A Human 3D Cardiomyocyte Risk Model to Study the Cardiotoxic Influence of X-rays and Other Noxae in Adults
Abstract
:1. Introduction
2. Materials and Methods
2.1. The Cell Lines and Culture Conditions
2.2. Differentiation of H9-cTNT into hPSC-CMC
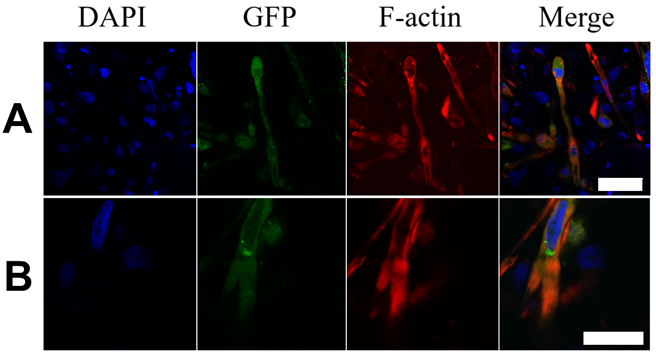
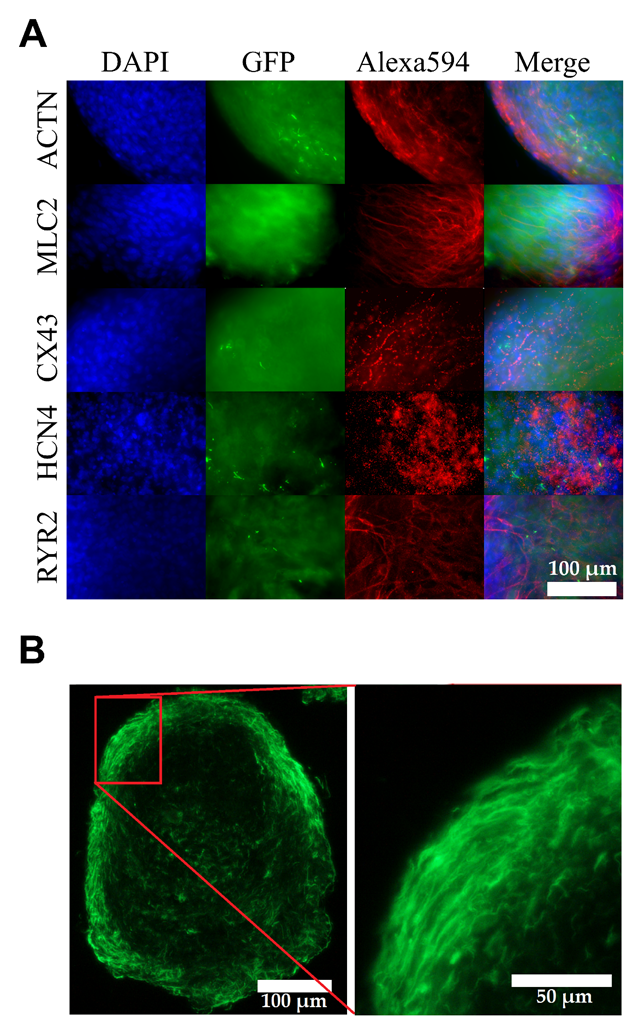
2.3. Immunocytochemical Analyses of Cardiac Markers
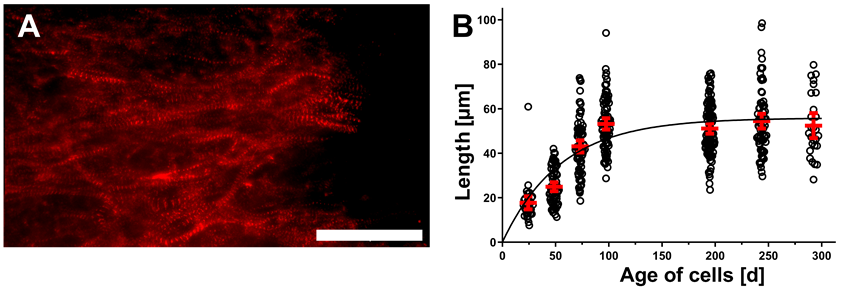
2.4. Measurements of α-Actinin Chain Lengths
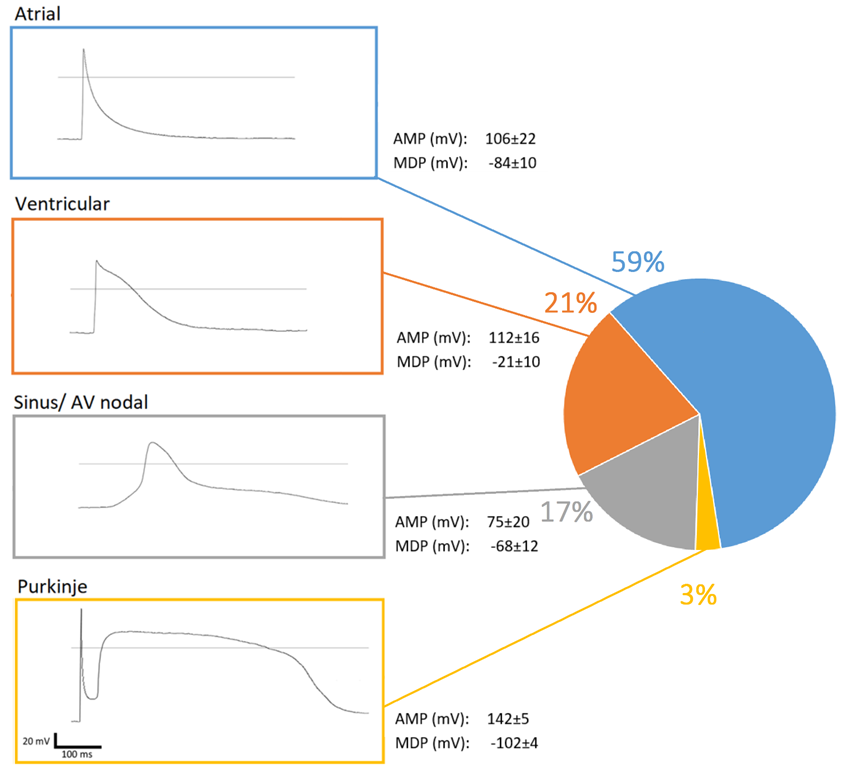
2.5. Patch-Clamp Analysis of Individual Cells
2.6. X-ray Irradiation of hPSC-CMC
2.7. Video-Based Analysis of hPSC-CMC
2.8. Interleukin 6 and 8 ELISA Assays
2.9. Size Measurements
2.10. Proteomic Analysis
3. Results
3.1. Prolonged Cultivation Leads to Maturation of CM within hPSC-CMC
3.2. Matured hPSC-CMC Exhibit Key-Markers of the Heart and Mirror the Myocardial Structure
3.3. Matured hPSC-CMC Contain All Major Cardiac Cell Types Featuring a Physiologic Cell Composition
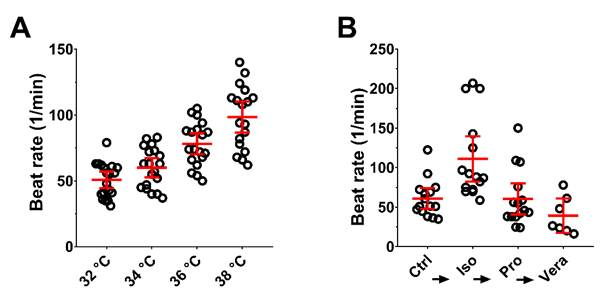
3.4. External Stimuli Alter the Contraction of hPSC-CMC in a Physiologic Manner
3.5. X-ray Irradiation Induces Subtle Pacing and Arrhythmic Manifestations in Matured hPSC-CMC
3.6. A Moderate Dose of 2 Gy X-rays Induces Neither a Interleukin Storm nor Hypertrophic Growth in Matured hPSC-CMC
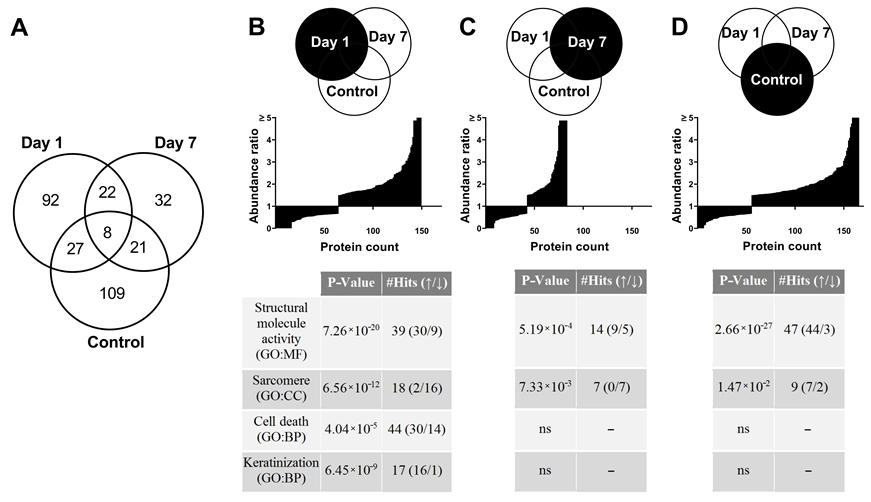
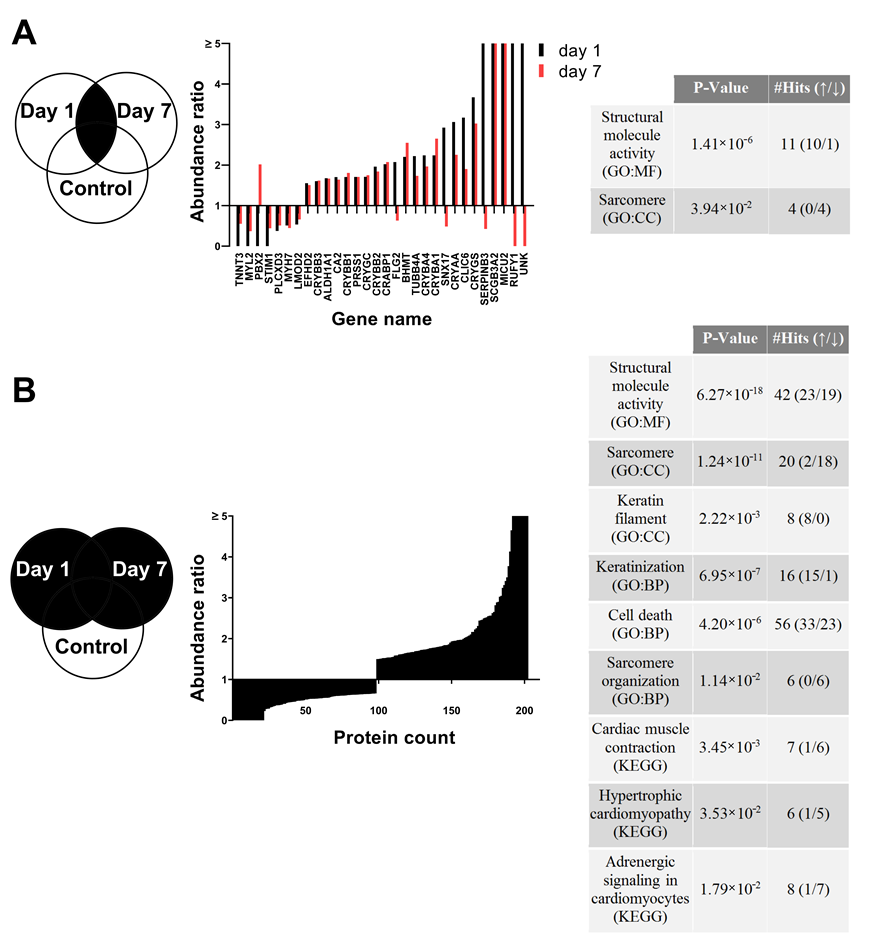
3.7. Proteome Changes after X-ray Irradiation Pointing towards a Fast Structural Remodeling and Disease-Related Signaling
4. Discussion
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Virani, S.S.; Alonso, A.; Benjamin, E.J.; Bittencourt, M.S.; Callaway, C.W.; Carson, A.P.; Chamberlain, A.M.; Chang, A.R.; Cheng, S.; Delling, F.N.; et al. Heart disease and stroke statistics—2020 update: A report from the American Heart Association. Circulation 2020, E139–E596. [Google Scholar] [CrossRef] [PubMed]
- Tapio, S. Pathology and biology of radiation-induced cardiac disease. J. Radiat. Res. 2016, 57, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Schultz-Hector, S.; Trott, K.R. Radiation-induced cardiovascular diseases: Is the epidemiologic evidence compatible with the radiobiologic data? Int. J. Radiat. Oncol. Biol. Phys. 2007, 67, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Baselet, B.; Rombouts, C.; Benotmane, A.M.; Baatout, S.; Aerts, A. Cardiovascular