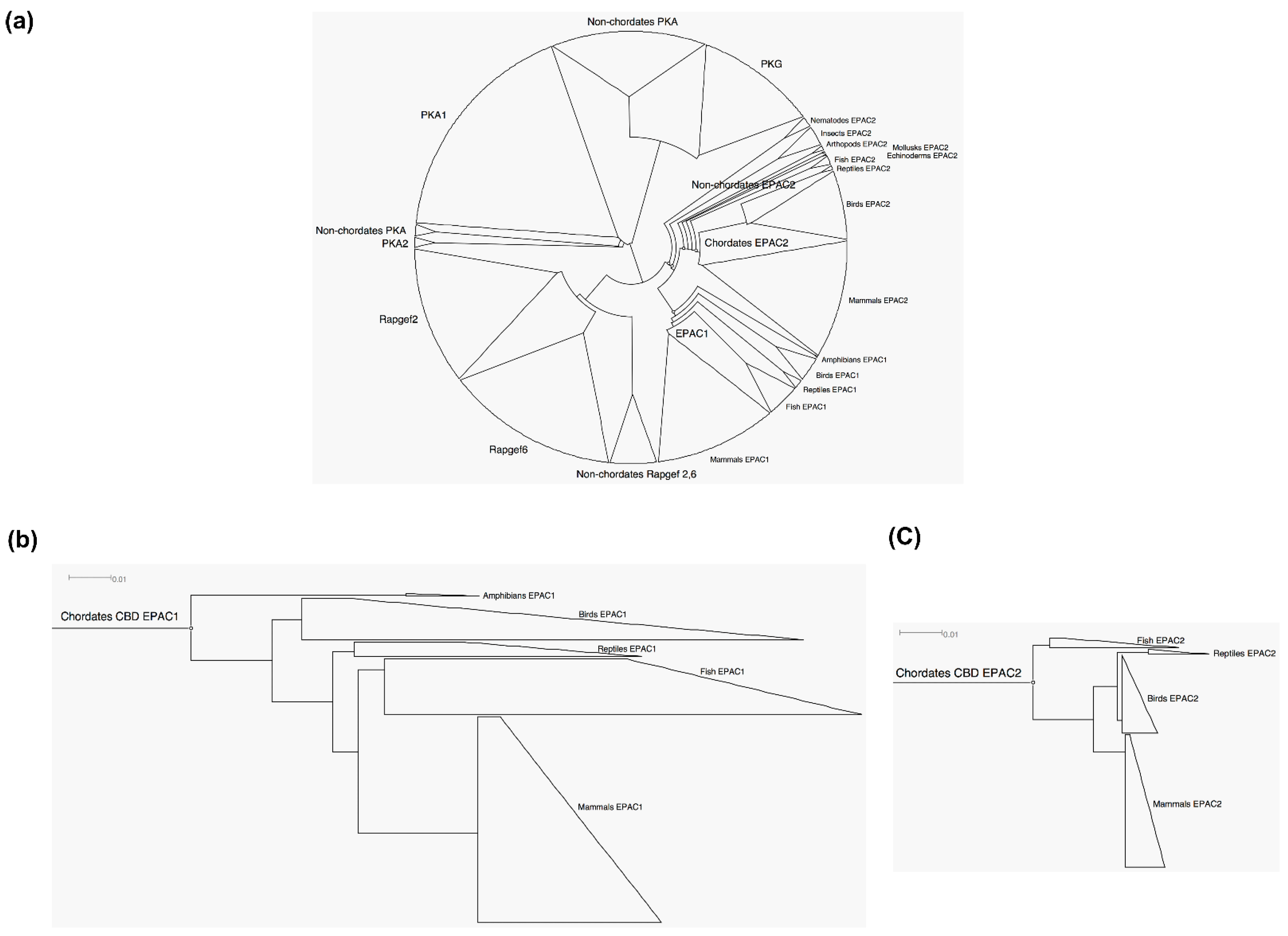
3.1. EPAC2 Is More Ancient and Conserved Than EPAC1
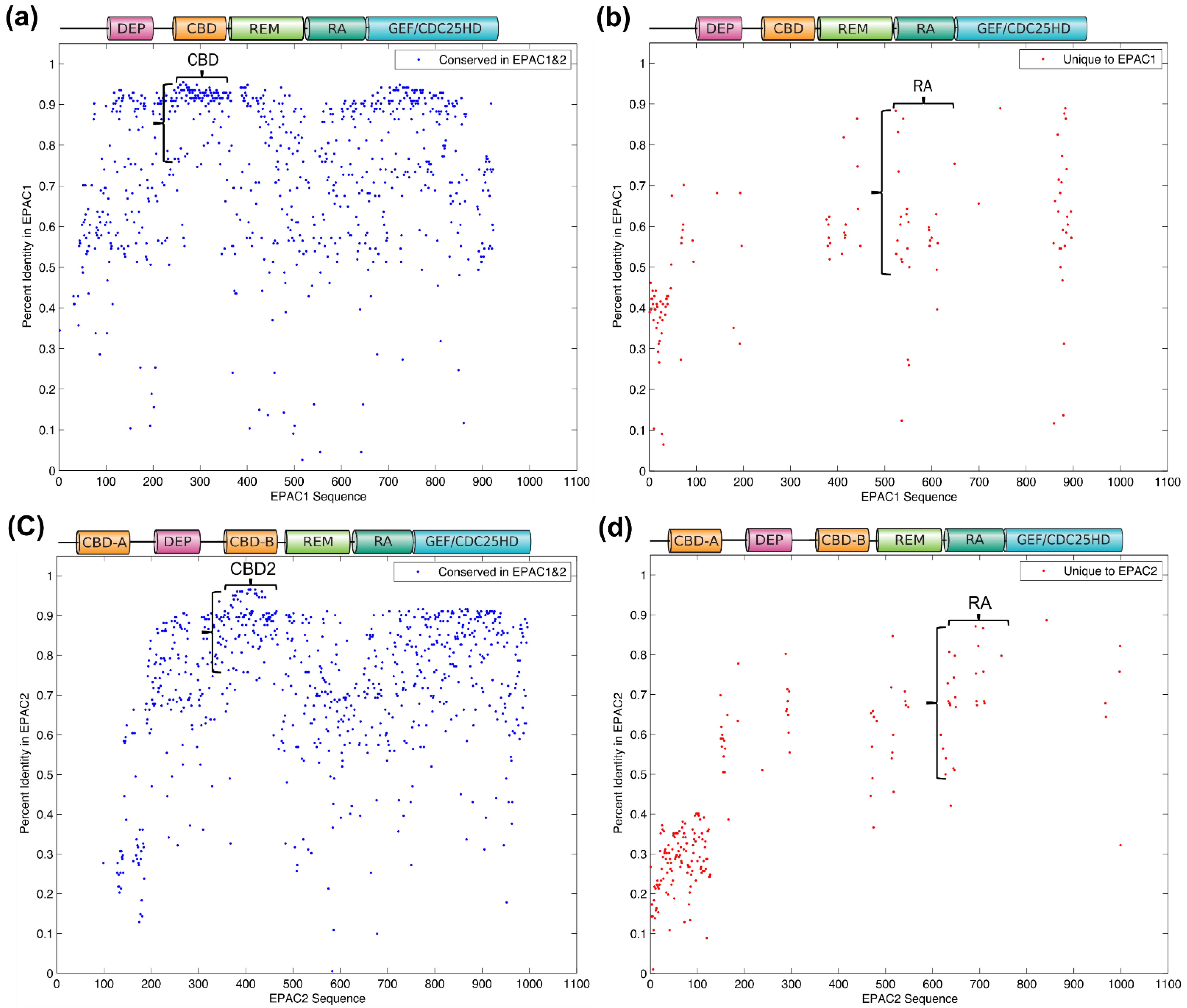
3.2. Common Ancestor and Co-Evolution of EPAC1 and EPAC2 CBD
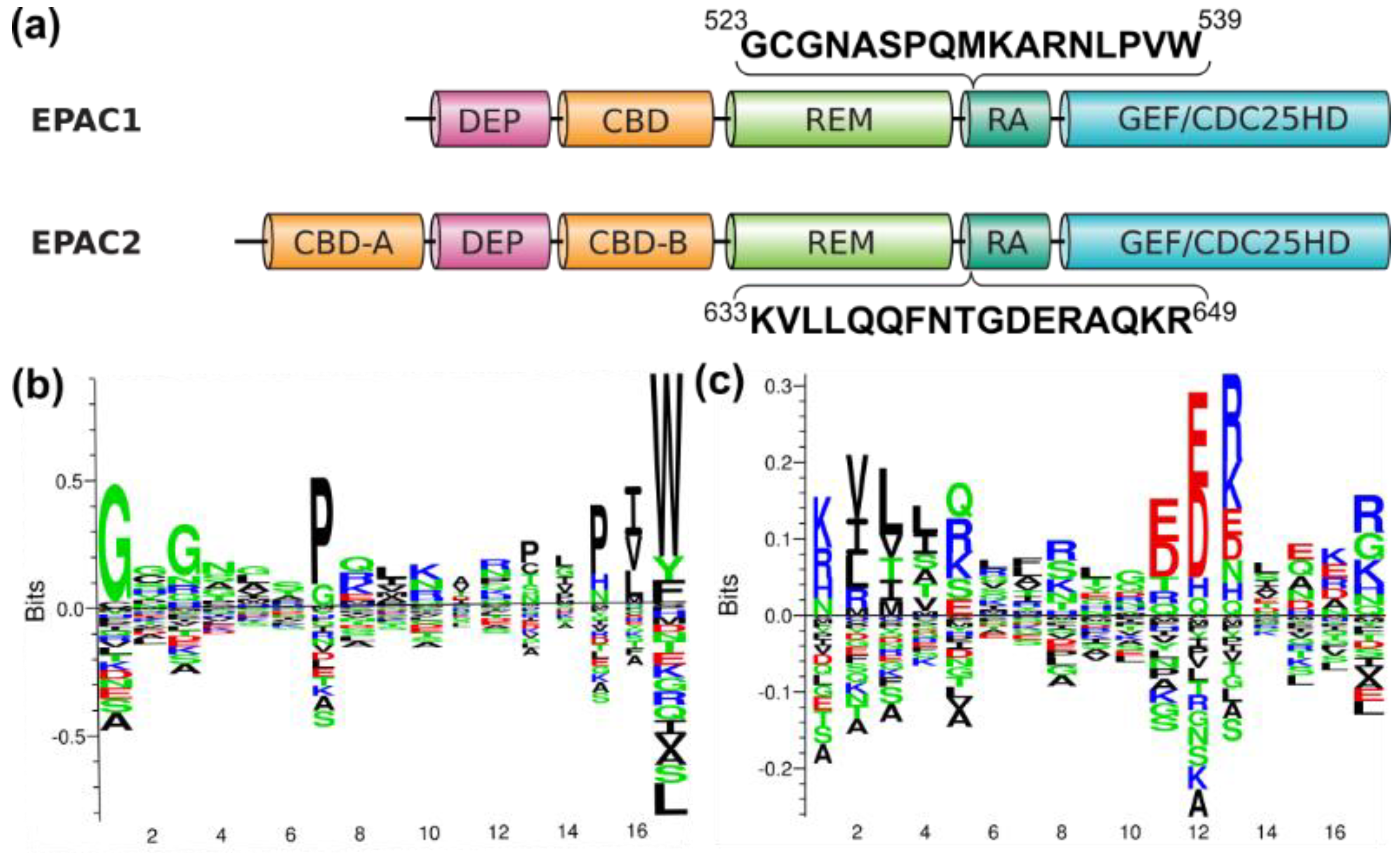
3.3. Identification of Isoform-Specific Sequence Motifs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Berman, H.M.; Eyck, L.F.T.; Goodsell, D.S.; Haste, N.M.; Kornev, A.; Taylor, S.S. The cAMP binding domain: An ancient signaling module. Proc. Natl. Acad. Sci. USA 2004, 102, 45–50. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Lee, J.C. Absolute requirement of cyclic nucleotide in the activation of the G141Q mutant cAMP receptor protein from Escherichia coli. J. Biol. Chem. 1994, 269, 30781–30784. [Google Scholar] [CrossRef]
- Cheng, X.; Ji, Z.; Tsalkova, T.; Mei, F. Epac and PKA: A tale of two intracellular cAMP receptors. Acta Biochim. Biophys. Sin. 2008, 40, 651–662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zufall, F.; Shepherd, G.M.; Barnstable, C.J. Cyclic Nucleotide Gated Channels as Regulators of Cns Development and Plasticity. Curr. Opin. Neurobiol. 1997, 7, 404–412. [Google Scholar] [CrossRef]
- Brand, T.; Poon, K.L.; Simrick, S.; Schindler, R.F.R. The Popeye Domain Containing Genes and cAMP Signaling. J. Cardiovasc. Dev. Dis. 2014, 1, 121–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krähling, A.M.; Alvarez, L.K.; Debowski, Q.; Van, M.; Gunkel, S.; Irsen, A.; Al-Amoudi, T.; Strünker, E.; Kremmer, E.; Krause, I.; et al. Cris-a Novel Camp-Binding Protein Controlling Spermiogenesis and the Development of Flagellar Bending. PLoS Genet. 2013, 9, e1003960. [Google Scholar] [CrossRef] [Green Version]
- Kannan, N.; Wu, J.; Anand, G.S.; Yooseph, S.; Neuwald, A.F.; Venter, J.C.; Taylor, S.S. Evolution of allostery in the cyclic nucleotide binding module. Genome Biol. 2007, 8, R264. [Google Scholar] [CrossRef] [Green Version]
- De Rooij, J.; Zwartkruis, F.J.T.; Verheijen, M.H.G.; Cool, R.; Nijman, S.M.B.; Wittinghofer, A.; Bos, J.L. Epac is a Rap1 guanine-nucleotide-exchange factor directly activated by cyclic AMP. Nature 1998, 396, 474–477. [Google Scholar] [CrossRef]
- Kawasaki, H.; Springett, G.M.; Mochizuki, N.; Toki, S.; Nakaya, M.; Matsuda, M.; Housman, D.E.; Graybiel, A.M. A Family of Camp-Binding Proteins That Directly Activate Rap1. Science 1998, 282, 2275–2279. [Google Scholar] [CrossRef] [Green Version]
- De Rooij, J.; Rehmann, H.; van Triest, M.; Cool, R.H.; Wittinghofer, A.; Bos, J.L. Mechanism of Regulation of the Epac Family of Camp-Dependent Rapgefs. J. Biol. Chem. 2000, 275, 20829–20836. [Google Scholar] [CrossRef] [Green Version]
