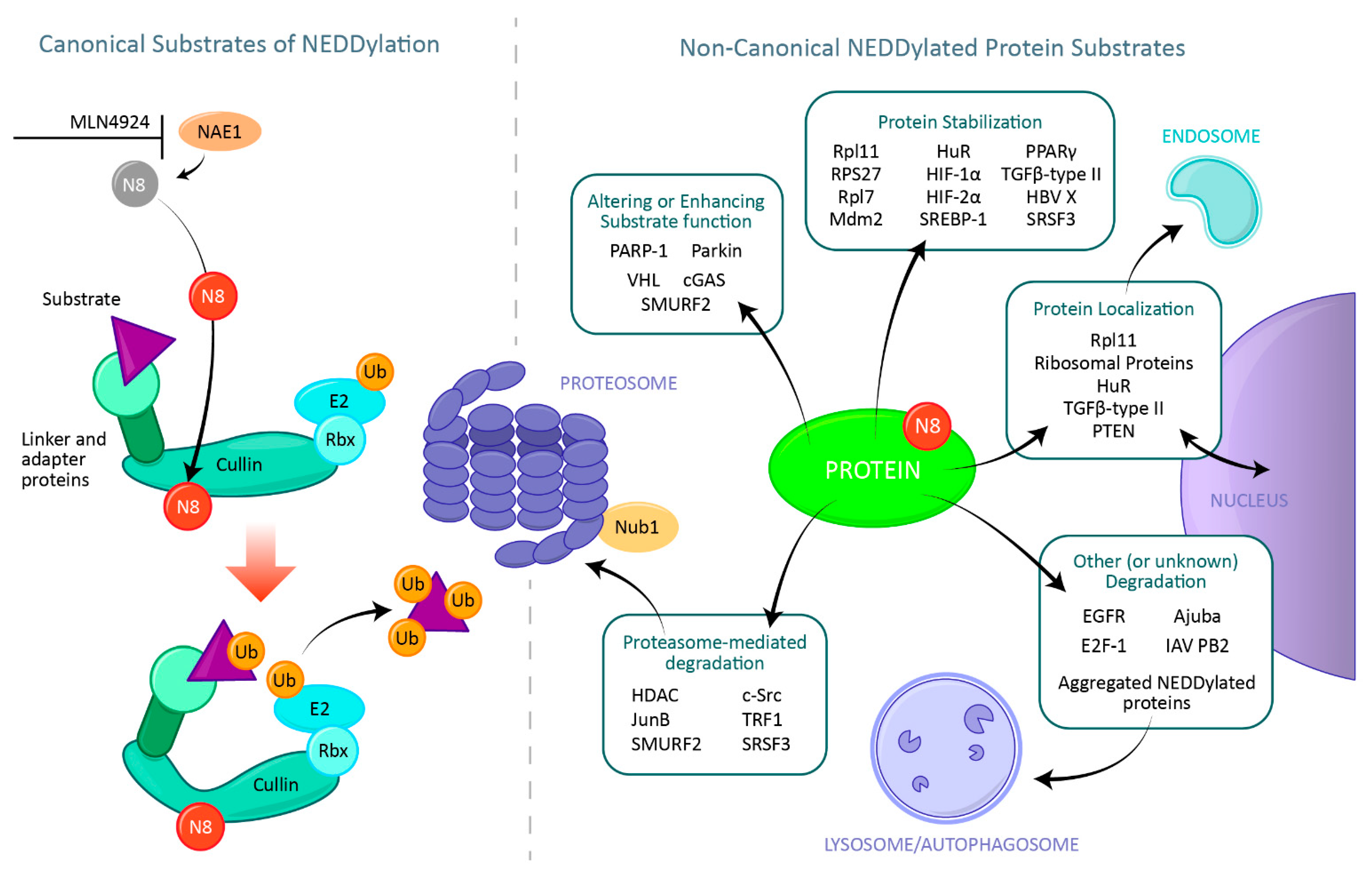
The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation
Abstract
Ha, ha, ha, thou entanglest thyself in thine own work like a silkworm-John Webster, The White Devil
1. Introduction
2. NEDD8 Modification of Cullins: The Canonical NEDD8 Function
3. Proteomic Analysis for NEDD8-Substrate Identification
4. Ribosomal Protein NEDDylation
5. Protein Degradation
6. Protein Stabilization
7. Alterations and Enhancement of a Substrates Function
8. Alterations to NEDD8 Levels in Cells under Physiological Conditions
9. NEDD8 Chains Bind to Non-Canonical Substrates
10. Conclusions and Future Perspectives
Funding
Conflicts of Interest
References
- Goldknopf, I.L.; Busch, H. Isopeptide linkage between nonhistone and histone 2A polypeptides of chromosomal conjugate-protein A24. Proc. Natl. Acad. Sci. USA 1977, 74, 864–868. [Google Scholar] [CrossRef] [PubMed]
- Hunt, L.T.; Dayhoff, M.O. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem. Biophys. Res. Commun. 1977, 74, 650–655. [Google Scholar] [CrossRef]
- Zhang, Y. Transcriptional regulation by histone ubiquitination and deubiquitination. Genes Dev. 2003, 17, 2733–2740. [Google Scholar] [CrossRef] [PubMed]
- Komander, D.; Rape, M. The Ubiquitin Code. Annu. Rev. Biochem. 2012, 81, 203–229. [Google Scholar] [CrossRef] [PubMed]
- Kerscher, O.; Felberbaum, R.; Hochstrasser, M. Modification of Proteins by Ubiquitin and Ubiquitin-Like Proteins. Annu. Rev. Cell Dev. Biol. 2006, 22, 159–180. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. EMBO Rep. 2008, 9, 859–864. [Google Scholar] [CrossRef] [PubMed]
- Enserink, J.M. Regulation of Cellular Processes by SUMO: Understudied Topics. In SUMO Regulation of Cellular Processes; Wilson, V.G., Ed.; Springer International Publishing: Cham, Switzerland, 2017; pp. 89–97. [Google Scholar] [CrossRef]
- Perng, Y.-C.; Lenschow, D.J. ISG15 in antiviral immunity and beyond. Nat. Rev. Microbiol. 2018, 16, 423–439. [Google Scholar] [CrossRef]
- Durfee, L.A.; Lyon, N.; Seo, K.; Huibregtse, J.M. The ISG15 Conjugation System Broadly Targets Newly Synthesized Proteins: Implications for the Antiviral Function of ISG15. Mol. Cell 2010, 38, 722–732. [Google Scholar] [CrossRef]
- Dubiel, W.; Chaithongyot, S.; Dubiel, D.; Naumann, M. The COP9 Signalosome: A Multi-DUB Complex. Biomolecules 2020, 10, 1082. [Google Scholar] [CrossRef]
- Chan, Y.; Yoon, J.; Wu, J.-T.; Kim, H.-J.; Pan, K.-T.; Yim, J.; Chien, C.-T. DEN1 deneddylates non-cullin proteins in vivo. J. Cell Sci. 2008, 121, 3218–3223. [Google Scholar] [CrossRef]
- Mendoza, H.M.; Shen, L.N.; Botting, C.; Lewis, A.; Chen, J.; Ink, B.; Hay, R.T. NEDP1, a highly conserved cysteine protease that deNEDDylates Cullins. J. Biol. Chem. 2003, 278, 25637–25643. [Google Scholar] [CrossRef] [PubMed]
- Soucy, T.A.; Smith, P.G.; Milhollen, M.A.; Berger, A.J.; Gavin, J.M.; Adhikari, S.; Brownell, J.E.; Burke, K.E.; Cardin, D.P.; Critchley, S.; et al. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 2009, 458, 732–736. [Google Scholar] [CrossRef] [PubMed]
- Gai, W.; Peng, Z.; Liu, C.H.; Zhang, L.; Jiang, H. Advances in Cancer Treatment by Targeting the Neddylation Pathway. Front. Cell Dev. Biol. 2021, 9. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Jia, L. Targeting Protein Neddylation for Cancer Therapy. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 297–315. [Google Scholar]