diseases related to ionizing radiation: The risk of low-dose exposure (Review). Int. J. Mol. Med. 2016, 38, 1623–1641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madan, R.; Benson, R.; Sharma, D.N.; Julka, P.K.; Rath, G.K. Radiation induced heart disease: Pathogenesis, management and review literature. J. Egypt. Natl. Cancer Inst. 2015, 27, 187–193. [Google Scholar] [CrossRef] [PubMed]
- Sanguinetti, M.C.; Tristani-Firouzi, M. hERG potassium channels and cardiac arrhythmia. Nature 2006, 440, 463–469. [Google Scholar] [CrossRef] [PubMed]
- Tamargo, J.; Caballero, R.; Núñez, L.; Gómez, R.; Vaquero, M.; Delpón, E. Genetically engineered mice as a model for studying cardiac arrhythmias. Front. Biosci. 2007, 12, 22–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Burridge, P.W.; Keller, G.; Gold, J.D.; Wu, J.C. Production of De Novo Cardiomyocytes: Human Pluripotent Stem Cell Differentiation and Direct Reprogramming. Cell Stem Cell 2012, 10, 16–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giacomelli, E.; Meraviglia, V.; Campostrini, G.; Cochrane, A.; Cao, X.; van Helden, R.W.J.; Krotenberg Garcia, A.; Mircea, M.; Kostidis, S.; Davis, R.P.; et al. Human-iPSC-derived cardiac stromal cells enhance maturation in 3D cardiac microtissues and reveal non-cardiomyocyte contributions to heart disease. Cell Stem Cell 2020, 26, 862–879. [Google Scholar] [CrossRef] [PubMed]
- Strauss, D.G.; Wu, W.W.; Li, Z.; Koerner, J.; Garnett, C. Translational Models and tools to reduce clinical trials and improve regulatory decision making for qtc and proarrhythmia risk (ICH E14/S7B Updates). Clin. Pharmacol. Ther. 2021, 109, 319–333. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Lei, W.; Hu, S. Cardiac organoid—A promising perspective of preclinical model. Stem Cell Res. Ther. 2021, 12, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Goldfracht, I.; Protze, S.; Shiti, A.; Setter, N.; Gruber, A.; Shaheen, N.; Nartiss, Y.; Keller, G.; Gepstein, L. Generating ring-shaped engineered heart tissues from ventricular and atrial human pluripotent stem cell-derived cardiomyocytes. Nat. Commun. 