- Niimura, M.; Miki, T.; Shibasaki, T.; Fujimoto, W.; Iwanaga, T.; Seino, S. Critical role of the N-terminal cyclic AMP-binding domain of Epac2 in its subcellular localization and function. J. Cell. Physiol. 2009, 219, 652–658. [Google Scholar] [CrossRef] [PubMed]
- Alenkvist, I.; Gandasi, N.; Barg, S.; Tengholm, A. Recruitment of Epac2A to Insulin Granule Docking Sites Regulates Priming for Exocytosis. Diabetes 2017, 66, 2610–2622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parnell, E.; Smith, B.O.; Yarwood, S.J. The cAMP sensors, EPAC1 and EPAC2, display distinct subcellular distributions despite sharing a common nuclear pore localisation signal. Cell. Signal. 2015, 27, 989–996. [Google Scholar] [CrossRef] [Green Version]
- Robichaux, W.G.; Cheng, X. Intracellular Camp Sensor Epac: Physiology, Pathophysiology, and Therapeutics Development. Physiol. Rev. 2018, 98, 919–1053. [Google Scholar] [CrossRef] [PubMed]
- Sugawara, K.; Shibasaki, T.; Takahashi, H.; Seino, S. Structure and Functional Roles of Epac2 (Rapgef4). Gene 2015, 575, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, M.; Dekker, F.; Maarsingh, H. Exchange Protein Directly Activated by cAMP (epac): A Multidomain cAMP Mediator in the Regulation of Diverse Biological Functions. Pharmacol. Rev. 2013, 65, 670–709. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, U.; Cheng, X. Exchange Protein Directly Activated by Camp Encoded by the Mammalian Rapgef3 Gene: Structure, Function and Therapeutics. Gene 2015, 570, 157–167. [Google Scholar] [CrossRef]
- Holz, G.G. Epac: A New Camp-Binding Protein in Support of Glucagon-Like Peptide-1 Receptor-Mediated Signal Transduction in the Pancreatic Beta-Cell. Diabetes 2004, 53, 5–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.-L.; Katoh, M.; Shibasaki, T.; Minami, K.; Sunaga, Y.; Takahashi, H.; Yokoi, N.; Iwasaki, M.; Miki, T.; Seino, S. The cAMP Sensor Epac2 Is a Direct Target of Antidiabetic Sulfonylurea Drugs. Science 2009, 325, 607–610. [Google Scholar] [CrossRef] [Green Version]
- Mukai, E.; Fujimoto, S.; Sato, H.; Oneyama, C.; Kominato, R.; Sato, Y.; Sasaki, M.; Nishi, Y.; Okada, M.; Inagaki, N. Exendin-4 Suppresses Src Activation and Reactive Oxygen Species Production in Diabetic Goto-Kakizaki Rat Islets in an Epac-Dependent Manner. Diabetes 2010, 60, 218–226. [Google Scholar] [CrossRef] [Green Version]
- Woolfrey, K.M.; Srivastava, D.P.; Photowala, H.; Yamashita, M.; Barbolina, M.V.; Cahill, M.; Xie, Z.; A Jones, K.; A Quilliam, L.; Prakriya, M.; et al. Epac2 induces synapse remodeling and depression and its disease-associated forms alter spines. Nat. Neurosci. 2009, 12, 1275–1284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penzes, P.; Woolfrey, K.M.; Srivastava, D.P. Epac2-mediated dendritic spine remodeling: Implications for disease. Mol. Cell. Neurosci. 