The N-Terminal Region of the Polo Kinase Cdc5 Is Required for Downregulation of the Meiotic Recombination Checkpoint
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plasmids
2.2. Strains
2.3. Meiotic Time Courses, Sporulation Efficiency and Spore Viability
2.4. Microscopy
2.5. Western Blotting
3. Results
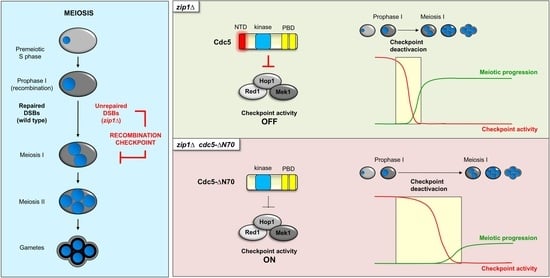
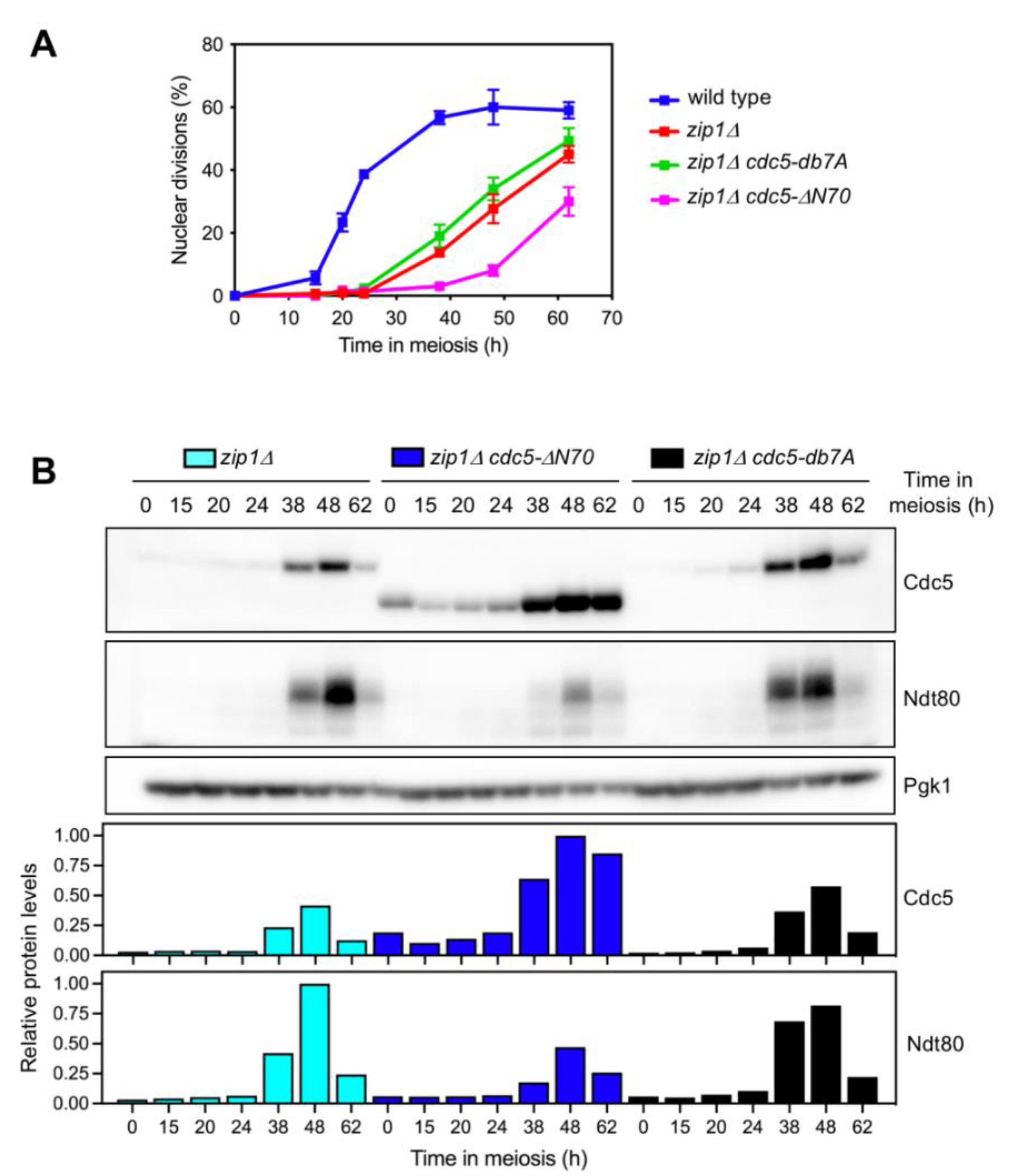
3.1. The N-Terminus of Cdc5 Is Required to Bypass the zip1Δ-Induced Meiotic Checkpoint
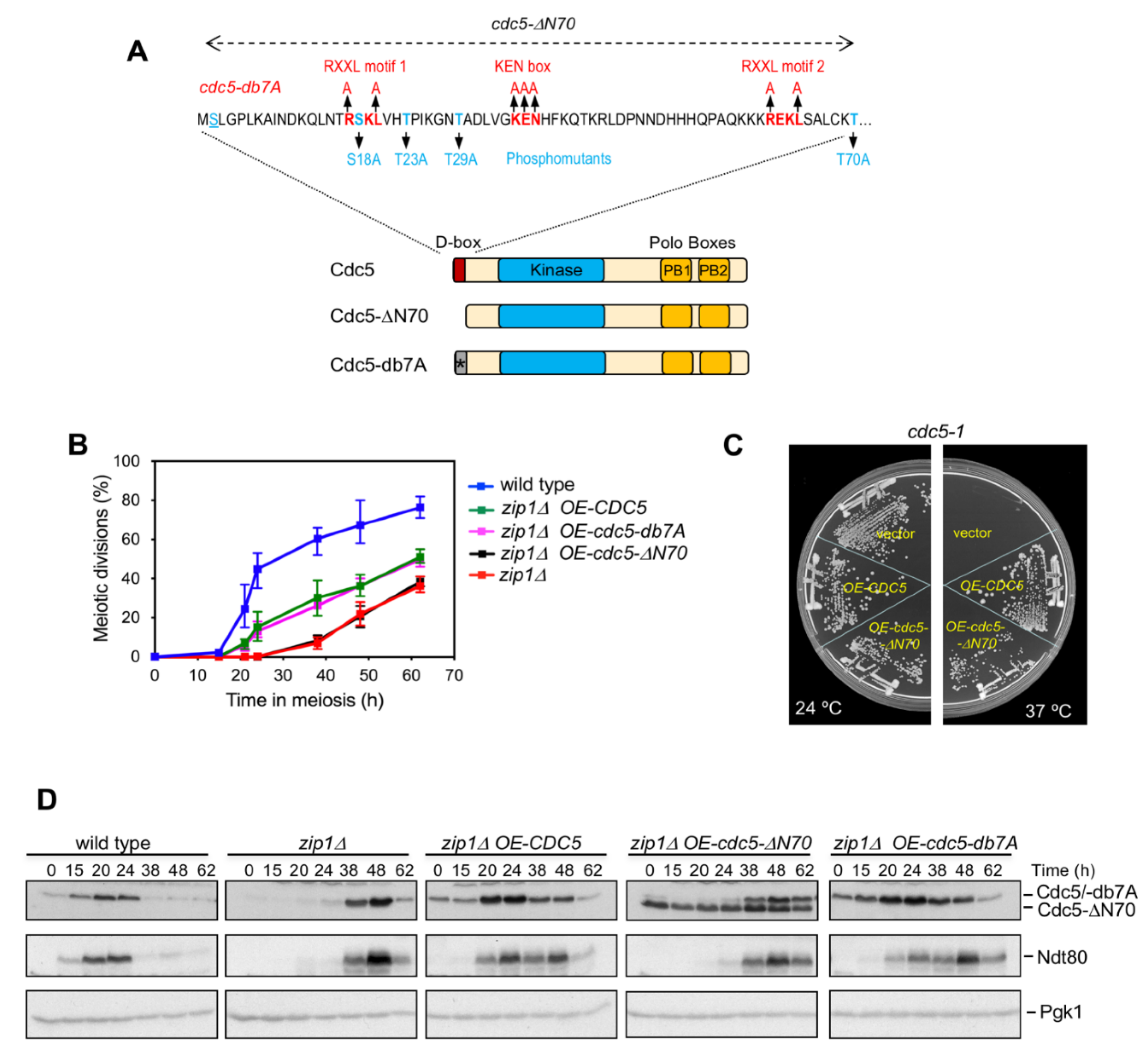
3.2. The Meiotic Checkpoint Function of Cdc5 N-Terminal Domain Is Independent of the Consensus Motifs for Targeted Degradation via APC/C
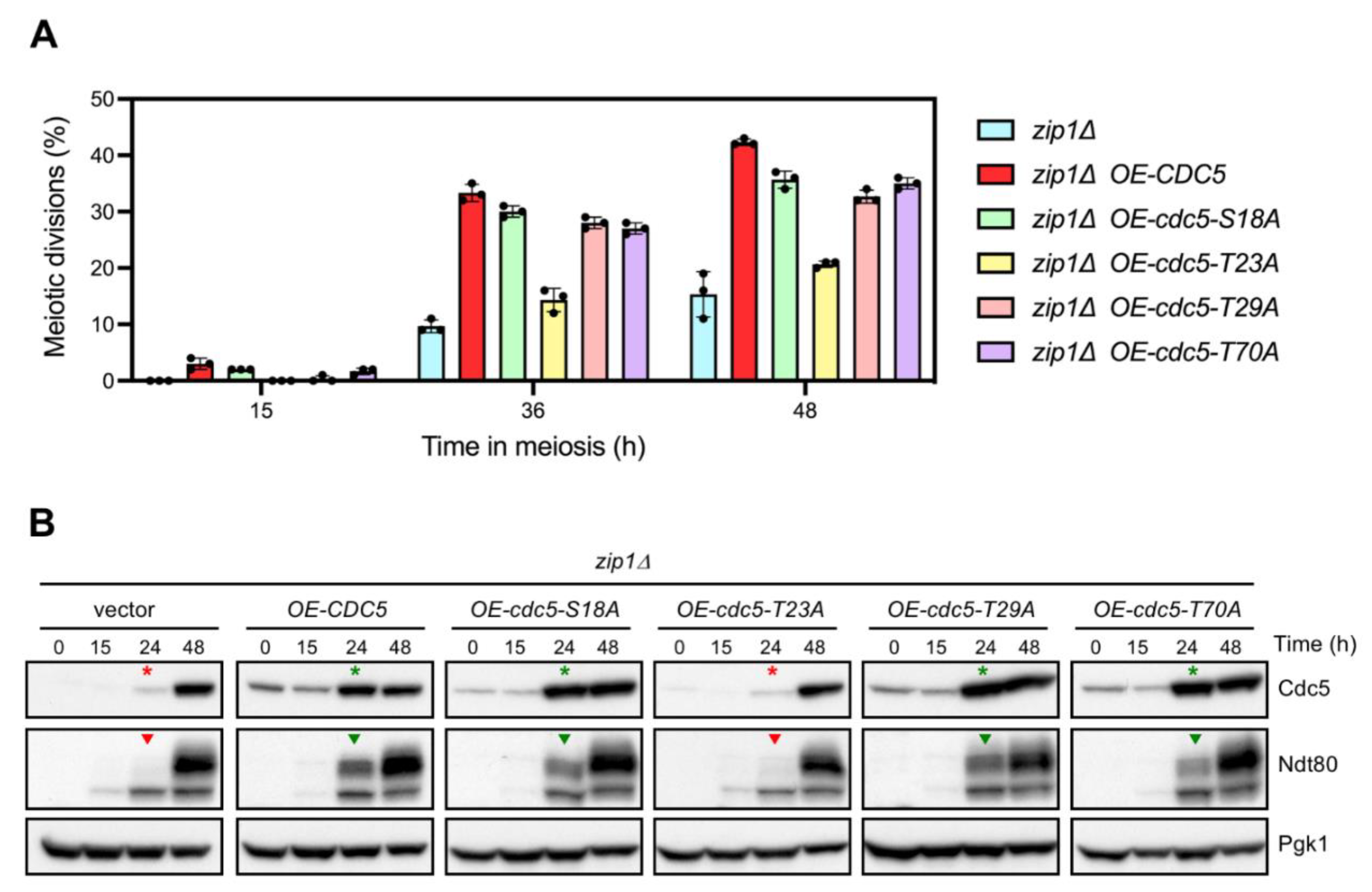
3.3. CDK Sites in the N-Terminal Domain of Cdc5 Are not Required for Its Meiotic Checkpoint Function
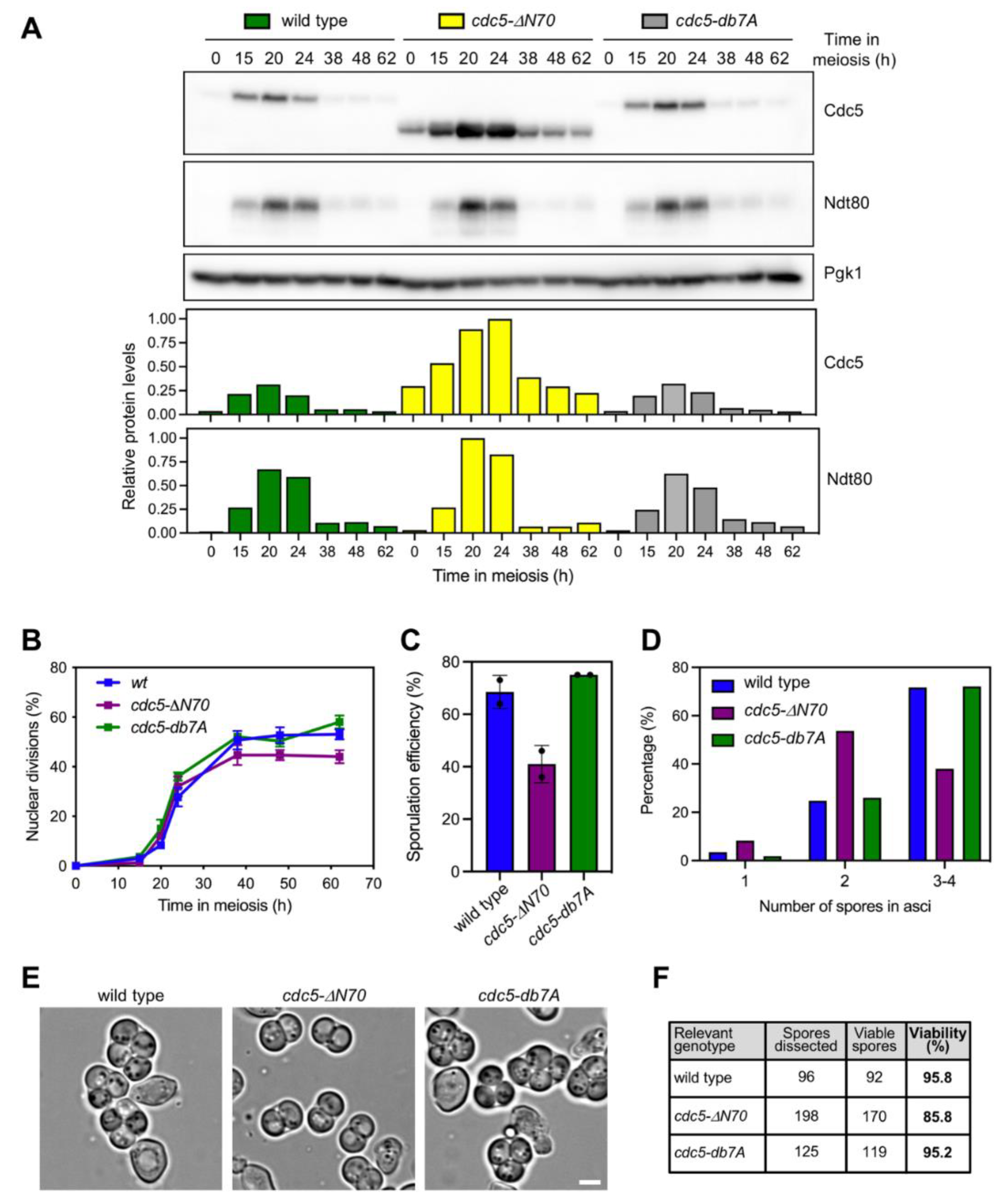
3.4. The N-Terminal Domain of Cdc5, but Not the APC/C Recognition Motifs, Is Required for Efficient Meiotic Progression and Sporulation
3.5. Persistent Checkpoint Activity in the Absence of Cdc5 N-Terminal Domain
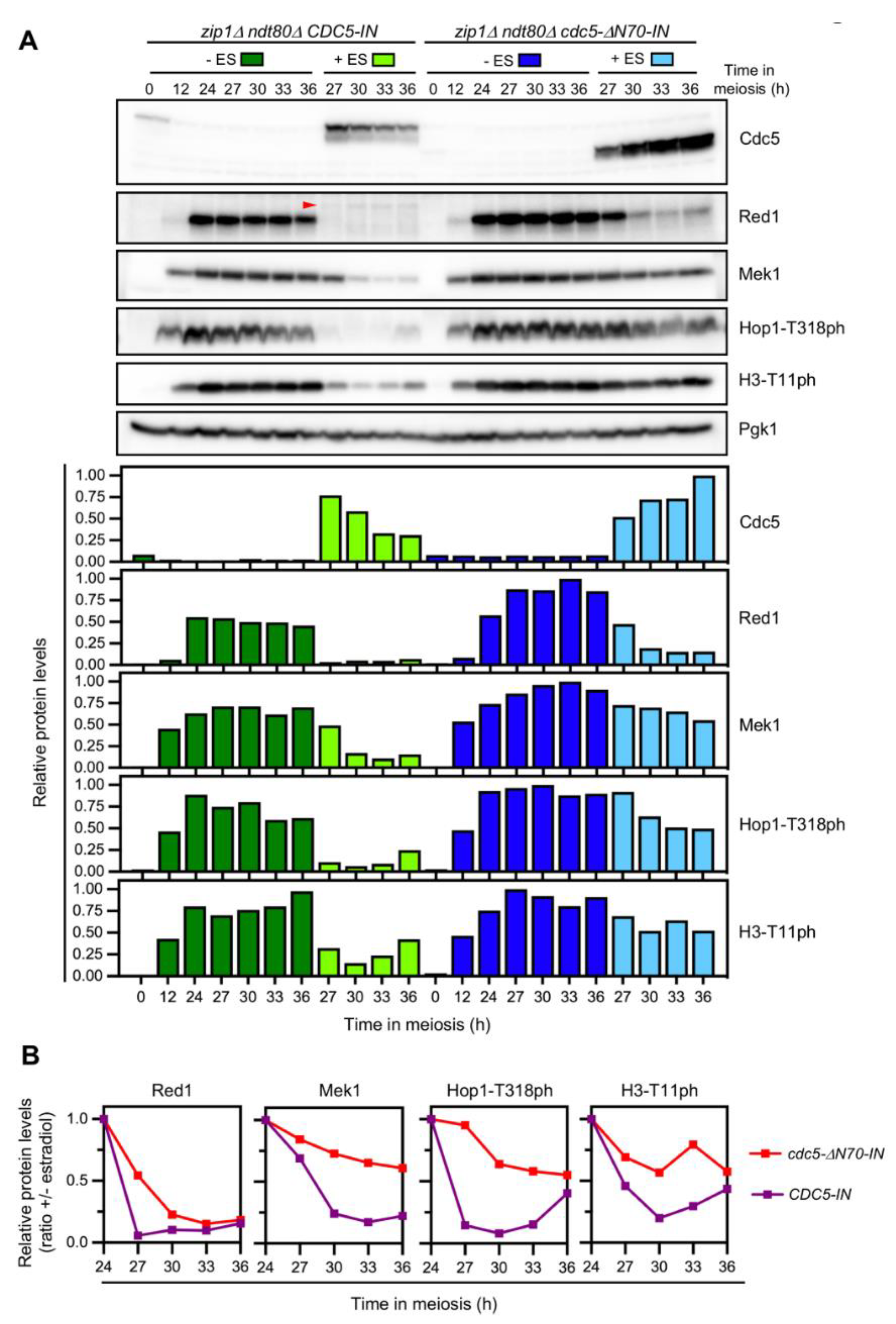
3.6. The N-Terminal Domain of Cdc5 Is Required for Efficient Checkpoint Downregulation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Archambault, V.; Lepine, G.; Kachaner, D. Understanding the Polo Kinase machine. Oncogene 2015, 34, 4799–4807. [Google Scholar] [CrossRef]
- Botchkarev, V.V., Jr.; Haber, J.E. Functions and regulation of the Polo-like kinase Cdc5 in the absence and presence of DNA damage. Curr. Genet. 2018, 64, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Duro, E.; Marston, A.L. From equator to pole: Splitting chromosomes in mitosis and meiosis. Genes Dev. 2015, 29, 109–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunter, N. Meiotic recombination: The essence of heredity. Cold Spring Harb. Perspect. Biol. 2015, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- San-Segundo, P.A.; Clemente-Blanco, A. Resolvases, dissolvases, and helicases in homologous recombination: Clearing the road for chromosome segregation. Genes 2020, 11, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaid, R.; Sharma, N.; Chauhan, S.; Deshta, A.; Dev, K.; Sourirajan, A. Functions of Polo-Like Kinases: A Journey From Yeast To Humans. Protein Pept. Lett. 2016, 23, 185–197. [Google Scholar] [CrossRef]
