Progressive Domain Segregation in Early Embryonic Development and Underlying Correlation to Genetic and Epigenetic Changes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Overall Segregation Ratio
2.2. 3D Chromatin Structure Modeling
2.3. Domain Segregation Ratio Calculation
2.4. Distance-Dependent Segregation Ratio Calculation
2.5. Open-Sea Methylation Difference Index (MDI) between Forest and Prairie
2.6. Overall Relative Segregation Ratios
2.7. Significantly More Segregated or More Mixed Domains
2.8. Compartment Index Calculation
2.9. Gene Function Analysis
2.10. Identification of Lineage-Specific Genes
2.11. Comparison of Segregation Extent between SINE and CpG Density
3. Results
3.1. Domain Segregation in Early Embryonic Development
3.2. Compartment Changes Related to Domain Segregation
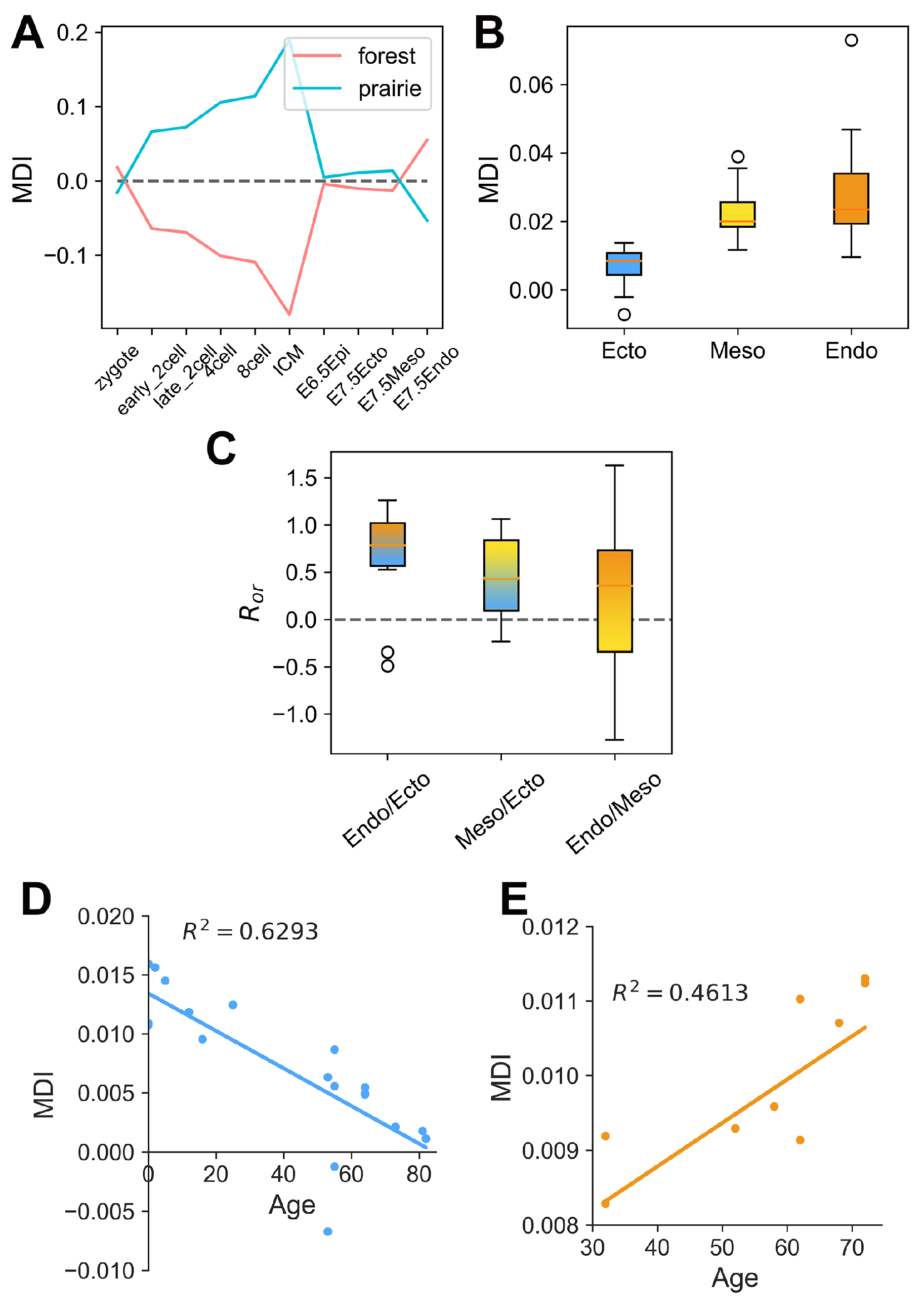
3.3. The Association between Domain Segregation and DNA Methylation
3.4. ZGA and Associated 3D Genome Architecture Change
3.5. Implantation-Related Domain Mixing in Differentiation
4. Discussion
4.1. Sequence-Based Chromatin Domain Segregation
4.2. The Association between Transcription Inhibition and Genome Architecture
4.3. Other Factors That May Relate to Chromatin Structure Change
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zheng, H.; Xie, W. The role of 3D genome organization in development and cell differentiation. Nat. Rev. Mol. Cell Biol. 2019, 20, 535–550. [Google Scholar] [CrossRef]
- Xu, Q.; Xie, W. Epigenome in early mammalian development: Inheritance, reprogramming and establishment. Trends Cell Biol. 2018, 28, 237–253. [Google Scholar] [CrossRef]
- Burton, A.; Torres-Padilla, M.-E. Chromatin dynamics in the regulation of cell fate allocation during early embryogenesis. Nat. Rev. Mol. Cell Biol. 2014, 15, 723–735. [Google Scholar] [CrossRef]
- Du, Z.; Zheng, H.; Huang, B.; Ma, R.; Wu, J.; Zhang, X.; He, J.; Xiang, Y.; Wang, Q.; Li, Y.; et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature 2017, 547, 232–235. [Google Scholar] [CrossRef]
- Ke, Y.; Xu, Y.; Chen, X.; Feng, S.; Liu, Z.; Sun, Y.; Yao, X.; Li, F.; Zhu, W.; Gao, L.; et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell 2017, 170, 367–381.e20. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ke, Y.; Wu, K.; Zhao, H.; Sun, Y.; Gao, L.; Liu, Z.; Zhang, J.; Tao, W.; Hou, Z.; et al. Key role for CTCF in establishing chromatin structure in human embryos. Nature 2019, 576, 306–310. [Google Scholar] [CrossRef]
- Morgan, H.D.; Santos, F.; Green, K.; Dean, W.; Reik, W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005, 14, R47–R58. [Google Scholar] [CrossRef] [Green Version]
- Burton, A.; Torres-Padilla, M.-E. Epigenetic reprogramming and development: A unique heterochromatin organization in the preimplantation mouse embryo. Brief. Funct. Genom. 2010, 9, 444–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Liu, X.; Gao, Y.; Yang, L.; Li, C.; Liu, W.; Chen, C.; Kou, X.; Zhao, Y.; Chen, J.; et al. Reprogramming of H3K9me3-dependent heterochromatin during mammalian embryo development. Nat. Cell Biol. 2018, 20, 620–631. [Google Scholar] [CrossRef]