- Lammer, D.; Mathias, N.; Laplaza, J.M.; Jiang, W.; Liu, Y.; Callis, J.; Goebl, M.; Estelle, M. Modification of yeast Cdc53p by the ubiquitin-related protein rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998, 12, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Liakopoulos, D.; Doenges, G.; Matuschewski, K.; Jentsch, S. A novel protein modification pathway related to the ubiquitin system. EMBO J. 1998, 17, 2208–2214. [Google Scholar] [CrossRef] [PubMed]
- Baek, K.; Krist, D.T.; Prabu, J.R.; Hill, S.; Klügel, M.; Neumaier, L.-M.; von Gronau, S.; Kleiger, G.; Schulman, B.A. NEDD8 nucleates a multivalent cullin–RING–UBE2D ubiquitin ligation assembly. Nature 2020, 578, 461–466. [Google Scholar] [CrossRef] [PubMed]
- Duda, D.M.; Borg, L.A.; Scott, D.C.; Hunt, H.W.; Hammel, M.; Schulman, B.A. Structural Insights into NEDD8 Activation of Cullin-RING Ligases: Conformational Control of Conjugation. Cell 2008, 134, 995–1006. [Google Scholar] [CrossRef] [PubMed]
- Saha, A.; Deshaies, R.J. Multimodal Activation of the Ubiquitin Ligase SCF by Nedd8 Conjugation. Mol. Cell 2008, 32, 21–31. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.-T.; Lin, H.-C.; Hu, Y.-C.; Chien, C.-T. Neddylation and deneddylation regulate Cul1 and Cul3 protein accumulation. Nat. Cell Biol. 2005, 7, 1014–1020. [Google Scholar] [CrossRef]
- Pierce, N.W.; Lee, J.E.; Liu, X.; Sweredoski, M.J.; Graham, R.L.J.; Larimore, E.A.; Rome, M.; Zheng, N.; Clurman, B.E.; Hess, S.; et al. Cand1 Promotes Assembly of New SCF Complexes through Dynamic Exchange of F Box Proteins. Cell 2013, 153, 206–215. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Deshaies, R.J.; Liu, X. Assembly and Regulation of CRL Ubiquitin Ligases. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 33–46. [Google Scholar] [CrossRef]
- Rusnac, D.-V.; Zheng, N. Structural Biology of CRL Ubiquitin Ligases. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 9–31. [Google Scholar] [CrossRef]
- Rao, F.; Lin, H.; Su, Y. Cullin-RING Ligase Regulation by the COP9 Signalosome: Structural Mechanisms and New Physiologic Players. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 47–60. [Google Scholar] [CrossRef]
- Shi, W.; Ding, R.; Zhou, P.P.; Fang, Y.; Wan, R.; Chen, Y.; Jin, J. Coordinated Actions Between p97 and Cullin-RING Ubiquitin Ligases for Protein Degradation. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 61–78. [Google Scholar] [CrossRef]
- Fu, L.; Cui, C.-P.; Zhang, L. Regulation of Stem Cells by Cullin-RING Ligase. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 79–98. [Google Scholar] [CrossRef]
- Liu, Y.; Tan, X. Viral Manipulations of the Cullin-RING Ubiquitin Ligases. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 99–110. [Google Scholar] [CrossRef]
- Nguyen, K.M.; Busino, L. The Biology of F-box Proteins: The SCF Family of E3 Ubiquitin Ligases. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 111–122. [Google Scholar] [CrossRef]
- Liu, J.; Peng, Y.; Zhang, J.; Long, J.; Liu, J.; Wei, W. Targeting SCF E3 Ligases for Cancer Therapies. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 123–146. [Google Scholar] [CrossRef]
- Nakagawa, T.; Nakayama, K.; Nakayama, K.I. Knockout Mouse Models Provide Insight into the Biological Functions of CRL1 Components. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 147–171. [Google Scholar] [CrossRef]
- Liu, X.; Zurlo, G.; Zhang, Q. The Roles of Cullin-2 E3 Ubiquitin Ligase Complex in Cancer. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 173–186. [Google Scholar] [CrossRef]
