Ariadne’s Thread in the Developing Cerebral Cortex: Mechanisms Enabling the Guiding Role of the Radial Glia Basal Process during Neuron Migration
Abstract
:1. Introduction
2. Radial Migration in the Cerebral Cortex
3. Radial Glia-Dependent Mechanisms Regulating Neuron Migration
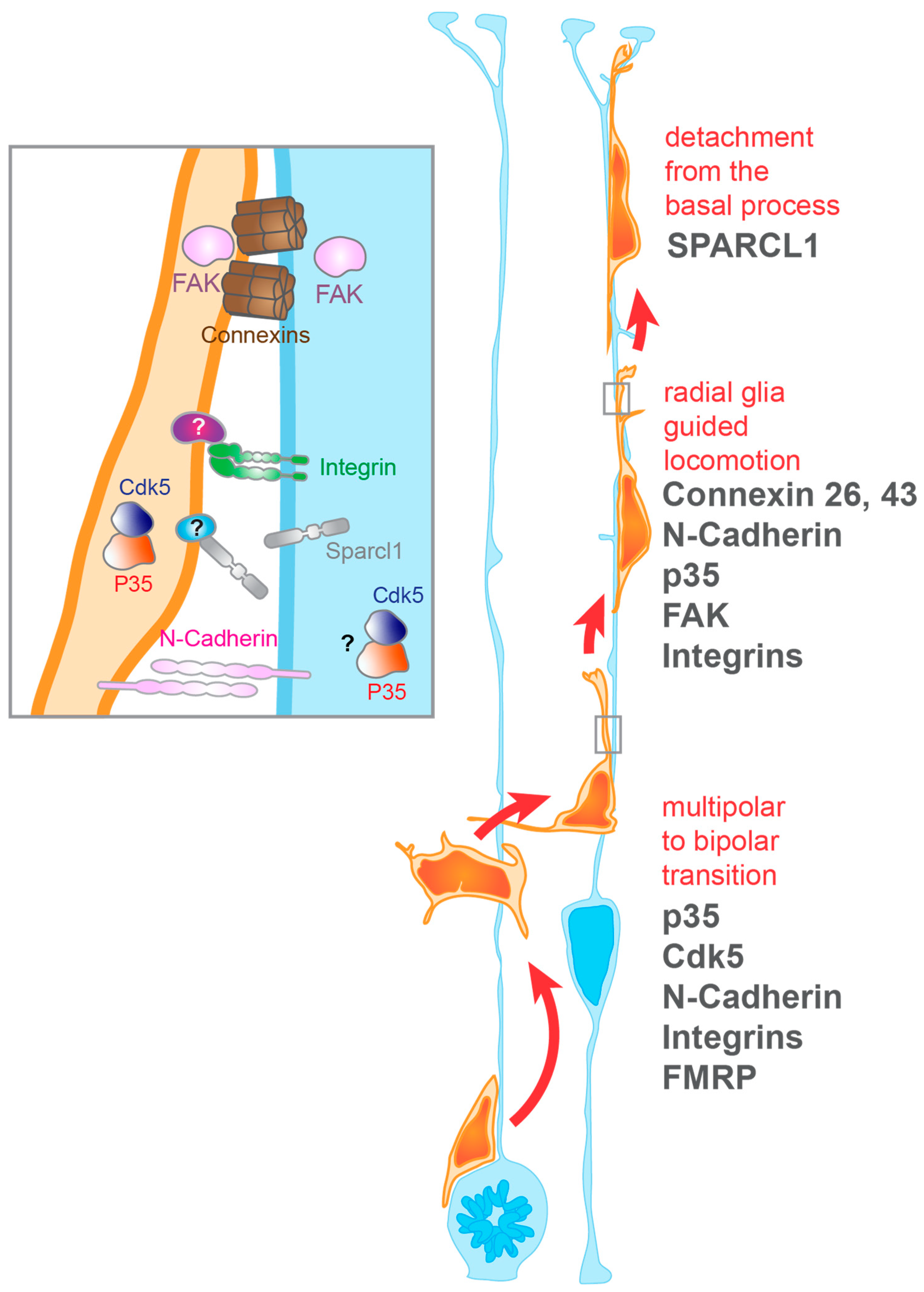
3.1. Establishment and Maintenance of Neuron-Radial Glia Interactions During Migration
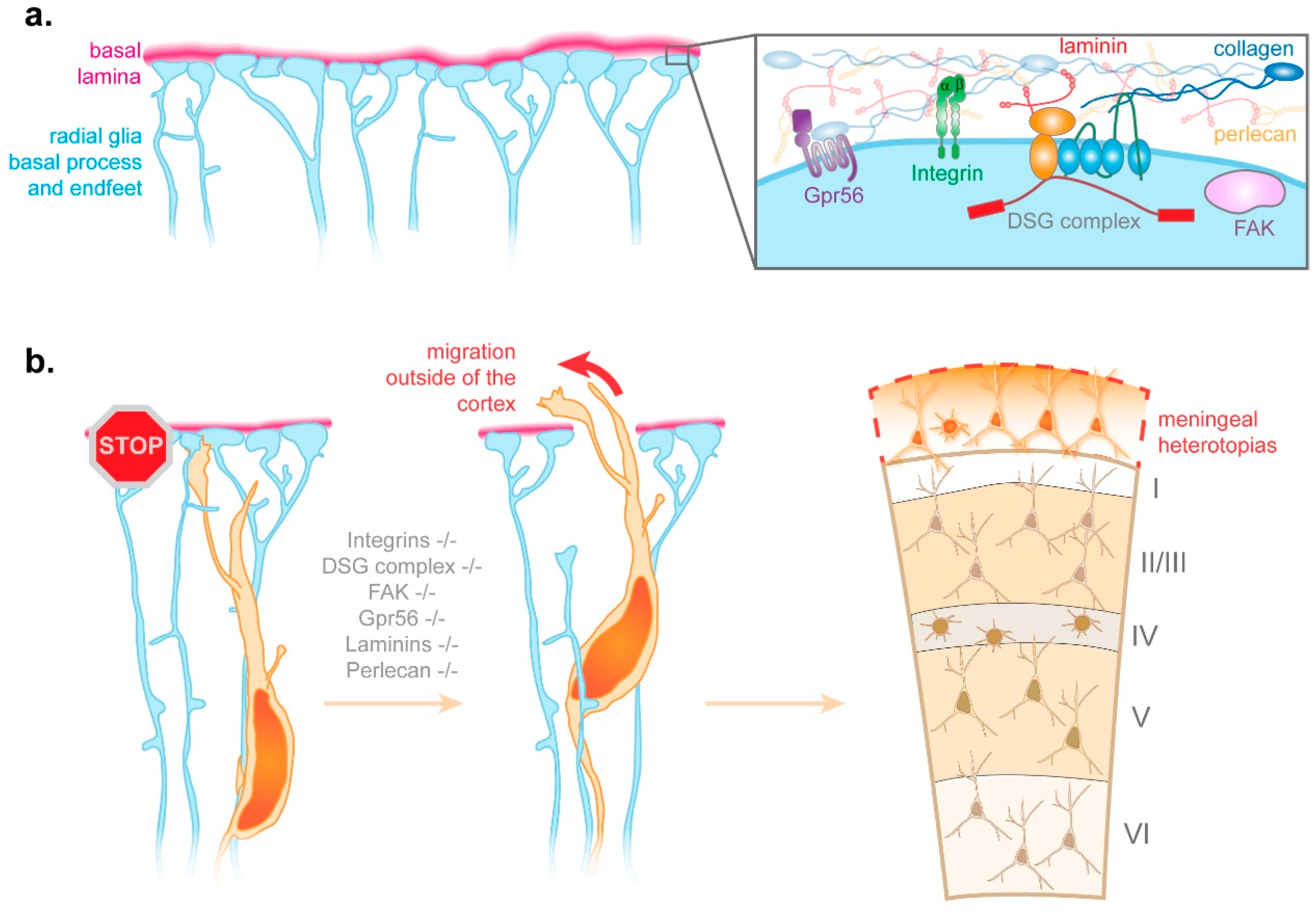
3.1.1. Integrins
3.1.2. N-Cadherin
3.1.3. Gap Junctions
3.1.4. p35
3.2. Termination of Neuronal Migration
3.2.1. SPARC Like-1
3.2.2. Radial Glia Endfeet-Basal Lamina Connection
4. Regulation of the Morphology of the Radial Glia Basal Process
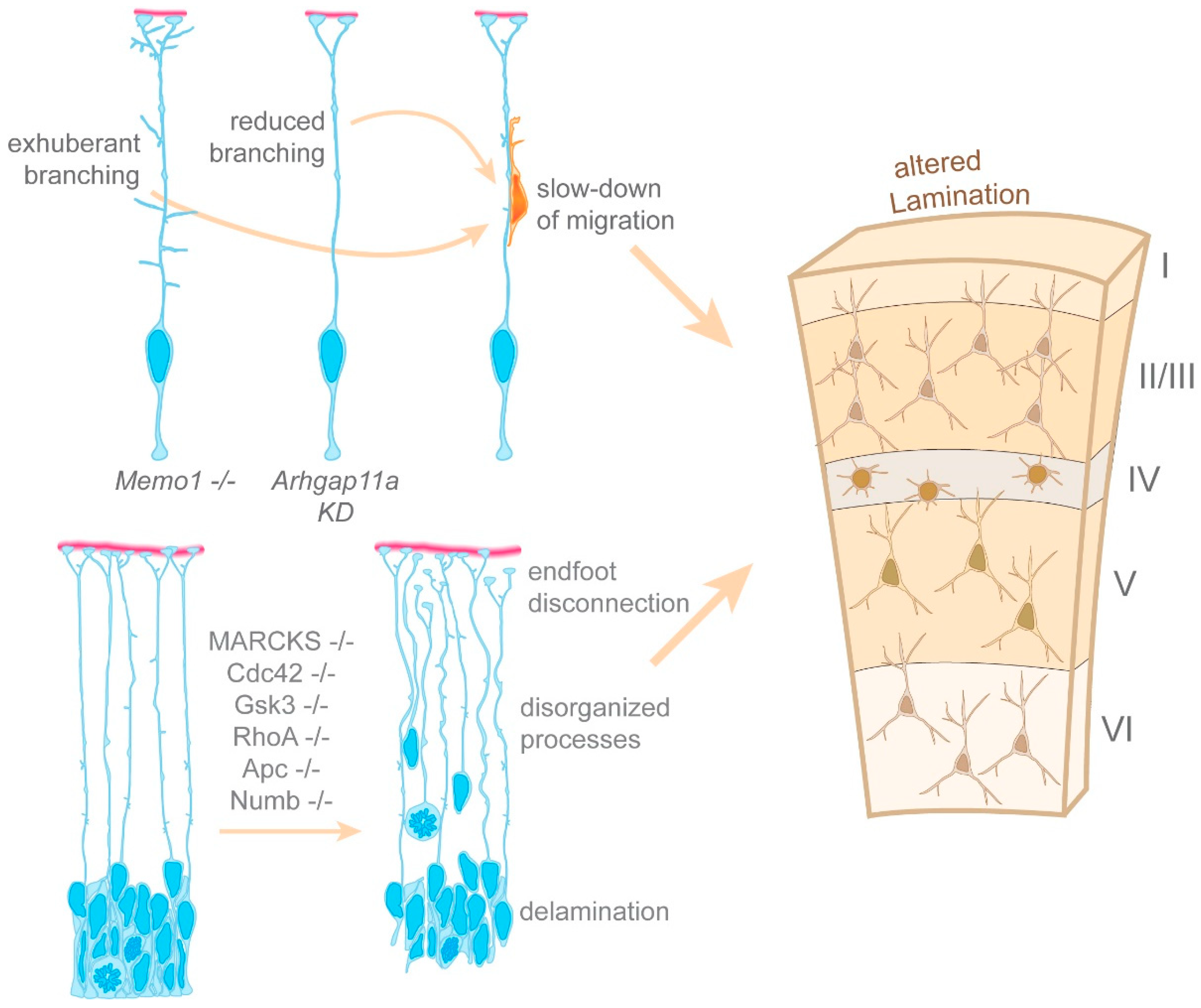
4.1. Generation of the Radial Glia Basal Process
4.2. The Morphology of the Radial Glia Basal Process Relies on a Microtubule Network
4.3. Radial Glia Utilize mRNA Transport and Local Protein Synthesis to Locally Deliver Key Regulators of Basal Process Morphology
4.4. Regulation of Cell Polarity in Radial Glia
5. Conclusions and Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Faheem, M.; Naseer, M.I.; Rasool, M.; Chaudhary, A.G.; Kumosani, T.A.; Ilyas, A.M.; Pushparaj, P.; Ahmed, F.; Algahtani, H.A.; Al-Qahtani, M.H.; et al. Molecular genetics of human primary microcephaly: An overview. BMC Med. Genom. 2015, 8 (Suppl. 1), S4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferraris, P.; Cochet, M.; Hamel, R.; Gladwyn-Ng, I.; Alfano, C.; Diop, F.; Garcia, D.; Talignani, L.; Montero-Menei, C.N.; Nougairede, A.; et al. Zika virus differentially infects human neural progenitor cells according to their state of differentiation and dysregulates neurogenesis through the Notch pathway. Emerg. Microbes Infect. 2019, 8, 1003–1016. [Google Scholar] [CrossRef] [PubMed]
- Souza, B.S.; Sampaio, G.L.; Pereira, C.S.; Campos, G.S.; Sardi, S.I.; Freitas, L.A.; Figueira, C.P.; Paredes, B.D.; Nonaka, C.K.; Azevedo, C.M.; et al. Zika virus infection induces mitosis abnormalities and apoptotic cell death of human neural progenitor cells. Sci. Rep. 2016, 6, 39775. [Google Scholar] [CrossRef]
- Parikshak, N.N.; Luo, R.; Zhang, A.; Won, H.; Lowe, J.K.; Chandran, V.; Horvath, S.; Geschwind, D.H. Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism. Cell 2013, 155, 1008–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Willsey, A.J.; Sanders, S.J.; Li, M.; Dong, S.; Tebbenkamp, A.T.; Muhle, R.A.; Reilly, S.K.; Lin, L.; Fertuzinhos, S.; Miller, J.A.; et al. Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism. Cell 2013, 155, 997–1007. [Google Scholar] [CrossRef] [Green Version]