- Liu, X.; Wang, C.; Liu, W.; Li, J.; Li, C.; Kou, X.; Chen, J.; Zhao, Y.; Gao, H.; Wang, H.; et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature 2016, 537, 558–562. [Google Scholar] [CrossRef]
- Dahl, J.A.; Jung, I.; Aanes, H.; Greggains, G.D.; Manaf, A.; Lerdrup, M.; Li, G.; Kuan, S.; Li, B.; Lee, A.Y.; et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537, 548–552. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zheng, H.; Huang, B.; Li, W.; Xiang, Y.; Peng, X.; Ming, J.; Wu, X.; Zhang, Y.; Xu, Q.; et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016, 537, 553–557. [Google Scholar] [CrossRef] [PubMed]
- Zheng, H.; Huang, B.; Zhang, B.; Xiang, Y.; Du, Z.; Xu, Q.; Li, Y.; Wang, Q.; Ma, J.; Peng, X.; et al. Resetting epigenetic memory by reprogramming of histone modifications in mammals. Mol. Cell 2016, 63, 1066–1079. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef]
- Lee, H.J.; Hore, T.A.; Reik, W. Reprogramming the methylome: Erasing memory and creating diversity. Cell Stem Cell 2014, 14, 710–719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, F.; Liu, Y.; Inoue, A.; Suzuki, T.; Zhao, K.; Zhang, Y. Establishing chromatin regulatory landscape during mouse preimplantation development. Cell 2016, 165, 1375–1388. [Google Scholar] [CrossRef] [Green Version]
- Jukam, D.; Shariati, S.A.M.; Skotheim, J.M. Zygotic genome activation in vertebrates. Dev. Cell 2017, 42, 316–332. [Google Scholar] [CrossRef]
- Arnold, S.J.; Robertson, E.J. Making a commitment: Cell lineage allocation and axis patterning in the early mouse embryo. Nat. Rev. Mol. Cell Biol. 2009, 10, 91–103. [Google Scholar] [CrossRef]
- Lawson, K.A.; Meneses, J.J.; Pedersen, R.A. Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 1991, 113, 891–911. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xiang, Y.; Yin, Q.; Du, Z.; Peng, X.; Wang, Q.; Fidalgo, M.; Xia, W.; Li, Y.; Zhao, Z.; et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 2018, 50, 96–105. [Google Scholar] [CrossRef]
- Liu, S.; Zhang, L.; Quan, H.; Tian, H.; Meng, L.; Yang, L.; Feng, H.; Gao, Y.Q. From 1D sequence to 3D chromatin dynamics and cellular functions: A phase separation perspective. Nucleic Acids Res. 2018, 46, 9367–9383. [Google Scholar] [CrossRef] [Green Version]
- Servant, N.; Varoquaux, N.; Lajoie, B.R.; Viara, E.; Chen, C.-J.; Vert, J.-P.; Heard, E.; Dekker, J.; Barillot, E. HiC-Pro: An optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015, 16, 259. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.J.; Meng, L.; Liu, S.; Zhang, L.; Cai, X.; Gao, Y.Q. Structural modeling of chromatin integrates genome features and reveals chromosome folding principle. Sci. Rep. 2017, 7, 2818. [Google Scholar] [CrossRef]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef] [Green Version]
- Svoboda, P.; Franke, V.; Schultz, R.M. Sculpting the transcriptome during the oocyte-to-embryo transition in mouse. Curr. Top. Dev. Biol. 2015, 113, 305–349. [Google Scholar] [CrossRef]