2011, 46, 368–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Y.; Shu, X.; Liu, D.; Shang, Y.; Wu, Y.; Pei, L.; Xu, X.; Tian, Q.; Zhang, J.; Qian, K.; et al. EPAC Null Mutation Impairs Learning and Social Interactions via Aberrant Regulation of miR-124 and Zif268 Translation. Neuron 2012, 73, 774–788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, D.P.; Woolfrey, K.M.; Jones, K.A.; Anderson, C.T.; Smith, K.R.; Russell, T.A.; Lee, H.; Yasvoina, M.; Wokosin, D.L.; Ozdinler, P.H.; et al. An Autism-Associated Variant of Epac2 Reveals a Role for Ras/Epac2 Signaling in Controlling Basal Dendrite Maintenance in Mice. PLoS Biol. 2012, 10, e1001350. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, D.P.; Jones, K.A.; Woolfrey, K.M.; Burgdorf, J.; Russell, T.A.; Kalmbach, A.; Lee, H.; Yang, C.; Bradberry, M.M.; Wokosin, D.; et al. Social, Communication, and Cortical Structural Impairments in Epac2-Deficient Mice. J. Neurosci. 2012, 32, 11864–11878. [Google Scholar] [CrossRef] [Green Version]
- Mei, F.C.; Qiao, J.B.; Tsygankova, O.M.; Meinkoth, J.L.; Quilliam, L.A.; Cheng, X.D. Differential Signaling of Cyclic Amp—Opposing Effects of Exchange Protein Directly Activated by Cyclic Amp and Camp-Dependent Protein Kinase on Protein Kinase B Activation. J. Biol. Chem. 2002, 277, 11497–11504. [Google Scholar] [CrossRef] [Green Version]
- Nijholt, I.M.; Dolga, A.M.; Ostroveanu, A.; Luiten, P.G.; Schmidt, M.; Eisel, U.L. Neuronal AKAP150 coordinates PKA and Epac-mediated PKB/Akt phosphorylation. Cell. Signal. 2008, 20, 1715–1724. [Google Scholar] [CrossRef]
- Oestreich, E.A.; Wang, H.S.; Malik, K.; Kaproth-Joslin, A.; Blaxall, B.C.; Kelley, G.G.; Dirksen, R.T.; Smrcka, A.V. Epac-Mediated Activation of Phospholipase Cε Plays a Critical Role in β-Adrenergic Receptor-Dependent Enhancement of Ca2+ Mobilization in Cardiac Myocytes. J. Biol. Chem. 2007, 282, 5488–5495. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, M.; Evellin, S.; Weernink, P.A.O.; Dorp, F.V.; Rehmann, H.; Lomasney, J.W.; Jakobs, K.H. A new phospholipase-C–calcium signalling pathway mediated by cyclic AMP and a Rap GTPase. Nature 2001, 3, 1020–1024. [Google Scholar] [CrossRef]
- Yang, W.; Mei, F.C.; Cheng, X. EPAC1 regulates endothelial annexin A2 cell surface translocation and plasminogen activation. FASEB J. 2018, 32, 2212–2222. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, U.; Patel, H.H.; Lai, N.C.; Aroonsakool, N.; Roth, D.M.; Insel, P.A. The cyclic AMP effector Epac integrates pro- and anti-fibrotic signals. Proc. Natl. Acad. Sci. USA 2008, 105, 6386–6391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almahariq, M.; Mei, F.C.; Wang, H.; Cao, A.T.; Yao, S.; Soong, L.; Sun, J.; Cong, Y.; Chen, J.; Cheng, X. Exchange protein directly activated by cAMP modulates regulatory T-cell-mediated immunosuppression. Biochem. J. 2015, 465, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Williams, K.W.; Gautron, L.; Elmquist, J.K. Induction of Leptin Resistance by Activation of Camp-Epac Signaling. Cell Metab. 