2020, 11, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braam, S.R.; Tertoolen, L.; van de Stolpe, A.; Meyer, T.; Passier, R.; Mummery, C.L. Prediction of drug-induced cardiotoxicity using human embryonic stem cell-derived cardiomyocytes. Stem Cell Res. 2010, 4, 107–116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clements, M.; Thomas, N. High-throughput multi-parameter profiling of electrophysiological drug effects in human embryonic stem cell derived cardiomyocytes using multi-electrode arrays. Toxicol. Sci. 2014, 140, 445–461. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denning, C.; Anderson, D. Cardiomyocytes from human embryonic stem cells as predictors of cardiotoxicity. Drug Discov. Today Ther. Strateg. 2008, 5, 223–232. [Google Scholar] [CrossRef]
- Denning, C.; Borgdorff, V.; Crutchley, J.; Firth, K.S.A.; George, V.; Kalra, S.; Kondrashov, A.; Hoang, M.D.; Mosqueira, D.; Patel, A.; et al. Cardiomyocytes from human pluripotent stem cells: From laboratory curiosity to industrial biomedical platform. Biochim. Biophys. Acta Mol. Cell Res. 2016, 1863, 1728–1748. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.H.; Ye, L. Maturation of pluripotent stemcell-derived cardiomyocytes: A critical step for drug development and cell therapy. J. Cardiovasc. Transl. Res. 2018, 11, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; McKeithan, W.L.; Serrano, R.; Kitani, T.; Burridge, P.W.; del Álamo, J.C.; Mercola, M.; Wu, J.C. Use of human induced pluripotent stem cell–derived cardiomyocytes to assess drug cardiotoxicity. Nat. Protoc. 2018, 13, 3018–3041. [Google Scholar] [CrossRef] [PubMed]
- Stella Stoter, A.M.; Hirt, M.N.; Stenzig, J.; Weinberger, F. Assessment of cardiotoxicity with stem cell-based strategies. Clin. Ther. 2020, 42, 1892–1910. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Pabon, L.; Murry, C.E. Engineering adolescence: Maturation of human pluripotent stem cell-derived cardiomyocytes. Circ. Res. 2014, 114, 511–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kadari, A.; Mekala, S.; Wagner, N.; Malan, D.; Köth, J.; Doll, K.; Stappert, L.; Eckert, D.; Peitz, M.; Matthes, J.; et al. Robust generation of cardiomyocytes from human iPS cells requires precise modulation of BMP and WNT signaling. Stem Cell Rev. Rep. 2015, 11, 560–569. [Google Scholar] [CrossRef] [PubMed]