- Sourirajan, A.; Lichten, M. Polo-like kinase Cdc5 drives exit from pachytene during budding yeast meiosis. Genes Dev. 2008, 22, 2627–2632. [Google Scholar] [CrossRef] [Green Version]
- Argunhan, B.; Leung, W.K.; Afshar, N.; Terentyev, Y.; Subramanian, V.V.; Murayama, Y.; Hochwagen, A.; Iwasaki, H.; Tsubouchi, T.; Tsubouchi, H. Fundamental cell cycle kinases collaborate to ensure timely destruction of the synaptonemal complex during meiosis. EMBO J. 2017, 36, 2488–2509. [Google Scholar] [CrossRef]
- Alonso-Ramos, P.; Álvarez-Melo, D.; Strouhalova, K.; Pascual-Silva, C.; Garside, G.B.; Arter, M.; Bermejo, T.; Grigaitis, R.; Wettstein, R.; Fernández-Díaz, M.; et al. The Cdc14 phosphatase controls resolution of recombination intermediates and crossover formation during meiosis. Int. J. Mol. Sci. 2021, 22, 9811. [Google Scholar] [CrossRef]
- Clyne, R.K.; Katis, V.L.; Jessop, L.; Benjamin, K.R.; Herskowitz, I.; Lichten, M.; Nasmyth, K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat. Cell Biol. 2003, 5, 480–485. [Google Scholar] [CrossRef]
- Lee, B.H.; Amon, A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science 2003, 300, 482–486. [Google Scholar] [CrossRef]
- Matos, J.; Lipp, J.J.; Bogdanova, A.; Guillot, S.; Okaz, E.; Junqueira, M.; Shevchenko, A.; Zachariae, W. Dbf4-dependent CDC7 kinase links DNA replication to the segregation of homologous chromosomes in meiosis I. Cell 2008, 135, 662–678. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, H.G.; Koshland, D. Chromosome morphogenesis: Condensin-dependent cohesin removal during meiosis. Cell 2005, 123, 397–407. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, V.V.; Hochwagen, A. The meiotic checkpoint network: Step-by-step through meiotic prophase. Cold Spring Harb. Perspect. Biol. 2014, 6, a016675. [Google Scholar] [CrossRef]