- Stoner, R.; Chow, M.L.; Boyle, M.P.; Sunkin, S.M.; Mouton, P.R.; Roy, S.; Wynshaw-Boris, A.; Colamarino, S.A.; Lein, E.S.; Courchesne, E. Patches of disorganization in the neocortex of children with autism. N. Engl. J. Med. 2014, 370, 1209–1219. [Google Scholar] [CrossRef] [Green Version]
- Sacco, R.; Cacci, E.; Novarino, G. Neural stem cells in neuropsychiatric disorders. Curr. Opin. Neurobiol. 2017, 48, 131–138. [Google Scholar] [CrossRef]
- Rakic, P. Mode of cell migration to the superficial layers of fetal monkey neocortex. J. Comp. Neurol. 1972, 145, 61–83. [Google Scholar] [CrossRef]
- Edmondson, J.C.; Hatten, M.E. Glial-guided granule neuron migration in vitro: A high-resolution time-lapse video microscopic study. J. Neurosci. 1987, 7, 1928–1934. [Google Scholar] [CrossRef]
- Noctor, S.C.; Flint, A.C.; Weissman, T.A.; Dammerman, R.S.; Kriegstein, A.R. Neurons derived from radial glial cells establish radial units in neocortex. Nature 2001, 409, 714–720. [Google Scholar] [CrossRef]
- Malatesta, P.; Hartfuss, E.; Götz, M. Isolation of radial glial cells by fluorescent-activated cell sorting reveals neuronal lineage. Development 2000, 127, 5253–5263. [Google Scholar]
- Kon, E.; Cossard, A.; Jossin, Y. Neuronal Polarity in the Embryonic Mammalian Cerebral Cortex. Front. Cell. Neurosci. 2017, 11, 163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torii, M.; Hashimoto-Torii, K.; Levitt, P.; Rakic, P. Integration of neuronal clones in the radial cortical columns by EphA and ephrin-A signalling. Nature 2009, 461, 524–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, C.G.; Peyre, E.; Nguyen, L. Cell migration promotes dynamic cellular interactions to control cerebral cortex morphogenesis. Nat. Rev. Neurosci. 2019, 20, 318–329. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, B.; Brunstrom, J.E.; Grutzendler, J.; Wong, R.O.; Pearlman, A.L. Two modes of radial migration in early development of the cerebral cortex. Nat. Neurosci. 2001, 4, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Hansen, D.V.; Lui, J.H.; Parker, P.R.; Kriegstein, A.R. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature 2010, 464, 554–561. [Google Scholar] [CrossRef] [PubMed]
- Smart, I.H.; Dehay, C.; Giroud, P.; Berland, M.; Kennedy, H. Unique morphological features of the proliferative zones and postmitotic compartments of the neural epithelium giving rise to striate and extrastriate cortex in the monkey. Cereb. Cortex 2002, 12, 37–53. [Google Scholar] [CrossRef]
- Meller, K. Scanning electron microscope studies on the development of the nervous system in vivo and in vitro. Int. Rev. Cytol. 1979, 56, 23–56. [Google Scholar] [CrossRef]
- Hatten, M.E. Riding the glial monorail: A common mechanism for glial-guided neuronal migration in different regions of the developing mammalian brain. Trends Neurosci. 1990, 13, 179–184. [Google Scholar] [CrossRef]