- Chen, R.-H. Cullin 3 and Its Role in Tumorigenesis. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 187–210. [Google Scholar] [CrossRef]
- Wang, P.; Song, J.; Ye, D. CRL3s: The BTB-CUL3-RING E3 Ubiquitin Ligases. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 211–223. [Google Scholar] [CrossRef]
- Zhou, P.; Yan, F. CRL4 Ubiquitin Pathway and DNA Damage Response. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 225–239. [Google Scholar] [CrossRef]
- Zhou, Z.; Song, X.; Wavelet, C.M.; Wan, Y. Cullin 4-DCAF Proteins in Tumorigenesis. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 241–259. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, Y. Cullin RING Ligase 5 (CRL-5): Neddylation Activation and Biological Functions. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 261–283. [Google Scholar] [CrossRef]
- Pan, Z.-Q. Cullin-RING E3 Ubiquitin Ligase 7 in Growth Control and Cancer. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 285–296. [Google Scholar] [CrossRef]
- Gong, L.; Cui, D.; Xiong, X.; Zhao, Y. Targeting Cullin-RING Ubiquitin Ligases and the Applications in PROTACs. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 317–347. [Google Scholar] [CrossRef]
- Zhou, H.; Lu, J.; Yang, C.-Y.; Sun, Y.; Wang, S. Targeting DCN1-UBC12 Protein-Protein Interaction for Regulation of Neddylation Pathway. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 349–362. [Google Scholar] [CrossRef]
- Mao, H.; Sun, Y. Neddylation-Independent Activities of MLN4924. In Cullin-RING Ligases and Protein Neddylation: Biology and Therapeutics; Sun, Y., Wei, W., Jin, J., Eds.; Springer: Singapore, 2020; pp. 363–372. [Google Scholar] [CrossRef]
- Jones, J.; Wu, K.; Yang, Y.; Guerrero, C.; Nillegoda, N.; Pan, Z.-Q.; Huang, L. A Targeted Proteomic Analysis of the Ubiquitin-Like Modifier Nedd8 and Associated Proteins. J. Proteome Res. 2008, 7, 1274–1287. [Google Scholar] [CrossRef] [PubMed]
- Xirodimas, D.P.; Sundqvist, A.; Nakamura, A.; Shen, L.; Botting, C.; Hay, R.T. Ribosomal proteins are targets for the NEDD8 pathway. EMBO Rep. 2008, 9, 280–286. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Santockyte, R.; Shen, R.-F.; Tekle, E.; Wang, G.; Yang, D.C.H.; Chock, P.B. A general approach for investigating enzymatic pathways and substrates for ubiquitin-like modifiers. Arch. Biochem. Biophys. 2006, 453, 70–74. [Google Scholar] [CrossRef] [PubMed]
- Norman, J.A.; Shiekhattar, R. Analysis of Nedd8-Associated Polypeptides: A Model for Deciphering the Pathway for Ubiquitin-like Modifications. Biochemistry 2006, 45, 3014–3019. [Google Scholar] [CrossRef] [PubMed]
- Enchev, R.I.; Schulman, B.A.; Peter, M. Protein neddylation: Beyond cullin–RING ligases. Nat. Rev. Mol. Cell Biol. 2015, 16, 30–44. [Google Scholar] [CrossRef] [PubMed]
- Coleman, K.E.; Békés, M.; Chapman, J.R.; Crist, S.B.; Jones, M.J.K.; Ueberheide, B.M.; Huang, T.T. SENP8 limits aberrant neddylation of NEDD8 pathway components to promote cullin-RING ubiquitin ligase function. eLife 2017, 6, e24325. [Google Scholar] [CrossRef] [PubMed]
- Vogl, A.M.; Phu, L.; Becerra, R.; Giusti, S.A.; Verschueren, E.; Hinkle, T.B.; Bordenave, M.D.; Adrian, M.; Heidersbach, A.; Yankilevich, P.; et al. Global site-specific neddylation profiling reveals that NEDDylated cofilin regulates actin dynamics. Nat. Struct. Mol. Biol. 2020, 27, 210–220. [Google Scholar] [CrossRef]