- Refolio, E.; Cavero, S.; Marcon, E.; Freire, R.; San-Segundo, P.A. The Ddc2/ATRIP checkpoint protein monitors meiotic recombination intermediates. J. Cell Sci. 2011, 124, 2488–2500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carballo, J.A.; Johnson, A.L.; Sedgwick, S.G.; Cha, R.S. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell 2008, 132, 758–770. [Google Scholar] [CrossRef] [Green Version]
- Lo, Y.H.; Chuang, C.N.; Wang, T.F. Pch2 prevents Mec1/Tel1-mediated Hop1 phosphorylation occurring independently of Red1 in budding yeast meiosis. PLoS ONE 2014, 9, e85687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Penedos, A.; Johnson, A.L.; Strong, E.; Goldman, A.S.; Carballo, J.A.; Cha, R.S. Essential and checkpoint functions of budding yeast ATM and ATR during meiotic prophase are facilitated by differential phosphorylation of a meiotic adaptor protein, Hop1. PLoS ONE 2015, 10, e0134297. [Google Scholar] [CrossRef] [Green Version]
- Woltering, D.; Baumgartner, B.; Bagchi, S.; Larkin, B.; Loidl, J.; de los Santos, T.; Hollingsworth, N.M. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell Biol. 2000, 20, 6646–6658. [Google Scholar] [CrossRef] [Green Version]
- Herruzo, E.; Lago-Maciel, A.; Baztán, S.; Santos, B.; Carballo, J.A.; San-Segundo, P.A. Pch2 orchestrates the meiotic recombination checkpoint from the cytoplasm. PLoS Genet. 2021, 17, e1009560. [Google Scholar] [CrossRef] [PubMed]
- Herruzo, E.; Ontoso, D.; González-Arranz, S.; Cavero, S.; Lechuga, A.; San-Segundo, P.A. The Pch2 AAA+ ATPase promotes phosphorylation of the Hop1 meiotic checkpoint adaptor in response to synaptonemal complex defects. Nucleic Acids Res. 2016, 44, 7722–7741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herruzo, E.; Santos, B.; Freire, R.; Carballo, J.A.; San-Segundo, P.A. Characterization of Pch2 localization determinants reveals a nucleolar-independent role in the meiotic recombination checkpoint. Chromosoma 2019, 128, 297–316. [Google Scholar] [CrossRef] [PubMed]