- Anton, E.S.; Kreidberg, J.A.; Rakic, P. Distinct functions of alpha3 and alpha(v) integrin receptors in neuronal migration and laminar organization of the cerebral cortex. Neuron 1999, 22, 277–289. [Google Scholar] [CrossRef] [Green Version]
- Belvindrah, R.; Graus-Porta, D.; Goebbels, S.; Nave, K.A.; Muller, U. Beta1 integrins in radial glia but not in migrating neurons are essential for the formation of cell layers in the cerebral cortex. J. Neurosci. 2007, 27, 13854–13865. [Google Scholar] [CrossRef] [PubMed]
- Kawauchi, T.; Sekine, K.; Shikanai, M.; Chihama, K.; Tomita, K.; Kubo, K.; Nakajima, K.; Nabeshima, Y.; Hoshino, M. Rab GTPases-dependent endocytic pathways regulate neuronal migration and maturation through N-cadherin trafficking. Neuron 2010, 67, 588–602. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Funahashi, Y.; Watanabe, T.; Takano, T.; Nakamuta, S.; Namba, T.; Kaibuchi, K. Radial Glial Cell-Neuron Interaction Directs Axon Formation at the Opposite Side of the Neuron from the Contact Site. J. Neurosci. 2015, 35, 14517–14532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawauchi, T. Cellullar insights into cerebral cortical development: Focusing on the locomotion mode of neuronal migration. Front. Cell. Neurosci. 2015, 9, 394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- La Fata, G.; Gartner, A.; Dominguez-Iturza, N.; Dresselaers, T.; Dawitz, J.; Poorthuis, R.B.; Averna, M.; Himmelreich, U.; Meredith, R.M.; Achsel, T.; et al. FMRP regulates multipolar to bipolar transition affecting neuronal migration and cortical circuitry. Nat. Neurosci. 2014, 17, 1693–1700. [Google Scholar] [CrossRef] [PubMed]
- Bassell, G.J.; Warren, S.T. Fragile X syndrome: Loss of local mRNA regulation alters synaptic development and function. Neuron 2008, 60, 201–214. [Google Scholar] [CrossRef] [Green Version]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef] [Green Version]
- Elias, L.A.; Wang, D.D.; Kriegstein, A.R. Gap junction adhesion is necessary for radial migration in the neocortex. Nature 2007, 448, 901–907. [Google Scholar] [CrossRef]
- Cina, C.; Maass, K.; Theis, M.; Willecke, K.; Bechberger, J.F.; Naus, C.C. Involvement of the cytoplasmic C-terminal domain of connexin43 in neuronal migration. J. Neurosci. 2009, 29, 2009–2021. [Google Scholar] [CrossRef] [Green Version]
- Valiente, M.; Ciceri, G.; Rico, B.; Marin, O. Focal adhesion kinase modulates radial glia-dependent neuronal migration through connexin-26. J. Neurosci. 2011, 31, 11678–11691. [Google Scholar] [CrossRef] [Green Version]