2011, 13, 331–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Mei, F.C.; Cheng, H.; Lao, D.H.; Hu, Y.; Wei, J.; Patrikeev, I.; Hao, D.; Stutz, S.J.; Dineley, K.T.; et al. Enhanced Leptin Sensitivity, Reduced Adiposity, and Improved Glucose Homeostasis in Mice Lacking Exchange Protein Directly Activated by Cyclic AMP Isoform 1. Mol. Cell. Biol. 2013, 33, 918–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namkoong, S.; Kim, C.-K.; Cho, Y.-L.; Kim, J.-H.; Lee, H.; Ha, K.-S.; Choe, J.; Kim, P.-H.; Won, M.-H.; Kwon, Y.-G.; et al. Forskolin increases angiogenesis through the coordinated cross-talk of PKA-dependent VEGF expression and Epac-mediated PI3K/Akt/eNOS signaling. Cell. Signal. 2009, 21, 906–915. [Google Scholar] [CrossRef]
- Jang, M.W.; Yun, S.P.; Park, J.H.; Ryu, J.M.; Lee, J.H.; Han, H.J. Cooperation of Epac1/Rap1/Akt and Pka in Prostaglandin E(2) -Induced Proliferation of Human Umbilical Cord Blood Derived Mesenchymal Stem Cells: Involvement of C-Myc and Vegf Expression. J. Cell. Physiol. 2012, 227, 3756–3767. [Google Scholar] [CrossRef]
- Liu, H.; Mei, F.C.; Yang, W.; Wang, H.; Wong, E.; Cai, J.; Toth, E.; Luo, P.; Li, Y.-M.; Zhang, W.; et al. Epac1 inhibition ameliorates pathological angiogenesis through coordinated activation of Notch and suppression of VEGF signaling. Sci. Adv. 2020, 6, eaay3566. [Google Scholar] [CrossRef] [Green Version]
- Métrich, M.; Lucas, A.; Gastineau, M.; Samuel, J.L.; Heymes, C.; Morel, E.; Lezoualc’h, F. Epac Mediates β-Adrenergic Receptor-Induced Cardiomyocyte Hypertrophy. Circ. Res. 2008, 102, 959–965. [Google Scholar] [CrossRef] [Green Version]
- Oestreich, E.A.; Malik, S.; Goonasekera, S.A.; Blaxall, B.C.; Kelley, G.G.; Dirksen, R.T.; Smrcka, A.V. Epac and Phospholipase Cepsilon Regulate Ca2+ Release in the Heart by Activation of Protein Kinase Cepsilon and Calcium-Calmodulin Kinase Ii. J. Biol. Chem. 2009, 284, 1514–1522. [Google Scholar] [CrossRef] [Green Version]
- Okumura, S.; Fujita, T.; Cai, W.; Jin, M.; Namekata, I.; Mototani, Y.; Jin, H.; Ohnuki, Y.; Tsuneoka, Y.; Kurotani, R.; et al. Epac1-dependent phospholamban phosphorylation mediates the cardiac response to stresses. J. Clin. Investig. 2014, 124, 2785–2801. [Google Scholar] [CrossRef] [Green Version]
- Pereira, L.; Cheng, H.; Lao, D.H.; Na, L.; Van Oort, R.J.; Brown, J.H.; Wehrens, X.; Chen, J.; Bers, D.M. Epac2 Mediates Cardiac β1-Adrenergic–Dependent Sarcoplasmic Reticulum Ca2+ Leak and Arrhythmia. Circulation 2013, 127, 913–922. [Google Scholar] [CrossRef] [Green Version]