- Lobato-Gil, S.; Heidelberger, J.B.; Maghames, C.; Bailly, A.; Brunello, L.; Rodriguez, M.S.; Beli, P.; Xirodimas, D.P. Proteome-wide identification of NEDD8 modification sites reveals distinct proteomes for canonical and atypical NEDDylation. Cell Rep. 2021, 34, 108635. [Google Scholar] [CrossRef]
- Jayabalan, A.K.; Sanchez, A.; Park, R.Y.; Yoon, S.P.; Kang, G.-Y.; Baek, J.-H.; Anderson, P.; Kee, Y.; Ohn, T. NEDDylation promotes stress granule assembly. Nat. Commun. 2016, 7, 12125. [Google Scholar] [CrossRef]
- Maghames, C.M.; Lobato-Gil, S.; Perrin, A.; Trauchessec, H.; Rodriguez, M.S.; Urbach, S.; Marin, P.; Xirodimas, D.P. NEDDylation promotes nuclear protein aggregation and protects the Ubiquitin Proteasome System upon proteotoxic stress. Nat. Commun. 2018, 9, 4376. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Fang, W.; Cui, Y.; Shi, H.; Chen, J.; Li, L.; Zhang, L.; Zhang, X. Neddylation promotes protein translocation between the cytoplasm and nucleus. Biochem. Biophys. Res. Commun. 2020, 529, 991–997. [Google Scholar] [CrossRef]
- Sundqvist, A.; Liu, G.; Mirsaliotis, A.; Xirodimas, D.P. Regulation of nucleolar signalling to p53 through NEDDylation of L11. EMBO Rep. 2009, 10, 1132–1139. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.-X.; Wang, Y.-G.; Xirodimas, D.P.; Dai, M.-S. Perturbation of 60 S Ribosomal Biogenesis Results in Ribosomal Protein L5- and L11-dependent p53 Activation. J. Biol. Chem. 2010, 285, 25812–25821. [Google Scholar] [CrossRef] [PubMed]
- Ebina, M.; Tsuruta, F.; Katoh, M.C.; Kigoshi, Y.; Someya, A.; Chiba, T. Myeloma Overexpressed 2 (Myeov2) Regulates L11 Subnuclear Localization through Nedd8 Modification. PLoS ONE 2013, 8, e65285. [Google Scholar] [CrossRef] [PubMed]
- Xiong, X.; Cui, D.; Bi, Y.; Sun, Y.; Zhao, Y. Neddylation modification of ribosomal protein RPS27L or RPS27 by MDM2 or NEDP1 regulates cancer cell survival. Faseb. J. 2020, 34, 13419–13429. [Google Scholar] [CrossRef] [PubMed]
- Oved, S.; Mosesson, Y.; Zwang, Y.; Santonico, E.; Shtiegman, K.; Marmor, M.D.; Kochupurakkal, B.S.; Katz, M.; Lavi, S.; Cesareni, G.; et al. Conjugation to Nedd8 instigates ubiquitylation and down-regulation of activated receptor tyrosine kinases. J. Biol. Chem. 2006, 281, 21640–21651. [Google Scholar] [CrossRef] [PubMed]
- Pandey, D.; Hori, D.; Kim, J.H.; Bergman, Y.; Berkowitz, D.E.; Romer, L.H. NEDDylation promotes endothelial dysfunction: A role for HDAC2. J. Mol. Cell. Cardiol. 2015, 81, 18–22. [Google Scholar] [CrossRef] [PubMed]
- Lai, Q.Y.; He, Y.Z.; Peng, X.W.; Zhou, X.; Liang, D.; Wang, L. Histone deacetylase 1 induced by neddylation inhibition contributes to drug resistance in acute myelogenous leukemia. Cell Commun. Signal. 2019, 17, 86. [Google Scholar] [CrossRef]
- Li, H.; Zhu, H.; Liu, Y.; He, F.; Xie, P.; Zhang, L. Itch promotes the neddylation of JunB and regulates JunB-dependent transcription. Cell. Signal. 2016, 28, 1186–1195. [Google Scholar] [CrossRef]
- Shu, J.; Liu, C.; Wei, R.; Xie, P.; He, S.; Zhang, L. Nedd8 targets ubiquitin ligase Smurf2 for neddylation and promote its degradation. Biochem. Biophys. Res. Commun. 2016, 474, 51–56. [Google Scholar] [CrossRef]
- Lee, G.-W.; Park, J.B.; Park, S.Y.; Seo, J.; Shin, S.-H.; Park, J.-W.; Kim, S.J.; Watanabe, M.; Chun, Y.-S. The E3 ligase C-CBL inhibits cancer cell migration by neddylating the proto-oncogene c-Src. Oncogene 2018, 37, 5552–5568. [Google Scholar] [CrossRef] [PubMed]