- Bedzhov, I.; Zernicka-Goetz, M. Self-organizing properties of mouse pluripotent cells initiate morphogenesis upon implantation. Cell 2014, 156, 1032–1044. [Google Scholar] [CrossRef] [Green Version]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Falk, M.; Feodorova, Y.; Naumova, N.; Imakaev, M.; Lajoie, B.R.; Leonhardt, H.; Joffe, B.; Dekker, J.; Fudenberg, G.; Solovei, I.; et al. Heterochromatin drives compartmentalization of inverted and conventional nuclei. Nature 2019, 570, 395–399. [Google Scholar] [CrossRef]
- Tian, H.; Yang, Y.; Liu, S.; Quan, H.; Gao, Y.Q. Toward an understanding of the relation between gene regulation and 3D genome organization. Quant. Biol. 2020, 8, 295–311. [Google Scholar] [CrossRef]
- Sandoval, J.; Heyn, H.; Moran, S.; Serra-Musach, J.; Pujana, M.A.; Bibikova, M.; Esteller, M. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics 2011, 6, 692–702. [Google Scholar] [CrossRef]
- Schultz, M.D.; He, Y.; Whitaker, J.W.; Hariharan, M.; Mukamel, E.A.; Leung, D.; Rajagopal, N.; Nery, J.R.; Urich, M.A.; Chen, H.; et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature 2015, 523, 212–216. [Google Scholar] [CrossRef]
- Lister, R.; Mukamel, E.A.; Nery, J.R.; Urich, M.; Puddifoot, C.A.; Johnson, N.D.; Lucero, J.; Huang, Y.; Dwork, A.J.; Schultz, M.D.; et al. Global epigenomic reconfiguration during mammalian brain development. Science 2013, 341, 1237905. [Google Scholar] [CrossRef] [Green Version]
- The ENCODE Project Consortium, An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [CrossRef]
- Schmitt, A.D.; Hu, M.; Jung, I.; Xu, Z.; Qiu, Y.; Tan, C.L.; Li, Y.; Lin, S.; Lin, Y.; Barr, C.L.; et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016, 17, 2042–2059. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Yang, Y.; Tian, H.; Quan, H.; Liu, S.; Zhang, L.; Gao, Y.Q. Domain segregated 3D chromatin structure and segmented DNA methylation in carcinogenesis. bioRvix 2020. [Google Scholar] [CrossRef]
- Zhou, W.; Dinh, H.Q.; Ramjan, Z.; Weisenberger, D.J.; Nicolet, C.M.; Shen, H.; Laird, P.W.; Berman, B.P. DNA methylation loss in late-replicating domains is linked to mitotic cell division. Nat. Genet. 2018, 50, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Ming, X.; Zhang, Z.; Zou, Z.; Lv, C.; Dong, Q.; He, Q.; Yi, Y.; Li, Y.; Wang, H.; Zhu, B. Kinetics and mechanisms of mitotic inheritance of DNA methylation and their roles in aging-associated methylome deterioration. Cell Res. 2020, 30, 980–996. [Google Scholar] [CrossRef] [PubMed]