- Lezoualc’h, F.; Fazal, L.; Laudette, M.; Conte, C. Cyclic AMP Sensor EPAC Proteins and Their Role in Cardiovascular Function and Disease. Circ. Res. 2016, 118, 881–897. [Google Scholar] [CrossRef]
- Cai, W.; Fujita, T.; Hidaka, Y.; Jin, H.; Suita, K.; Prajapati, R.; Liang, C.; Umemura, M.; Yokoyama, U.; Sato, M.; et al. Disruption of Epac1 protects the heart from adenylyl cyclase type 5-mediated cardiac dysfunction. Biochem. Biophys. Res. Commun. 2016, 475, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Morel, E.; Marcantoni, A.; Gastineau, M.; Birkedal, R.; Rochais, F.; Garnier, A.; Lompré, A.M.; Vandecasteele, G.; Lezoualc’h, F. Camp-Binding Protein Epac Induces Cardiomyocyte Hypertrophy. Circ. Res. 2005, 97, 1296–1304. [Google Scholar] [CrossRef]
- Almahariq, M.; Mei, F.C.; Cheng, X. Cyclic AMP sensor EPAC proteins and energy homeostasis. Trends Endocrinol. Metab. 2013, 25, 60–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Onodera, Y.; Nam, J.M.; Bissell, M.J. Increased Sugar Uptake Promotes Oncogenesis Via Epac/Rap1 and O-Glcnac Pathways. J. Clin. Investig. 2014, 124, 367–384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almahariq, M.; Mei, F.C.; Cheng, X. The pleiotropic role of exchange protein directly activated by cAMP 1 (EPAC1) in cancer: Implications for therapeutic intervention. Acta Biochim. Biophys. Sin. 2015, 48, 75–81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehbe, N.; Slika, H.; Mesmar, J.; Nasser, S.A.; Pintus, G.; Baydoun, S.; Badran, A.; Kobeissy, F.; Eid, A.H.; Baydoun, E. The Role of Epac in Cancer Progression. Int. J. Mol. Sci. 2020, 21, 6489. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Prasad, P.; Jash, E.; Saini, M.; Husain, A.; Goldman, A.; Sehrawat, S. Insights into exchange factor directly activated by cAMP (EPAC) as potential target for cancer treatment. Mol. Cell. Biochem. 2018, 447, 77–92. [Google Scholar] [CrossRef]
- Hucho, T.B.; Dina, O.A.; Levine, J.D. Epac Mediates a Camp-to-Pkc Signaling in Inflammatory Pain: An Isolectin B4(+) Neuron-Specific Mechanism. J. Neurosci. 2005, 25, 6119–6126. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eijkelkamp, N.; Wang, H.; Garza-Carbajal, A.; Willemen, H.L.; Zwartkruis, F.J.; Wood, J.N.; Dantzer, R.; Kelley, K.W.; Heijnen, C.J.; Kavelaars, A. Low Nociceptor Grk2 Prolongs Prostaglandin E2 Hyperalgesia Via Biased Camp Signaling to Epac/Rap1, Protein Kinase Cepsilon, and Mek/Erk. J. Neurosci. 2010, 30, 12806–12815. [Google Scholar] [CrossRef]