- Kamitani, T.; Kito, K.; Fukuda-Kamitani, T.; Yeh, E.T. Targeting of NEDD8 and its conjugates for proteasomal degradation by NUB1. J. Biol. Chem. 2001, 276, 46655–46660. [Google Scholar] [CrossRef] [PubMed]
- Kito, K.; Yeh, E.T.; Kamitani, T. NUB1, a NEDD8-interacting protein, is induced by interferon and down-regulates the NEDD8 expression. J. Biol. Chem. 2001, 276, 20603–20609. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Yang, H.; Zhao, J.; Zhang, Y.-H.; Song, A.-X.; Hu, H.-Y. NEDD8 Ultimate Buster-1 Long (NUB1L) Protein Promotes Transfer of NEDD8 to Proteasome for Degradation through the P97UFD1/NPL4 Complex. J. Biol. Chem. 2013, 288, 31339–31349. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Kawashima, H.; Yeh, E.T.; Kamitani, T. Regulation of the NEDD8 conjugation system by a splicing variant, NUB1L. J. Biol. Chem. 2003, 278, 32905–32913. [Google Scholar] [CrossRef] [PubMed]
- Hipp, M.S.; Raasi, S.; Groettrup, M.; Schmidtke, G. NEDD8 ultimate buster-1L interacts with the ubiquitin-like protein FAT10 and accelerates its degradation. J. Biol. Chem. 2004, 279, 16503–16510. [Google Scholar] [CrossRef]
- Schmidtke, G.; Kalveram, B.; Groettrup, M. Degradation of FAT10 by the 26S proteasome is independent of ubiquitylation but relies on NUB1L. FEBS Lett. 2009, 583, 591–594. [Google Scholar] [CrossRef] [PubMed]
- Schmidtke, G.; Kalveram, B.; Weber, E.; Bochtler, P.; Lukasiak, S.; Hipp, M.S.; Groettrup, M. The UBA domains of NUB1L are required for binding but not for accelerated degradation of the ubiquitin-like modifier FAT10. J. Biol. Chem. 2006, 281, 20045–20054. [Google Scholar] [CrossRef]
- Tanji, K.; Tanaka, T.; Kamitani, T. Interaction of NUB1 with the proteasome subunit S5a. Biochem. Biophys. Res. Commun. 2005, 337, 116–120. [Google Scholar] [CrossRef] [PubMed]
- Rani, N.; Aichem, A.; Schmidtke, G.; Kreft, S.G.; Groettrup, M. FAT10 and NUB1L bind to the VWA domain of Rpn10 and Rpn1 to enable proteasome-mediated proteolysis. Nat. Commun. 2012, 3, 749. [Google Scholar] [CrossRef] [PubMed]
- Jeong, Y.Y.; Her, J.; Chung, I.K. NEDD8 ultimate buster-1 regulates the abundance of TRF1 at telomeres by promoting its proteasomal degradation. FEBS Lett. 2016, 590, 1776–1790. [Google Scholar] [CrossRef][Green Version]
- Lu, B.; Al-Ramahi, I.; Valencia, A.; Wang, Q.; Berenshteyn, F.; Yang, H.; Gallego-Flores, T.; Ichcho, S.; Lacoste, A.; Hild, M.; et al. Identification of NUB1 as a suppressor of mutant Huntington toxicity via enhanced protein clearance. Nat. Neurosci. 2013, 16, 562–570. [Google Scholar] [CrossRef] [PubMed]
- Tanji, K.; Tanaka, T.; Mori, F.; Kito, K.; Takahashi, H.; Wakabayashi, K.; Kamitani, T. NUB1 suppresses the formation of Lewy body-like inclusions by proteasomal degradation of synphilin-1. Am. J. Pathol. 2006, 169, 553–565. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ma, W.; Li, H.; Hou, N.; Wang, X.; Kim, I.M.; Li, F.; Su, H. NEDD8 Ultimate Buster 1 Long (NUB1L) Protein Suppresses Atypical Neddylation and Promotes the Proteasomal Degradation of Misfolded Proteins. J. Biol. Chem. 2015, 290, 23850–23862. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Jiang, K.; Lin, G.; Liu, P.; Yan, Y.; Ye, T.; Yao, G.; Barr, M.P.; Liang, D.; Wang, Y.; et al. Ajuba inhibits hepatocellular carcinoma cell growth via targeting of β-catenin and YAP signaling and is regulated by E3 ligase Hakai through neddylation. J. Exp. Clin. Cancer Res. 2018, 37, 165. [Google Scholar] [CrossRef]