- Ohshima, T.; Hirasawa, M.; Tabata, H.; Mutoh, T.; Adachi, T.; Suzuki, H.; Saruta, K.; Iwasato, T.; Itohara, S.; Hashimoto, M.; et al. Cdk5 is required for multipolar-to-bipolar transition during radial neuronal migration and proper dendrite development of pyramidal neurons in the cerebral cortex. Development 2007, 134, 2273–2282. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gupta, A.; Sanada, K.; Miyamoto, D.T.; Rovelstad, S.; Nadarajah, B.; Pearlman, A.L.; Brunstrom, J.; Tsai, L.H. Layering defect in p35 deficiency is linked to improper neuronal-glial interaction in radial migration. Nat. Neurosci. 2003, 6, 1284–1291. [Google Scholar] [CrossRef] [PubMed]
- Rakic, S.; Kanatani, S.; Hunt, D.; Faux, C.; Cariboni, A.; Chiara, F.; Khan, S.; Wansbury, O.; Howard, B.; Nakajima, K.; et al. Cdk5 phosphorylation of ErbB4 is required for tangential migration of cortical interneurons. Cereb. Cortex 2015, 25, 991–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogawa, M.; Miyata, T.; Nakajimat, K.; Yagyu, K.; Seike, M.; Ikenaka, K.; Yamamoto, H.; Mikoshibat, K. The reeler gene-associated antigen on cajal-retzius neurons is a crucial molecule for laminar organization of cortical neurons. Neuron 1995, 14, 899–912. [Google Scholar] [CrossRef] [Green Version]
- Gongidi, V.; Ring, C.; Moody, M.; Brekken, R.; Sage, E.H.; Rakic, P.; Anton, E.S. SPARC-like 1 regulates the terminal phase of radial glia-guided migration in the cerebral cortex. Neuron 2004, 41, 57–69. [Google Scholar] [CrossRef] [Green Version]
- Halfter, W.; Dong, S.; Yip, Y.P.; Willem, M.; Mayer, U. A critical function of the pial basement membrane in cortical histogenesis. J. Neurosci. 2002, 22, 6029–6040. [Google Scholar] [CrossRef]
- Haubst, N.; Georges-Labouesse, E.; De Arcangelis, A.; Mayer, U.; Gotz, M. Basement membrane attachment is dispensable for radial glial cell fate and for proliferation, but affects positioning of neuronal subtypes. Development 2006, 133, 3245–3254. [Google Scholar] [CrossRef] [Green Version]
- Pilaz, L.-J.; Joshi, K.; Liu, J.; Tsunekawa, Y.; Alsina, F.C.; Sethi, S.; Suzuki, I.K.; Vanderhaeghen, P.; Polleux, F.; Silver, D.L. Subcellular mRNA localization and local translation of Arhgap11a in radial glial cells regulates cortical development. bioRxiv 2020. [Google Scholar] [CrossRef]
- Myshrall, T.D.; Moore, S.A.; Ostendorf, A.P.; Satz, J.S.; Kowalczyk, T.; Nguyen, H.; Daza, R.A.; Lau, C.; Campbell, K.P.; Hevner, R.F. Dystroglycan on radial glia end feet is required for pial basement membrane integrity and columnar organization of the developing cerebral cortex. J. Neuropathol. Exp. Neurol. 2012, 71, 1047–1063. [Google Scholar] [CrossRef] [Green Version]