- Hollingsworth, N.M.; Gaglione, R. The meiotic-specific Mek1 kinase in budding yeast regulates interhomolog recombination and coordinates meiotic progression with double-strand break repair. Curr. Genet. 2019, 65, 631–641. [Google Scholar] [CrossRef]
- Callender, T.L.; Laureau, R.; Wan, L.; Chen, X.; Sandhu, R.; Laljee, S.; Zhou, S.; Suhandynata, R.T.; Prugar, E.; Gaines, W.A.; et al. Mek1 down regulates Rad51 activity during yeast meiosis by phosphorylation of Hed1. PLoS Genet. 2016, 12, e1006226. [Google Scholar] [CrossRef]
- Niu, H.; Wan, L.; Busygina, V.; Kwon, Y.; Allen, J.A.; Li, X.; Kunz, R.C.; Kubota, K.; Wang, B.; Sung, P.; et al. Regulation of meiotic recombination via Mek1-mediated Rad54 phosphorylation. Mol. Cell 2009, 36, 393–404. [Google Scholar] [CrossRef] [Green Version]
- Kniewel, R.; Murakami, H.; Liu, Y.; Ito, M.; Ohta, K.; Hollingsworth, N.M.; Keeney, S. Histone H3 threonine 11 phosphorylation is catalyzed directly by the meiosis-specific kinase Mek1 and provides a molecular readout of Mek1 activity in vivo. Genetics 2017, 207, 1313–1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cavero, S.; Herruzo, E.; Ontoso, D.; San-Segundo, P.A. Impact of histone H4K16 acetylation on the meiotic recombination checkpoint in Saccharomyces cerevisiae. Microb. Cell 2016, 3, 606–620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Gaglione, R.; Leong, T.; Bednor, L.; de Los Santos, T.; Luk, E.; Airola, M.; Hollingsworth, N.M. Mek1 coordinates meiotic progression with DNA break repair by directly phosphorylating and inhibiting the yeast pachytene exit regulator Ndt80. PLoS Genet. 2018, 14, e1007832. [Google Scholar] [CrossRef]
- González-Arranz, S.; Cavero, S.; Morillo-Huesca, M.; Andújar, E.; Pérez-Alegre, M.; Prado, F.; San-Segundo, P. Functional Impact of the H2A.Z Histone Variant During Meiosis in Saccharomyces cerevisiae. Genetics 2018, 209, 997–1015. [Google Scholar] [CrossRef] [Green Version]