- Loftus, S.J.; Liu, G.; Carr, S.M.; Munro, S.; La Thangue, N.B. NEDDylation regulates E2F-1-dependent transcription. EMBO Rep. 2012, 13, 811–818. [Google Scholar] [CrossRef] [PubMed]
- Vijayasimha, K.; Tran, M.V.; Leestemaker-Palmer, A.L.; Dolan, B.P. Direct Conjugation of NEDD8 to the N-Terminus of a Model Protein Can Induce Degradation. Cells 2021, 10, 854. [Google Scholar] [CrossRef]
- Ghosh, D.K.; Roy, A.; Ranjan, A. HYPK scaffolds the Nedd8 and LC3 proteins to initiate the formation of autophagosome around the poly-neddylated huntingtin exon1 aggregates. bioRxiv 2019, 780379. [Google Scholar] [CrossRef]
- Kim, S.; Kwon, M.; Hwang, Y.; Yoon, J.; Park, S.; Kang, H.C. Stress-induced NEDDylation promotes cytosolic protein aggregation through HDAC6 in a p62-dependent manner. iScience 2021, 24, 102146. [Google Scholar] [CrossRef]
- Xirodimas, D.P.; Saville, M.K.; Bourdon, J.-C.; Hay, R.T.; Lane, D.P. Mdm2-Mediated NEDD8 Conjugation of p53 Inhibits Its Transcriptional Activity. Cell 2004, 118, 83–97. [Google Scholar] [CrossRef]
- Watson, I.R.; Li, B.K.; Roche, O.; Blanch, A.; Ohh, M.; Irwin, M.S. Chemotherapy induces NEDP1-mediated destabilization of MDM2. Oncogene 2010, 29, 297–304. [Google Scholar] [CrossRef] [PubMed]
- Embade, N.; Fernández-Ramos, D.; Varela-Rey, M.; Beraza, N.; Sini, M.; de Juan, V.G.; Woodhoo, A.; Martínez-López, N.; Rodríguez-Iruretagoyena, B.; Bustamante, F.J.; et al. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 2012, 55, 1237–1248. [Google Scholar] [CrossRef] [PubMed]
- Cannito, S.; Foglia, B.; Villano, G.; Turato, C.; Delgado, T.C.; Morello, E.; Pin, F.; Novo, E.; Napione, L.; Quarta, S.; et al. SerpinB3 Differently Up-Regulates Hypoxia Inducible Factors -1α and -2α in Hepatocellular Carcinoma: Mechanisms Revealing Novel Potential Therapeutic Targets. Cancers 2019, 11, 1933. [Google Scholar] [CrossRef]
- Ryu, J.-H.; Li, S.-H.; Park, H.-S.; Park, J.-W.; Lee, B.; Chun, Y.-S. Hypoxia-inducible Factor α Subunit Stabilization by NEDD8 Conjugation Is Reactive Oxygen Species-dependent. J. Biol. Chem. 2011, 286, 6963–6970. [Google Scholar] [CrossRef] [PubMed]
- Heo, M.J.; Kang, S.H.; Kim, Y.S.; Lee, J.M.; Yu, J.; Kim, H.-R.; Lim, H.; Kim, K.M.; Jung, J.; Jeong, L.S.; et al. UBC12-mediated SREBP-1 neddylation worsens metastatic tumor prognosis. Int. J. Cancer 2020, 147, 2550–2563. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, Y.-L.; Qiu, G.; Pian, L.; Guo, L.; Cao, H.; Liu, J.; Zhao, Y.; Li, X.; Xu, Z.; et al. Hepatic neddylation targets and stabilizes electron transfer flavoproteins to facilitate fatty acid β-oxidation. Proc. Natl. Acad. Sci. USA 2020, 117, 2473–2483. [Google Scholar] [CrossRef] [PubMed]
- Park, H.S.; Ju, U.I.; Park, J.W.; Song, J.Y.; Shin, D.H.; Lee, K.H.; Jeong, L.S.; Yu, J.; Lee, H.W.; Cho, J.Y.; et al. PPARγ neddylation essential for adipogenesis is a potential target for treating obesity. Cell Death Differ. 2016, 23, 1296–1311. [Google Scholar] [CrossRef] [PubMed]
- Zuo, W.; Huang, F.; Chiang, Y.J.; Li, M.; Du, J.; Ding, Y.; Zhang, T.; Lee, H.W.; Jeong, L.S.; Chen, Y.; et al. c-Cbl-Mediated Neddylation Antagonizes Ubiquitination and Degradation of the TGF-β Type II Receptor. Mol. Cell 2013, 49, 499–510. [Google Scholar] [CrossRef]
- Zhang, T.; Ye, Z.; Yang, X.; Qin, Y.; Hu, Y.; Tong, X.; Lai, W.; Ye, X. NEDDylation of PB2 Reduces Its Stability and Blocks the Replication of Influenza A Virus. Sci. Rep. 2017, 7, 43691. [Google Scholar] [CrossRef]