- Radakovits, R.; Barros, C.S.; Belvindrah, R.; Patton, B.; Müller, U. Regulation of radial glial survival by signals from the meninges. J. Neurosci. 2009, 29, 7694–7705. [Google Scholar] [CrossRef] [Green Version]
- Beggs, H.E.; Schahin-Reed, D.; Zang, K.; Goebbels, S.; Nave, K.A.; Gorski, J.; Jones, K.R.; Sretavan, D.; Reichardt, L.F. FAK deficiency in cells contributing to the basal lamina results in cortical abnormalities resembling congenital muscular dystrophies. Neuron 2003, 40, 501–514. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Jin, Z.; Koirala, S.; Bu, L.; Xu, L.; Hynes, R.O.; Walsh, C.A.; Corfas, G.; Piao, X. GPR56 regulates pial basement membrane integrity and cortical lamination. J. Neurosci. 2008, 28, 5817–5826. [Google Scholar] [CrossRef] [Green Version]
- Pilaz, L.J.; Silver, D.L. Moving messages in the developing brain-emerging roles for mRNA transport and local translation in neural stem cells. FEBS Lett. 2017, 591, 1526–1539. [Google Scholar] [CrossRef] [Green Version]
- Pilaz, L.J.; Lennox, A.L.; Rouanet, J.P.; Silver, D.L. Dynamic mRNA Transport and Local Translation in Radial Glial Progenitors of the Developing Brain. Curr. Biol. 2016, 26, 3383–3392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsunekawa, Y.; Kikkawa, T.; Osumi, N. Asymmetric inheritance of Cyclin D2 maintains proliferative neural stem/progenitor cells: A critical event in brain development and evolution. Dev. Growth Differ. 2014, 56, 349–357. [Google Scholar] [CrossRef] [PubMed]
- Tsunekawa, Y.; Britto, J.M.; Takahashi, M.; Polleux, F.; Tan, S.S.; Osumi, N. Cyclin D2 in the basal process of neural progenitors is linked to non-equivalent cell fates. EMBO J. 2012, 31, 1879–1892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Duan, M.; Song, L.; Zhang, W.; Hu, X.; Zhao, S.; Chen, S. Morphological changes of radial glial cells during mouse embryonic development. Brain Res. 2015, 1599, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Kosodo, Y.; Toida, K.; Dubreuil, V.; Alexandre, P.; Schenk, J.; Kiyokage, E.; Attardo, A.; Mora-Bermudez, F.; Arii, T.; Clarke, J.D.; et al. Cytokinesis of neuroepithelial cells can divide their basal process before anaphase. EMBO J. 2008, 27, 3151–3163. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, L.; Bershteyn, M.; Paredes, M.F.; Kriegstein, A.R. Dynamic behaviour of human neuroepithelial cells in the developing forebrain. Nat. Commun. 2017, 8, 14167. [Google Scholar] [CrossRef] [Green Version]
- Noctor, S.C.; Martinez-Cerdeño, V.; Ivic, L.; Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004, 7, 136–144. [Google Scholar] [CrossRef]
- Shitamukai, A.; Konno, D.; Matsuzaki, F. Oblique radial glial divisions in the developing mouse neocortex induce self-renewing progenitors outside the germinal zone that resemble primate outer subventricular zone progenitors. J. Neurosci. 2011, 31, 3683–3695. [Google Scholar] [CrossRef] [PubMed]