- Leu, J.Y.; Roeder, G.S. The pachytene checkpoint in S. cerevisiae depends on Swe1-mediated phosphorylation of the cyclin-dependent kinase Cdc28. Mol. Cell 1999, 4, 805–814. [Google Scholar] [CrossRef]
- Prugar, E.; Burnett, C.; Chen, X.; Hollingsworth, N.M. Coordination of double strand break repair and meiotic progression in yeast by a Mek1-Ndt80 negative feedback loop. Genetics 2017, 206, 497–512. [Google Scholar] [CrossRef] [Green Version]
- Charles, J.F.; Jaspersen, S.L.; Tinker-Kulberg, R.L.; Hwang, L.; Szidon, A.; Morgan, D.O. The Polo-related kinase Cdc5 activates and is destroyed by the mitotic cyclin destruction machinery in S. cerevisiae. Curr. Biol. 1998, 8, 497–507. [Google Scholar] [CrossRef] [Green Version]
- Shirayama, M.; Zachariae, W.; Ciosk, R.; Nasmyth, K. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 1998, 17, 1336–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Visintin, C.; Tomson, B.N.; Rahal, R.; Paulson, J.; Cohen, M.; Taunton, J.; Amon, A.; Visintin, R. APC/C-Cdh1-mediated degradation of the Polo kinase Cdc5 promotes the return of Cdc14 into the nucleolus. Genes Dev. 2008, 22, 79–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cooper, K.F.; Mallory, M.J.; Egeland, D.B.; Jarnik, M.; Strich, R. Ama1p is a meiosis-specific regulator of the anaphase promoting complex/cyclosome in yeast. Proc. Natl. Acad. Sci. USA 2000, 97, 14548–14553. [Google Scholar] [CrossRef] [Green Version]
- Okaz, E.; Arguello-Miranda, O.; Bogdanova, A.; Vinod, P.K.; Lipp, J.J.; Markova, Z.; Zagoriy, I.; Novak, B.; Zachariae, W. Meiotic prophase requires proteolysis of M phase regulators mediated by the meiosis-specific APC/CAma1. Cell 2012, 151, 603–618. [Google Scholar] [CrossRef] [Green Version]
- Jaspersen, S.L.; Charles, J.F.; Tinker-Kulberg, R.L.; Morgan, D.O. A late mitotic regulatory network controlling cyclin destruction in Saccharomyces cerevisiae. Mol. Biol. Cell 1998, 9, 2803–2817. [Google Scholar] [CrossRef] [Green Version]