- Wang, H.; Heijnen, C.J.; van Velthoven, C.; Willemen, H.L.; Ishikawa, Y.; Zhang, X.; Sood, A.K.; Vroon, A.; Eijkelkamp, N.; Kavelaars, A. Balancing GRK2 and EPAC1 levels prevents and relieves chronic pain. J. Clin. Investig. 2013, 123, 5023–5034. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Li, G.; Chen, Y.; Mae Huang, L.Y. Epac-Pkcalpha Signaling in Purinergic P2x3r-Mediated Hyperalgesia after Inflammation. Pain 2016, 157, 1541–1550. [Google Scholar] [CrossRef]
- Gong, B.; Shelite, T.; Mei, F.C.; Ha, T.; Hu, Y.; Xu, G.; Chang, Q.; Wakamiya, M.; Ksiazek, T.G.; Boor, P.J.; et al. Exchange protein directly activated by cAMP plays a critical role in bacterial invasion during fatal rickettsioses. Proc. Natl. Acad. Sci. USA 2013, 110, 19615–19620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tao, X.; Mei, F.; Agrawal, A.; Peters, C.J.; Ksiazek, T.G.; Cheng, X.; Tseng, C.-T.K. Blocking of Exchange Proteins Directly Activated by cAMP Leads to Reduced Replication of Middle East Respiratory Syndrome Coronavirus. J. Virol. 2014, 88, 3902–3910. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Robichaux, W.G.; Wang, Z.; Mei, F.C.; Cai, M.; Du, G.; Chen, J.; Cheng, X. Inhibition of Epac1 suppresses mitochondrial fission and reduces neointima formation induced by vascular injury. Sci. Rep. 2016, 6, 36552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kato, Y.; Yokoyama, U.; Yanai, C.; Ishige, R.; Kurotaki, D.; Umemura, M.; Fujita, T.; Kubota, T.; Okumura, S.; Sata, M.; et al. Epac1 Deficiency Attenuated Vascular Smooth Muscle Cell Migration and Neointimal Formation. Arterioscler. Thromb. Vasc. Biol. 2015, 35, 2617–2625. [Google Scholar] [CrossRef] [Green Version]
- Yokoyama, U.; Minamisawa, S.; Quan, H.; Akaike, T.; Jin, M.; Otsu, K.; Ulucan, C.; Wang, X.; Baljinnyam, E.; Takaoka, M.; et al. Epac1 is upregulated during neointima formation and promotes vascular smooth muscle cell migration. Am. J. Physiol. Circ. Physiol. 2008, 295, H1547–H1555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robichaux, W.G.; Mei, F.C.; Yang, W.; Wang, H.; Sun, H.; Zhou, Z.; Milewicz, D.M.; Teng, B.B.; Cheng, X. Epac1 (Exchange Protein Directly Activated by cAMP 1) Upregulates LOX-1 (Oxidized Low-Density Lipoprotein Receptor 1) to Promote Foam Cell Formation and Atherosclerosis Development. Arterioscler. Thromb. Vasc. Biol. 2020, 40, e322–e335. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Papadopoulos, J.S.; Agarwala, R. COBALT: Constraint-based alignment tool for multiple protein sequences. Bioinformatics 2007, 23, 1073–1079. [Google Scholar] [CrossRef] [Green Version]
- Desper, R.; Gascuel, O. Fast and Accurate Phylogeny Reconstruction Algorithms Based on the Minimum-Evolution Principle. J. Comput. Biol. 2002, 9, 687–705. [Google Scholar] [CrossRef]