- Nitsch, S.; Braun, F.; Ritter, S.; Scholz, M.; Schroeder, I.S. Functional video-based analysis of 3D cardiac structures generated from human embryonic stem cells. Stem Cell Res. 2018, 29, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewski, J.R.; Zougman, A.; Nagaraj, N.; Mann, M. Universal sample preparation method for proteome analysis. Nat. Methods 2009, 6, 359–362. [Google Scholar] [CrossRef] [PubMed]
- Grosche, A.; Hauser, A.; Lepper, M.F.; Mayo, R.; Von Toerne, C.; Merl-Pham, J.; Hauck, S.M. The proteome of native adult Müller glial cells from Murine retina. Mol. Cell. Proteom. 2016, 15, 462–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hladik, D.; Dalke, C.; Von Toerne, C.; Hauck, S.M.; Azimzadeh, O.; Philipp, J.; Ung, M.C.; Schlattl, H.; Rößler, U.; Graw, J.; et al. Creb signaling mediates dose-dependent radiation response in the murine hippocampus two years after total body exposure. J. Proteome Res. 2019, 19, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Navarro, P.; Trevisan-Herraz, M.; Bonzon-Kulichenko, E.; Núñez, E.; Martínez-Acedo, P.; Pérez-Hernández, D.; Jorge, I.; Mesa, R.; Calvo, E.; Carrascal, M.; et al. General statistical framework for quantitative proteomics by stable isotope labeling. J. Proteome Res. 2014, 13, 1234–1247. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Wei, J.; Zheng, Q.; Meng, L.; Xin, Y.; Yin, X.; Jiang, X. Radiation-induced heart disease: A review of classification, mechanism and prevention. Int. J. Biol. Sci. 2019, 15, 2128–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fritsche, E.; Haarmann-Stemmann, T.; Kapr, J.; Galanjuk, S.; Hartmann, J.; Mertens, P.R.; Kämpfer, A.A.M.; Schins, R.P.F.; Tigges, J.; Koch, K. Stem Cells for Next Level Toxicity Testing in the 21st Century. Small 2021, 17, 1–31. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Yamanaka, S. Induced Pluripotent Stem Cells 10 Years Later. Circ. Res. 2017, 120, 1958–1968. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wilson, G.F.; Soerens, A.G.; Koonce, C.H.; Yu, J.; Palecek, S.P.; Thomson, J.A.; Kamp, T.J. Functional Cardiomyocytes Derived From Human Induced Pluripotent Stem Cells. Circ. Res. 2009, 104, 564–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.-K.; Ng, K.-M.; Lai, W.-H.; Chan, Y.-C.; Lau, Y.-M.; Lian, Q.; Tse, H.-F.; Siu, C.-W. Calcium Homeostasis in Human Induced Pluripotent Stem Cell-Derived Cardiomyocytes. Stem Cell Rev. Rep. 2011, 7, 976–986. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsa, E.; Burridge, P.W.; Wu, J.C. Human stem cells for modeling heart disease and for drug discovery. Sci. Transl. Med. 2014, 6, 239ps6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robertson, C.; Tran, D.D.; George, S.C. Concise review: Maturation phases of human pluripotent stem cell-derived cardiomyocytes. Stem Cells 2013, 31, 829–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lundy, S.D.; Zhu, W.Z.; Regnier, M.; Laflamme, M.A. Structural and functional maturation of cardiomyocytes derived from human pluripotent stem cells. Stem Cells Dev. 2013, 22, 1991–2002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shih, H.T. Anatomy of the action potential in the heart. Texas Hear. Inst. J. 1994, 21, 30–41. [Google Scholar]
- Klimas, A.; Ambrosi, C.M.; Yu, J.; Williams, J.C.; Bien, H.; Entcheva, E. OptoDyCE as an automated system for high-throughput all-optical dynamic cardiac electrophysiology. Nat. Commun. 2016, 7, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frieß, J.L.; Heselich, A.; Ritter, S.; Haber, A.; Kaiser, N.; Layer, P.G.; Thielemann, C. Electrophysiologic and cellular characteristics of cardiomyocytes after X-ray irradiation. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2015, 777, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Becker, B.V.; Seeger, T.; Beiert, T.; Antwerpen, M.; Palnek, A.; Port, M.; Ullmann, R. Impact of Ionizing Radiation on Electrophysiological Behavior of Human-induced Ipsc-derived Cardiomyocytes on Multielectrode Arrays. Health Phys. 2018, 115, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Sala, L.; Bellin, M.; Mummery, C.L. Integrating cardiomyocytes from human pluripotent stem cells in safety pharmacology: Has the time come? Br. J. Pharmacol. 2017, 174, 3749–3765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gibhardt, C.S.; Roth, B.; Schroeder, I.; Fuck, S.; Becker, P.; Jakob, B.; Fournier, C.; Moroni, A.; Thiel, G. X-ray irradiation activates K+ channels via H2O2 signaling. Sci. Rep. 2015, 5, 13861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Azimzadeh, O.; Scherthan, H.; Sarioglu, H.; Barjaktarovic, Z.; Conrad, M.; Vogt, A.; Calzada-Wack, J.; Neff, F.; Aubele, M.; Buske, C.; et al. Rapid proteomic remodeling of cardiac tissue caused by total body ionizing radiation. Proteomics 2011, 11, 3299–3311. [Google Scholar] [CrossRef] [PubMed]