- Mortensen, E.M.; Haas, W.; Gygi, M.; Gygi, S.P.; Kellogg, D.R. Cdc28-dependent regulation of the Cdc5/Polo kinase. Curr. Biol. 2005, 15, 2033–2037. [Google Scholar] [CrossRef] [Green Version]
- Acosta, I.; Ontoso, D.; San-Segundo, P.A. The budding yeast polo-like kinase Cdc5 regulates the Ndt80 branch of the meiotic recombination checkpoint pathway. Mol. Biol. Cell 2011, 22, 3478–3490. [Google Scholar] [CrossRef] [Green Version]
- Pfleger, C.M.; Kirschner, M.W. The KEN box: An APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000, 14, 655–665. [Google Scholar]
- Pfleger, C.M.; Lee, E.; Kirschner, M.W. Substrate recognition by the Cdc20 and Cdh1 components of the anaphase-promoting complex. Genes Dev. 2001, 15, 2396–2407. [Google Scholar] [CrossRef] [Green Version]
- Arnold, L.; Hockner, S.; Seufert, W. Insights into the cellular mechanism of the yeast ubiquitin ligase APC/C-Cdh1 from the analysis of in vivo degrons. Mol. Biol. Cell 2015, 26, 843–858. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Rodriguez, J.A.; Moyano, Y.; Jativa, S.; Queralt, E. Mitotic Exit Function of Polo-like Kinase Cdc5 Is Dependent on Sequential Activation by Cdk1. Cell Rep. 2016, 15, 2050–2062. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson-Lavy, K.J.; Brandeis, M. Phosphorylation of Cdc5 regulates its accumulation. Cell Div. 2011, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simpson-Lavy, K.J.; Sajman, J.; Zenvirth, D.; Brandeis, M. APC/CCdh1 specific degradation of Hsl1 and Clb2 is required for proper stress responses of S. cerevisiae. Cell Cycle 2009, 8, 3003–3009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tung, K.S.; Hong, E.J.; Roeder, G.S. The pachytene checkpoint prevents accumulation and phosphorylation of the meiosis-specific transcription factor Ndt80. Proc. Natl. Acad. Sci. USA 2000, 97, 12187–12192. [Google Scholar] [CrossRef] [Green Version]
- Carlile, T.M.; Amon, A. Meiosis I is established through division-specific translational control of a cyclin. Cell 2008, 133, 280–291. [Google Scholar] [CrossRef] [Green Version]