- Benda, P.; Lightbody, J.; Sato, G.; Levine, L.; Sweet, W. Differentiated rat glial cell strain in tissue culture. Science 1968, 161, 370–371. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Berlin, Y.; Hart, R.P.; Grumet, M. Microtubules are critical for radial glial morphology: Possible regulation by MAPs and MARKs. Glia 2003, 44, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Tsai, J.W.; Lian, W.N.; Kemal, S.; Kriegstein, A.R.; Vallee, R.B. Kinesin 3 and cytoplasmic dynein mediate interkinetic nuclear migration in neural stem cells. Nat. Neurosci. 2010, 13, 1463–1471. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, N.; Plestant, C.; Yabuno-Nakagawa, K.; Li, J.; Lee, J.; Huang, C.W.; Lee, A.; Krupa, O.; Adhikari, A.; Thompson, S.; et al. Memo1-Mediated Tiling of Radial Glial Cells Facilitates Cerebral Cortical Development. Neuron 2019, 103, 836–852.e5. [Google Scholar] [CrossRef]
- Coquand, L.; Victoria, G.S.; Tata, A.; Brault, J.B.; Guimiot, F.; Fraisier, V.; Baffet, A.D. A dendritic-like microtubule network is organized from swellings of the basal fiber in neural progenitors. bioRxiv 2020. [Google Scholar] [CrossRef]
- Valenzuela, A.; Meservey, L.; Nguyen, H.; Fu, M.M. Golgi Outposts Nucleate Microtubules in Cells with Specialized Shapes. Trends Cell Biol. 2020, 30, 792–804. [Google Scholar] [CrossRef]
- Taverna, E.; Mora-Bermudez, F.; Strzyz, P.J.; Florio, M.; Icha, J.; Haffner, C.; Norden, C.; Wilsch-Brauninger, M.; Huttner, W.B. Non-canonical features of the Golgi apparatus in bipolar epithelial neural stem cells. Sci. Rep. 2016, 6, 21206. [Google Scholar] [CrossRef] [Green Version]
- Buxbaum, A.R.; Haimovich, G.; Singer, R.H. In the right place at the right time: Visualizing and understanding mRNA localization. Nat. Rev. Mol. Cell Biol. 2015, 16, 95–109. [Google Scholar] [CrossRef]
- Müller, P.M.; Rademacher, J.; Bagshaw, R.D.; Alp, K.M.; Giudice, G.; Heinrich, L.E.; Barth, C.; Eccles, R.L.; Sanchez-Castro, M.; Brandenburg, L.; et al. Spatial Organization of Rho GTPase signaling by RhoGEF/RhoGAP proteins. bioRxiv 2018. [Google Scholar] [CrossRef]
- Florio, M.; Heide, M.; Pinson, A.; Brandl, H.; Albert, M.; Winkler, S.; Wimberger, P.; Huttner, W.B.; Hiller, M. Evolution and cell-type specificity of human-specific genes preferentially expressed in progenitors of fetal neocortex. Elife 2018, 7. [Google Scholar] [CrossRef] [PubMed]