- Borsos, M.; Perricone, S.M.; Schauer, T.; Pontabry, J.; de Luca, K.L.; de Vries, S.S.; Ruiz-Morales, E.R.; Torres-Padilla, M.-E.; Kind, J. Genome–lamina interactions are established de novo in the early mouse embryo. Nature 2019, 569, 729–733. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2008, 4, 44–57. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012, 16, 284–287. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Liu, C.; Biechele, S.; Zhu, Q.; Song, L.; Lanner, F.; Jing, N.; Rossant, J. Location of transient ectodermal progenitor potential in mouse development. Development 2013, 140, 4533–4543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, J.Y.; Chang, L.; Li, T.; Wang, T.; Yin, Y.; Zhan, G.; Han, X.; Zhang, K.; Tao, Y.; Percharde, M.; et al. Homotypic clustering of L1 and B1/Alu repeats compartmentalizes the 3D genome. Cell Res. 2021, 31, 613–630. [Google Scholar] [CrossRef] [PubMed]
- Quan, H.; Yang, Y.; Liu, S.; Tian, H.; Xue, Y.; Gao, Y.Q. Chromatin structure changes during various processes from a DNA sequence view. Curr. Opin. Struct. Biol. 2020, 62, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chandra, T.; Ewels, P.A.; Schoenfelder, S.; Furlan-Magaril, M.; Wingett, S.W.; Kirschner, K.; Thuret, J.-Y.; Andrews, S.; Fraser, P.; Reik, W. Global reorganization of the nuclear landscape in senescent cells. Cell Rep. 2015, 10, 471–483. [Google Scholar] [CrossRef] [PubMed]
- DEEP Consortium; Salhab, A.; Nordström, K.; Gasparoni, G.; Kattler, K.; Ebert, P.; Ramirez, F.; Arrigoni, L.; Müller, F.; Polansky, J.K.; et al. A comprehensive analysis of 195 DNA methylomes reveals shared and cell-specific features of partially methylated domains. Genome Biol. 2018, 19, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsichlaki, E.; FitzHarris, G. Nucleus downscaling in mouse embryos is regulated by cooperative developmental and geometric programs. Sci. Rep. 2016, 6, 28040. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, R.N.; Chen, P.; Levy, D.L. Recent advances in understanding nuclear size and shape. Nucleus 2016, 7, 167–186. [Google Scholar] [CrossRef] [Green Version]
- Anifandis, G.; Messini, C.I.; Dafopoulos, K.; Messinis, I.E. Genes and conditions controlling mammalian pre- and post-implantation embryo development. Curr. Genomics 2015, 16, 32–46. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Quan, H.; Tian, H.; Liu, S.; Xue, Y.; Zhang, Y.; Xie, W.; Gao, Y.Q. Progressive Domain Segregation in Early Embryonic Development and Underlying Correlation to Genetic and Epigenetic Changes. Cells 2021, 10, 2521. https://doi.org/10.3390/cells10102521
Quan H, Tian H, Liu S, Xue Y, Zhang Y, Xie W, Gao YQ. Progressive Domain Segregation in Early Embryonic Development and Underlying Correlation to Genetic and Epigenetic Changes. Cells. 2021; 10(10):2521. https://doi.org/10.3390/cells10102521
Chicago/Turabian StyleQuan, Hui, Hao Tian, Sirui Liu, Yue Xue, Yu Zhang, Wei Xie, and Yi Qin Gao. 2021. "Progressive Domain Segregation in Early Embryonic Development and Underlying Correlation to Genetic and Epigenetic Changes" Cells 10, no. 10: 2521. https://doi.org/10.3390/cells10102521
APA StyleQuan, H., Tian, H., Liu, S., Xue, Y., Zhang, Y., Xie, W., & Gao, Y. Q. (2021). Progressive Domain Segregation in Early Embryonic Development and Underlying Correlation to Genetic and Epigenetic Changes. Cells, 10(10), 2521. https://doi.org/10.3390/cells10102521