- Liu, N.; Zhang, J.; Yang, X.; Jiao, T.; Zhao, X.; Li, W.; Zhu, J.; Yang, P.; Jin, J.; Peng, J.; et al. HDM2 promotes NEDDylation of HBV HBx to enhance its stability and function. J. Virol. 2017, 91, e00340-17. [Google Scholar] [CrossRef]
- Kumar, D.; Das, M.; Sauceda, C.; Ellies, L.G.; Kuo, K.; Parwal, P.; Kaur, M.; Jih, L.; Bandyopadhyay, G.K.; Burton, D.; et al. Degradation of splicing factor SRSF3 contributes to progressive liver disease. J. Clin. Investig. 2019, 129, 4477–4491. [Google Scholar] [CrossRef] [PubMed]
- Stickle, N.H.; Chung, J.; Klco, J.M.; Hill, R.P.; Kaelin, W.G.; Ohh, M. pVHL Modification by NEDD8 Is Required for Fibronectin Matrix Assembly and Suppression of Tumor Development. Mol. Cell. Biol. 2004, 24, 3251–3261. [Google Scholar] [CrossRef] [PubMed]
- Russell, R.C.; Ohh, M. NEDD8 acts as a ‘molecular switch’ defining the functional selectivity of VHL. EMBO Rep. 2008, 9, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Balasubramaniam, M.; Parcon, P.A.; Bose, C.; Liu, L.; Jones, R.A.; Farlow, M.R.; Mrak, R.E.; Barger, S.W.; Griffin, W.S.T. Interleukin-1β drives NEDD8 nuclear-to-cytoplasmic translocation, fostering parkin activation via NEDD8 binding to the P-ubiquitin activating site. J. Neuroinflamm. 2019, 16, 275. [Google Scholar] [CrossRef] [PubMed]
- Choo, Y.S.; Vogler, G.; Wang, D.; Kalvakuri, S.; Iliuk, A.; Tao, W.A.; Bodmer, R.; Zhang, Z. Regulation of parkin and PINK1 by neddylation. Hum. Mol. Genet. 2012, 21, 2514–2523. [Google Scholar] [CrossRef]
- Um, J.W.; Han, K.A.; Im, E.; Oh, Y.; Lee, K.; Chung, K.C. Neddylation positively regulates the ubiquitin E3 ligase activity of parkin. J. Neurosci. Res. 2012, 90, 1030–1042. [Google Scholar] [CrossRef]
- Li, C.; Zhang, L.; Qian, D.; Cheng, M.; Hu, H.; Hong, Z.; Cui, Y.; Yu, H.; Wang, Q.; Zhu, J.; et al. RNF111-facilitated neddylation potentiates cGAS-mediated antiviral innate immune response. PLoS Pathog. 2021, 17, e1009401. [Google Scholar] [CrossRef]
- Segovia, J.A.; Tsai, S.-Y.; Chang, T.-H.; Shil, N.K.; Weintraub, S.T.; Short, J.D.; Bose, S. Nedd8 regulates inflammasome-dependent caspase-1 activation. Mol. Cell. Biol. 2015, 35, 582–597. [Google Scholar] [CrossRef] [PubMed]
- Leidecker, O.; Matic, I.; Mahata, B.; Pion, E.; Xirodimas, D.P. The ubiquitin E1 enzyme Ube1 mediates NEDD8 activation under diverse stress conditions. Cell Cycle 2012, 11, 1142–1150. [Google Scholar] [CrossRef]
- Hjerpe, R.; Thomas, Y.; Chen, J.; Zemla, A.; Curran, S.; Shpiro, N.; Dick, L.R.; Kurz, T. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 2012, 441, 927–939. [Google Scholar] [CrossRef]
- Zou, T.; Zhang, J. Diverse and pivotal roles of neddylation in metabolism and immunity. FEBS J. 2021. [Google Scholar] [CrossRef] [PubMed]
- Zeng, Y.; Iv, Y.-S.; Pan, Q.-H.; Zhou, Y.-G.; Li, H. An overactive neddylation pathway serves as a therapeutic target and MLN4924 enhances the anticancer activity of cisplatin in pancreatic cancer. Oncol. Lett. 2019, 18, 2724–2732. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shi, C.C.; Zhang, H.P.; Li, G.Q.; Li, S.S. MLN4924 suppresses neddylation and induces cell cycle arrest, senescence, and apoptosis in human osteosarcoma. Oncotarget 2016, 7, 45263–45274. [Google Scholar] [CrossRef] [PubMed]