- Wong, H.H.; Lin, J.Q.; Strohl, F.; Roque, C.G.; Cioni, J.M.; Cagnetta, R.; Turner-Bridger, B.; Laine, R.F.; Harris, W.A.; Kaminski, C.F.; et al. RNA Docking and Local Translation Regulate Site-Specific Axon Remodeling In Vivo. Neuron 2017, 95, 852–868.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katayama, K.; Melendez, J.; Baumann, J.M.; Leslie, J.R.; Chauhan, B.K.; Nemkul, N.; Lang, R.A.; Kuan, C.Y.; Zheng, Y.; Yoshida, Y. Loss of RhoA in neural progenitor cells causes the disruption of adherens junctions and hyperproliferation. Proc. Natl. Acad. Sci. USA 2011, 108, 7607–7612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cappello, S.; Bohringer, C.R.; Bergami, M.; Conzelmann, K.K.; Ghanem, A.; Tomassy, G.S.; Arlotta, P.; Mainardi, M.; Allegra, M.; Caleo, M.; et al. A radial glia-specific role of RhoA in double cortex formation. Neuron 2012, 73, 911–924. [Google Scholar] [CrossRef] [Green Version]
- Cappello, S.; Attardo, A.; Wu, X.; Iwasato, T.; Itohara, S.; Wilsch-Brauninger, M.; Eilken, H.M.; Rieger, M.A.; Schroeder, T.T.; Huttner, W.B.; et al. The Rho-GTPase cdc42 regulates neural progenitor fate at the apical surface. Nat. Neurosci. 2006, 9, 1099–1107. [Google Scholar] [CrossRef]
- Yokota, Y.; Eom, T.Y.; Stanco, A.; Kim, W.Y.; Rao, S.; Snider, W.D.; Anton, E.S. Cdc42 and Gsk3 modulate the dynamics of radial glial growth, inter-radial glial interactions and polarity in the developing cerebral cortex. Development 2010, 137, 4101–4110. [Google Scholar] [CrossRef] [Green Version]
- Weimer, J.M.; Yokota, Y.; Stanco, A.; Stumpo, D.J.; Blackshear, P.J.; Anton, E.S. MARCKS modulates radial progenitor placement, proliferation and organization in the developing cerebral cortex. Development 2009, 136, 2965–2975. [Google Scholar] [CrossRef] [Green Version]
- Yokota, Y.; Kim, W.Y.; Chen, Y.; Wang, X.; Stanco, A.; Komuro, Y.; Snider, W.; Anton, E.S. The adenomatous polyposis coli protein is an essential regulator of radial glial polarity and construction of the cerebral cortex. Neuron 2009, 61, 42–56. [Google Scholar] [CrossRef] [Green Version]
- Rasin, M.R.; Gazula, V.R.; Breunig, J.J.; Kwan, K.Y.; Johnson, M.B.; Liu-Chen, S.; Li, H.S.; Jan, L.Y.; Jan, Y.N.; Rakic, P.; et al. Numb and Numbl are required for maintenance of cadherin-based adhesion and polarity of neural progenitors. Nat. Neurosci. 2007, 10, 819–827. [Google Scholar] [CrossRef]
- Li, H.S.; Wang, D.; Shen, Q.; Schonemann, M.D.; Gorski, J.A.; Jones, K.R.; Temple, S.; Jan, L.Y.; Jan, Y.N. Inactivation of Numb and Numblike in embryonic dorsal forebrain impairs neurogenesis and disrupts cortical morphogenesis. Neuron 2003, 40, 1105–1118. [Google Scholar] [CrossRef] [Green Version]
- Martinez-Martinez, M.A.; Ciceri, G.; Espinos, A.; Fernandez, V.; Marin, O.; Borrell, V. Extensive branching of radially-migrating neurons in the mammalian cerebral cortex. J. Comp. Neurol. 2019, 527, 1558–1576. [Google Scholar] [CrossRef] [PubMed]