- Huson, D.H.; Richter, D.C.; Rausch, C.; Dezulian, T.; Franz, M.; Rupp, R. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinform. 2007, 8, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Thomsen, M.C.; Nielsen, M. Seq2logo: A Method for Construction and Visualization of Amino Acid Binding Motifs and Sequence Profiles Including Sequence Weighting, Pseudo Counts and Two-Sided Representation of Amino Acid Enrichment and Depletion. Nucleic Acids Res. 2012, 40, W281–W287. [Google Scholar] [CrossRef] [Green Version]
- Warren, W.C.; Hillier, L.W.; Graves, J.A.M.; Birney, E.; Ponting, C.P.; Grützner, F.; Belov, K.; Miller, W.; Clarke, L.; Chinwalla, A.T.; et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature 2008, 453, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Gloerich, M.; Bos, J.L. Regulating Rap small G-proteins in time and space. Trends Cell Biol. 2011, 21, 615–623. [Google Scholar] [CrossRef] [PubMed]
- De Rooij, J.; Boenink, N.M.; van Triest, M.; Cool, R.; Wittinghofer, A.; Bos, J.L. PDZ-GEF1, a Guanine Nucleotide Exchange Factor Specific for Rap1 and Rap2. J. Biol. Chem. 1999, 274, 38125–38130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuiperij, H.B.; de Rooij, J.; Rehmann, H.; van Triest, M.; Wittinghofer, A.; Bos, J.L.; Zwartkruis, F.J. Characterisation of PDZ-GEFs, a family of guanine nucleotide exchange factors specific for Rap1 and Rap2. Biochim. Biophys. Acta 2003, 1593, 141–149. [Google Scholar] [CrossRef] [Green Version]
- Evans, S.D.; Hughes, I.V.; Gehling, J.G.; Droser, M.L. Discovery of the Oldest Bilaterian from the Ediacaran of South Australia. Proc. Natl. Acad. Sci. USA 2020, 117, 7845–7850. [Google Scholar] [CrossRef]
- Qiao, J.; Mei, F.C.; Popov, V.L.; Vergara, L.A.; Cheng, X. Cell Cycle-dependent Subcellular Localization of Exchange Factor Directly Activated by cAMP. J. Biol. Chem. 2002, 277, 26581–26586. [Google Scholar] [CrossRef] [Green Version]
- HHochbaum, D.; Barila, G.; Ribeiro-Neto, F.; Altschuler, D.L. Radixin Assembles Camp Effectors Epac and Pka into a Functional Camp Compartment: Role in Camp-Dependent Cell Proliferation. J. Biol. Chem. 2001, 286, 859–866. [Google Scholar] [CrossRef] [Green Version]
- Tsalkova, T.; Mei, F.C.; Li, S.; Chepurny, O.G.; Leech, C.A.; Liu, T.; Holz, G.G.; Woods, V.L.; Cheng, X. Isoform-Specific Antagonists of Exchange Proteins Directly Activated by Camp. Proc. Natl. Acad. Sci. USA 2012, 109, 18613–18618. [Google Scholar] [CrossRef] [Green Version]
- Taylor, S.S.; Ilouz, R.; Zhang, P.; Kornev, A. Assembly of allosteric macromolecular switches: Lessons from PKA. Nat. Rev. Mol. Cell Biol. 2012, 13, 646–658. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.; Weng, X.; Hofer, F.; Martin, G.S.; Kirn, S.-H. Three-dimensional structure of the Ras-interacting domain of RalGDS. Nat. Genet. 1997, 4, 609–615. [Google Scholar] [CrossRef] [Green Version]