- Tian, D.-W.; Wu, Z.-L.; Jiang, L.-M.; Gao, J.; Wu, C.-L.; Hu, H.-L. Neural precursor cell expressed, developmentally downregulated 8 promotes tumor progression and predicts poor prognosis of patients with bladder cancer. Cancer Sci. 2019, 110, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Zhang, M.; He, S.; Lu, K.; Chen, Y.; Xing, G.; Lu, Y.; Liu, P.; Li, Y.; Wang, S.; et al. The covalent modifier Nedd8 is critical for the activation of Smurf1 ubiquitin ligase in tumorigenesis. Nat. Commun. 2014, 5, 3733. [Google Scholar] [CrossRef] [PubMed]
- Xie, P.; Peng, Z.; Chen, Y.; Li, H.; Du, M.; Tan, Y.; Zhang, X.; Lu, Z.; Cui, C.-P.; Liu, C.H.; et al. Neddylation of PTEN regulates its nuclear import and promotes tumor development. Cell Res. 2021, 31, 291–311. [Google Scholar] [CrossRef]
- McGrail, D.J.; Garnett, J.; Yin, J.; Dai, H.; Shih, D.J.H.; Lam, T.N.A.; Li, Y.; Sun, C.; Li, Y.; Schmandt, R.; et al. Proteome Instability Is a Therapeutic Vulnerability in Mismatch Repair-Deficient Cancer. Cancer Cell 2020, 37, 371–386. [Google Scholar] [CrossRef]
- Goldman, M.J.; Craft, B.; Hastie, M.; Repečka, K.; McDade, F.; Kamath, A.; Banerjee, A.; Luo, Y.; Rogers, D.; Brooks, A.N.; et al. Visualizing and interpreting cancer genomics data via the Xena platform. Nat. Biotechnol. 2020, 38, 675–678. [Google Scholar] [CrossRef]
- Hua, W.; Li, C.; Yang, Z.; Li, L.; Jiang, Y.; Yu, G.; Zhu, W.; Liu, Z.; Duan, S.; Chu, Y.; et al. Suppression of glioblastoma by targeting the overactivated protein neddylation pathway. Neuro-oncology 2015, 17, 1333–1343. [Google Scholar] [CrossRef]
- Ma, T.; Chen, Y.; Zhang, F.; Yang, C.-Y.; Wang, S.; Yu, X. RNF111-dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell 2013, 49, 897–907. [Google Scholar] [CrossRef]
- Bailly, A.P.; Perrin, A.; Serrano-Macia, M.; Maghames, C.; Leidecker, O.; Trauchessec, H.; Martinez-Chantar, M.L.; Gartner, A.; Xirodimas, D.P. The Balance between Mono- and NEDD8-Chains Controlled by NEDP1 upon DNA Damage Is a Regulatory Module of the HSP70 ATPase Activity. Cell Rep. 2019, 29, 212–224. [Google Scholar] [CrossRef] [PubMed]
- Keuss, M.J.; Hjerpe, R.; Hsia, O.; Gourlay, R.; Burchmore, R.; Trost, M.; Kurz, T. Unanchored tri-NEDD8 inhibits PARP-1 to protect from oxidative stress-induced cell death. EMBO J. 2019, 38, e100024. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Cui, Q.; Wang, X.; Li, B.; Zhao, D.; Xia, Q.; Zhao, P. Functions and substrates of NEDDylation during cell cycle in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 2017, 90, 101–112. [Google Scholar] [CrossRef] [PubMed]
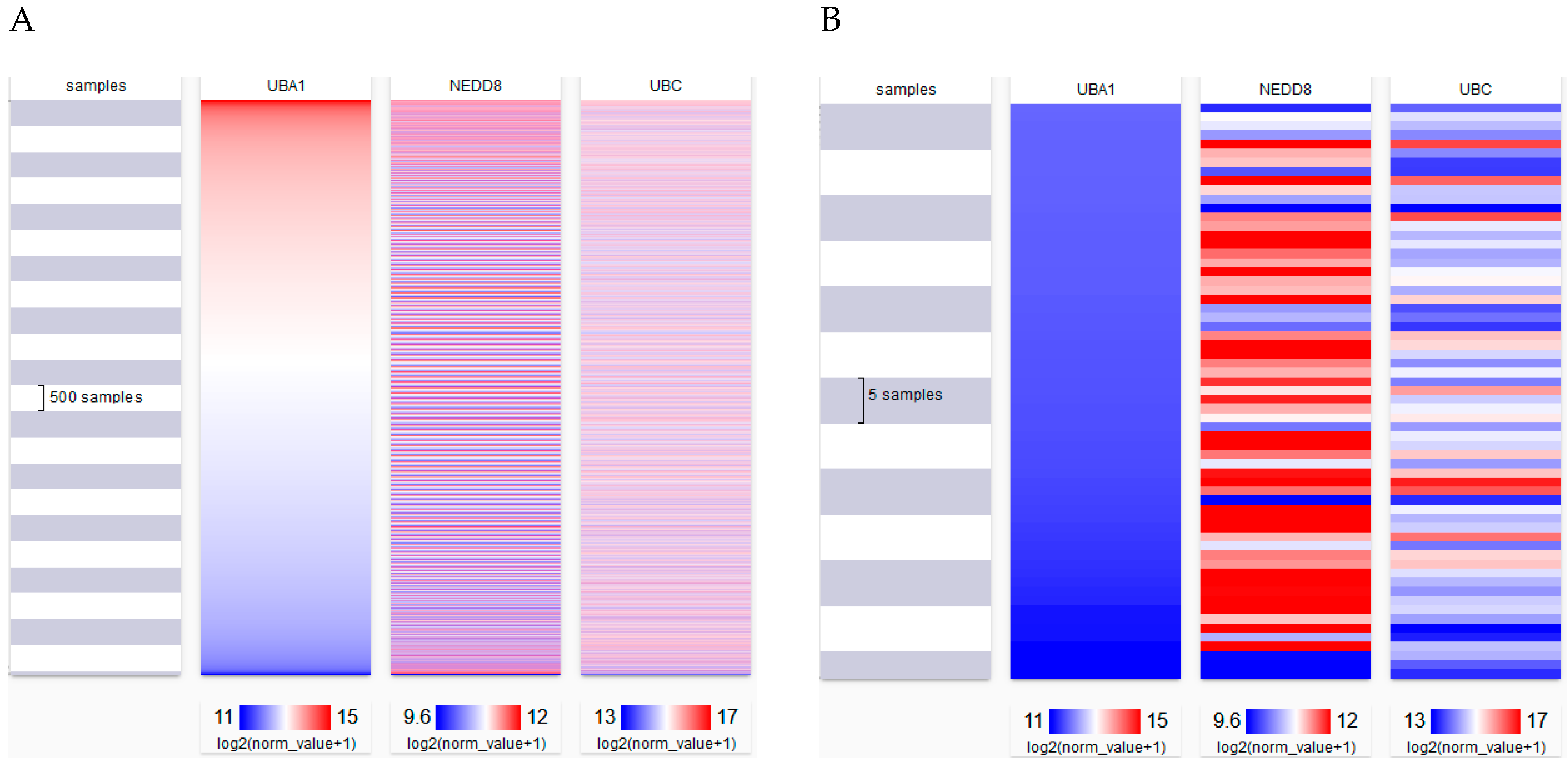
| TCGA Set | R Value | p Value |
|---|---|---|
| Pan-Cancer | 0.1034 | 1.172 × 10−27 |
| Colon and Rectal | −0.1414 | 0.003146 |
| Colon | −0.1422 | 0.009796 |
| Glioblastoma | −0.5184 | 3.319 × 10−13 |
| Head and Neck | −0.1160 | 0.005715 |
| Lung Adenocarcinoma | −0.1983 | 0.000001616 |
| Lung Cancer | −0.1608 | 5.586 × 10−8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vijayasimha, K.; Dolan, B.P. The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation. Cells 2021, 10, 2660. https://doi.org/10.3390/cells10102660
Vijayasimha K, Dolan BP. The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation. Cells. 2021; 10(10):2660. https://doi.org/10.3390/cells10102660
Chicago/Turabian StyleVijayasimha, Kartikeya, and Brian P. Dolan. 2021. "The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation" Cells 10, no. 10: 2660. https://doi.org/10.3390/cells10102660
APA StyleVijayasimha, K., & Dolan, B. P. (2021). The Many Potential Fates of Non-Canonical Protein Substrates Subject to NEDDylation. Cells, 10(10), 2660. https://doi.org/10.3390/cells10102660