- Dimauro, I.; Antonioni, A.; Mercatelli, N.; Caporossi, D. The role of αB-crystallin in skeletal and cardiac muscle tissues. Cell Stress Chaperones 2018, 23, 491–505. [Google Scholar] [CrossRef] [PubMed]
- Tsikitis, M.; Galata, Z.; Mavroidis, M.; Psarras, S.; Capetanaki, Y. Intermediate filaments in cardiomyopathy. Biophys. Rev. 2018, 10, 1007–1031. [Google Scholar] [CrossRef] [PubMed]
- Hirota, H.; Yoshida, K.; Kishimoto, T.; Taga, T. Continuous activation of gp130, a signal-transducing receptor component for interleukin 6-related cytokines, causes myocardial hypertrophy in mice. Proc. Natl. Acad. Sci. USA 1995, 92, 4862–4866. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taunk, N.K.; Haffty, B.G.; Kostis, J.B.; Goyal, S. Radiation-induced heart disease: Pathologic abnormalities and putative mechanisms. Front. Oncol. 2015, 5, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koutroumpakis, E.; Palaskas, N.L.; Lin, S.H.; Abe, J.I.; Liao, Z.; Banchs, J.; Deswal, A.; Yusuf, S.W. Modern Radiotherapy and Risk of Cardiotoxicity. Chemotherapy 2020, 65, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Darby, S.C.; Cutter, D.J.; Boerma, M.; Constine, L.S.; Fajardo, L.F.; Kodama, K.; Mabuchi, K.; Marks, L.B.; Mettler, F.A.; Pierce, L.J.; et al. Radiation-Related Heart Disease: Current Knowledge and Future Prospects. Int. J. Radiat. Oncol. Biol. Phys. 2010, 76, 656–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Darby, S.C.; Ewertz, M.; McGale, P.; Bennet, A.M.; Blom-Goldman, U.; Brønnum, D.; Correa, C.; Cutter, D.; Gagliardi, G.; Gigante, B.; et al. Risk of Ischemic Heart Disease in Women after Radiotherapy for Breast Cancer. N. Engl. J. Med. 2013, 368, 987–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lühr, A.; von Neubeck, C.; Pawelke, J.; Seidlitz, A.; Peitzsch, C.; Bentzen, S.M.; Bortfeld, T.; Debus, J.; Deutsch, E.; Langendijk, J.A.; et al. “Radiobiology of Proton Therapy”: Results of an international expert workshop. Radiother. Oncol. 2018, 128, 56–67. [Google Scholar] [CrossRef] [PubMed]