- Attner, M.A.; Miller, M.P.; Ee, L.S.; Elkin, S.K.; Amon, A. Polo kinase Cdc5 is a central regulator of meiosis I. Proc. Natl. Acad. Sci. USA 2013, 110, 14278–14283. [Google Scholar] [CrossRef] [Green Version]
- Galander, S.; Barton, R.E.; Kelly, D.A.; Marston, A.L. Spo13 prevents premature cohesin cleavage during meiosis. Wellcome Open Res. 2019, 4, 29. [Google Scholar] [CrossRef]
- Klapholz, S.; Esposito, R.E. Isolation of SPO12-1 and SPO13-1 from a natural variant of yeast that undergoes a single meiotic division. Genetics 1980, 96, 567–588. [Google Scholar] [CrossRef]
- Shirk, K.; Jin, H.; Giddings, T.H., Jr.; Winey, M.; Yu, H.G. The Aurora kinase Ipl1 is necessary for spindle pole body cohesion during budding yeast meiosis. J. Cell Sci. 2011, 124, 2891–2896. [Google Scholar] [CrossRef] [Green Version]
- Neiman, A.M. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 2011, 189, 737–765. [Google Scholar] [CrossRef] [Green Version]
- Alfaro, E.; López-Jiménez, P.; González-Martínez, J.; Malumbres, M.; Suja, J.A.; Gomez, R. PLK1 regulates centrosome migration and spindle dynamics in male mouse meiosis. EMBO Rep. 2021, 22, e51030. [Google Scholar] [CrossRef]
- Niu, H.; Wan, L.; Baumgartner, B.; Schaefer, D.; Loidl, J.; Hollingsworth, N.M. Partner choice during meiosis is regulated by Hop1-promoted dimerization of Mek1. Mol. Biol. Cell 2005, 16, 5804–5818. [Google Scholar] [CrossRef] [Green Version]
- Ontoso, D.; Acosta, I.; van Leeuwen, F.; Freire, R.; San-Segundo, P.A. Dot1-dependent histone H3K79 methylation promotes activation of the Mek1 meiotic checkpoint effector kinase by regulating the Hop1 adaptor. PLoS Genet. 2013, 9, e1003262. [Google Scholar] [CrossRef] [Green Version]
- Bailis, J.M.; Roeder, G.S. Pachytene exit controlled by reversal of Mek1-dependent phosphorylation. Cell 2000, 101, 211–221. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.J.; Lin, F.M.; Chuang, M.J.; Shen, H.J.; Wang, T.F. Genetic requirements and meiotic function of phosphorylation of the yeast axial element protein Red1. Mol. Cell Biol. 2011, 31, 912–923. [Google Scholar] [CrossRef] [Green Version]