- Cortay, V.; Delaunay, D.; Patti, D.; Gautier, E.; Doerflinger, N.; Giroud, P.; Knoblauch, K.; Huissoud, C.; Kennedy, H.; Dehay, C. Radial Migration Dynamics Is Modulated in a Laminar and Area-Specific Manner During Primate Corticogenesis. Front. Cell Dev. Biol. 2020, 8, 588814. [Google Scholar] [CrossRef] [PubMed]
- Florio, M.; Albert, M.; Taverna, E.; Namba, T.; Brandl, H.; Lewitus, E.; Haffner, C.; Sykes, A.; Wong, F.K.; Peters, J.; et al. Human-specific gene ARHGAP11B promotes basal progenitor amplification and neocortex expansion. Science 2015, 347, 1465–1470. [Google Scholar] [CrossRef] [PubMed]
- Heide, M.; Haffner, C.; Murayama, A.; Kurotaki, Y.; Shinohara, H.; Okano, H.; Sasaki, E.; Huttner, W.B. Human-specific ARHGAP11B increases size and folding of primate neocortex in the fetal marmoset. Science 2020, 369, 546–550. [Google Scholar] [CrossRef] [PubMed]
- Namba, T.; Doczi, J.; Pinson, A.; Xing, L.; Kalebic, N.; Wilsch-Brauninger, M.; Long, K.R.; Vaid, S.; Lauer, J.; Bogdanova, A.; et al. Human-Specific ARHGAP11B Acts in Mitochondria to Expand Neocortical Progenitors by Glutaminolysis. Neuron 2020, 105, 867–881.e9. [Google Scholar] [CrossRef]
- Pollen, A.A.; Nowakowski, T.J.; Chen, J.; Retallack, H.; Sandoval-Espinosa, C.; Nicholas, C.R.; Shuga, J.; Liu, S.J.; Oldham, M.C.; Diaz, A.; et al. Molecular identity of human outer radial glia during cortical development. Cell 2015, 163, 55–67. [Google Scholar] [CrossRef] [Green Version]
- Betizeau, M.; Cortay, V.; Patti, D.; Pfister, S.; Gautier, E.; Bellemin-Menard, A.; Afanassieff, M.; Huissoud, C.; Douglas, R.J.; Kennedy, H.; et al. Precursor diversity and complexity of lineage relationships in the outer subventricular zone of the primate. Neuron 2013, 80, 442–457. [Google Scholar] [CrossRef] [Green Version]
- Lancaster, M.A.; Renner, M.; Martin, C.A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meyerink, B.L.; Tiwari, N.K.; Pilaz, L.-J. Ariadne’s Thread in the Developing Cerebral Cortex: Mechanisms Enabling the Guiding Role of the Radial Glia Basal Process during Neuron Migration. Cells 2021, 10, 3. https://doi.org/10.3390/cells10010003
Meyerink BL, Tiwari NK, Pilaz L-J. Ariadne’s Thread in the Developing Cerebral Cortex: Mechanisms Enabling the Guiding Role of the Radial Glia Basal Process during Neuron Migration. Cells. 2021; 10(1):3. https://doi.org/10.3390/cells10010003
Chicago/Turabian StyleMeyerink, Brandon L., Neeraj K. Tiwari, and Louis-Jan Pilaz. 2021. "Ariadne’s Thread in the Developing Cerebral Cortex: Mechanisms Enabling the Guiding Role of the Radial Glia Basal Process during Neuron Migration" Cells 10, no. 1: 3. https://doi.org/10.3390/cells10010003