- Hughson, R.L.; Helm, A.; Durante, M. Heart in space: Effect of the extraterrestrial environment on the cardiovascular system. Nat. Rev. Cardiol. 2018, 15, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, P.; Jackson, C.B.; Ozhathil, L.C.; Agarkova, I.; Galindo, C.L.; Sawyer, D.B.; Suter, T.M.; Zuppinger, C. 3D Co-culture of hiPSC-Derived Cardiomyocytes With Cardiac Fibroblasts Improves Tissue-Like Features of Cardiac Spheroids. Front. Mol. Biosci. 2020, 7, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]


| Day | Basal Medium | Supplements |
|---|---|---|
| 0 | RPMI1640 B27 without insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | CHIR99021 BMP4 |
| 1 | RPMI1640 B27 without insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | CHIR99021 |
| 2 | RPMI1640 B27 without insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | - |
| 3–6 | RPMI1640 B27 without insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | 10 μM XAV939 |
| 7 | RPMI1640 B27 without insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | - |
| 8–13 | RPMI 1640 without glucose 50 µg/mL penicillin and 5 µg/mL streptomycin | 4 mM sodium l-lactate |
| 14–18 | RPMI1640 B27 with insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | - |
| 19–300 | RPMI1640 B27 without insulin 50 μg/mL l-ascorbic acid 50 µg/mL penicillin and 5 µg/mL streptomycin | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smit, T.; Schickel, E.; Azimzadeh, O.; von Toerne, C.; Rauh, O.; Ritter, S.; Durante, M.; Schroeder, I.S. A Human 3D Cardiomyocyte Risk Model to Study the Cardiotoxic Influence of X-rays and Other Noxae in Adults. Cells 2021, 10, 2608. https://doi.org/10.3390/cells10102608
Smit T, Schickel E, Azimzadeh O, von Toerne C, Rauh O, Ritter S, Durante M, Schroeder IS. A Human 3D Cardiomyocyte Risk Model to Study the Cardiotoxic Influence of X-rays and Other Noxae in Adults. Cells. 2021; 10(10):2608. https://doi.org/10.3390/cells10102608
Chicago/Turabian StyleSmit, Timo, Esther Schickel, Omid Azimzadeh, Christine von Toerne, Oliver Rauh, Sylvia Ritter, Marco Durante, and Insa S. Schroeder. 2021. "A Human 3D Cardiomyocyte Risk Model to Study the Cardiotoxic Influence of X-rays and Other Noxae in Adults" Cells 10, no. 10: 2608. https://doi.org/10.3390/cells10102608
APA StyleSmit, T., Schickel, E., Azimzadeh, O., von Toerne, C., Rauh, O., Ritter, S., Durante, M., & Schroeder, I. S. (2021). A Human 3D Cardiomyocyte Risk Model to Study the Cardiotoxic Influence of X-rays and Other Noxae in Adults. Cells, 10(10), 2608. https://doi.org/10.3390/cells10102608