- Almawi, A.W.; Langlois-Lemay, L.; Boulton, S.; Rodríguez González, J.; Melacini, G.; D’Amours, D.; Guarne, A. Distinct surfaces on Cdc5/PLK Polo-box domain orchestrate combinatorial substrate recognition during cell division. Sci. Rep. 2020, 10, 3379. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, A.; Adam, C.; Rauh, F.; Duroc, Y.; Ranjha, L.; Lombard, B.; Mu, X.; Wintrebert, M.; Loew, D.; Guarne, A.; et al. Exo1 recruits Cdc5 polo kinase to MutLgamma to ensure efficient meiotic crossover formation. Proc. Natl. Acad. Sci. USA 2020, 117, 30577–30588. [Google Scholar] [CrossRef] [PubMed]
- Ishiguro, K.; Kim, J.; Fujiyama-Nakamura, S.; Kato, S.; Watanabe, Y. A new meiosis-specific cohesin complex implicated in the cohesin code for homologous pairing. EMBO Rep. 2011, 12, 267–275. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
González-Arranz, S.; Acosta, I.; Carballo, J.A.; Santos, B.; San-Segundo, P.A. The N-Terminal Region of the Polo Kinase Cdc5 Is Required for Downregulation of the Meiotic Recombination Checkpoint. Cells 2021, 10, 2561. https://doi.org/10.3390/cells10102561
González-Arranz S, Acosta I, Carballo JA, Santos B, San-Segundo PA. The N-Terminal Region of the Polo Kinase Cdc5 Is Required for Downregulation of the Meiotic Recombination Checkpoint. Cells. 2021; 10(10):2561. https://doi.org/10.3390/cells10102561
Chicago/Turabian StyleGonzález-Arranz, Sara, Isabel Acosta, Jesús A. Carballo, Beatriz Santos, and Pedro A. San-Segundo. 2021. "The N-Terminal Region of the Polo Kinase Cdc5 Is Required for Downregulation of the Meiotic Recombination Checkpoint" Cells 10, no. 10: 2561. https://doi.org/10.3390/cells10102561
APA StyleGonzález-Arranz, S., Acosta, I., Carballo, J. A., Santos, B., & San-Segundo, P. A. (2021). The N-Terminal Region of the Polo Kinase Cdc5 Is Required for Downregulation of the Meiotic Recombination Checkpoint. Cells, 10(10), 2561. https://doi.org/10.3390/cells10102561