- Ponting, C.P.; Benjamin, D.R. A Novel Family of Ras-Binding Domains. Trends Biochem. Sci. 1996, 21, 422–425. [Google Scholar] [CrossRef]
- Rehmann, H.; Das, J.; Knipscheer, P.; Wittinghofer, A.; Bos, J.L. Structure of the cyclic-AMP-responsive exchange factor Epac2 in its auto-inhibited state. Nature 2006, 439, 625–628. [Google Scholar] [CrossRef] [PubMed]
- Rehmann, H.; Arias-Palomo, E.; Hadders, M.A.; Schwede, F.; Llorca, O.; Bos, J.L. Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B. Nature 2008, 455, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Tunyasuvunakool, K.; Adler, J.; Wu, Z.; Green, T.; Zielinski, M.; Žídek, A.; Bridgland, A.; Cowie, A.; Meyer, C.; Laydon, A.; et al. Highly accurate protein structure prediction for the human proteome. Nature 2021, 596, 590–596. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Asuri, S.; Rebhun, J.F.; Castro, A.F.; Paranavitana, N.C.; Quilliam, L.A. The RAP1 Guanine Nucleotide Exchange Factor Epac2 Couples Cyclic AMP and Ras Signals at the Plasma Membrane. J. Biol. Chem. 2006, 281, 2506–2514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Takahashi, M.; Li, Y.; Song, S.; Dillon, T.J.; Shinde, U.; Stork, P.J.S. Ras Is Required for the Cyclic AMP-Dependent Activation of Rap1 via Epac2. Mol. Cell. Biol. 2008, 28, 7109–7125. [Google Scholar] [CrossRef] [Green Version]
- Berthouze-Duquesnes, M.; Lucas, A.; Saulière, A.; Sin, Y.Y.; Laurent, A.C.; Galés, C.; Baillie, G.; Lezoualc’h, F. Specific Interactions between Epac1, β-Arrestin2 and Pde4d5 Regulate Β-Adrenergic Receptor Subtype Differential Effects on Cardiac Hypertrophic Signaling. Cell. Signal. 2013, 25, 970–980. [Google Scholar] [CrossRef]
- Liu, C.; Takahashi, M.; Li, Y.; Dillon, T.J.; Kaech, S.; Stork, P.J.S. The Interaction of Epac1 and Ran Promotes Rap1 Activation at the Nuclear Envelope. Mol. Cell. Biol. 2010, 30, 3956–3969. [Google Scholar] [CrossRef] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ni, Z.; Cheng, X. Origin and Isoform Specific Functions of Exchange Proteins Directly Activated by cAMP: A Phylogenetic Analysis. Cells 2021, 10, 2750. https://doi.org/10.3390/cells10102750
Ni Z, Cheng X. Origin and Isoform Specific Functions of Exchange Proteins Directly Activated by cAMP: A Phylogenetic Analysis. Cells. 2021; 10(10):2750. https://doi.org/10.3390/cells10102750
Chicago/Turabian StyleNi, Zhuofu, and Xiaodong Cheng. 2021. "Origin and Isoform Specific Functions of Exchange Proteins Directly Activated by cAMP: A Phylogenetic Analysis" Cells 10, no. 10: 2750. https://doi.org/10.3390/cells10102750
APA StyleNi, Z., & Cheng, X. (2021). Origin and Isoform Specific Functions of Exchange Proteins Directly Activated by cAMP: A Phylogenetic Analysis. Cells, 10(10), 2750. https://doi.org/10.3390/cells10102750





