Abstract
The Broad-complex Tramtrack and Bric-a-brac (BTB) domain participates in plant responses to biotic and abiotic stresses, however its role is unknown in pepper plants. CaBPM4 has meprin and TRAF homology (MATH) and BTB domains at its N- and C-termini, respectively, and it contains a 1589-bp full-length cDNA that encodes a protein containing 403 amino acids. In this study, the pepper gene CaBPM4 (Capsicum annuum BTB-POZ and MATH domain protein) was cloned, and its role in responses to Phytophthora capsici, cold, drought, and salt stress were characterized. The results of quantitative RT-PCR revealed that CaBPM4 was down-regulated under P. capsici infection, salicylic acid, H2O2, and abscisic acid treatments, while abiotic stresses, including salt, cold, and drought, enhanced its transcript level. Furthermore, CaBPM4 silencing significantly impaired resistance to P. capsici, apparently by altering the transcript level of defense-related genes CaPR1, CaDEF1, and CaSAR82 and reducing root activity. However, CaBPM4-silenced plants exhibited remarkably increased peroxidase activity and decreased malondialdehyde concentrations, indicating that CaBPM4 may enhance resistance to salt and drought stress. Further study should focus on the mechanism by which CaBPM4 regulates the defense response to P. capsici infection and abiotic stresses.
1. Introduction
Plants typically encounter several biotic and abiotic stresses, including cold [1], salinity [2], drought [3], and pathogens [4]. Various environmental stresses induce physiological problems in plants and lead to various types of damages. To defend against stress-induced injuries, plants have evolved many defense mechanisms, including transcriptional regulation [5]. Defense mechanisms can positively or negatively regulate reactive oxygen species (ROS) produced by environmental stresses to achieve long-term physiological adaption to adverse environmental conditions [6]. The effect of transcription factors (TFs) on gene expression plays an essential role in various cellular processes that include signaling transduction and cellular stress responses [7]. The numerous transcription factors (TFs) that regulate gene expression by binding to target gene promoters provide mechanisms of well developmental and physiological control that can be shaped by natural selection, thereby enabling adaptive responses to environmental stress [8]. For example, CaWRKY40 plays an essential role in regulating the response of pepper plants to Ralstonia solanacearum infection, and CaWRKYd also regulates hypersensitive and defense responses to pathogen infection [9,10]. MYB [11] and bZIP [12,13] also play vital roles in protecting plants from environmental stresses.
Broad-complex Tramtrack and Bric-a-brac (BTB) domain, also known as POX virus and Zinc finger (POZ) domain, is evolutionarily conserved and reported to be involved in transcriptional regulation [14,15] and targeting proteins for degradation [16]. BTB domain was first discovered in Drosophila [15], and since then, its vital roles in hormone-induced signaling transduction and plant responses to biotic and abiotic stresses have been revealed [17,18]. For example, in soybean, GmBTB expression was induced strongly by Phytophthora sojae infection [19]. The potato (Solanum tuberosum) BTB-containing protein NRL1 can interact with the positive immunity regulator NbSWAP70 in Nicotiana benthamiana and also suppress plant immunity against Phytophthora infestans infections [20]. Expression of Arabidopsis NPR1 in wheat, transgenic tobacco, rice, and tomato enhanced plant resistance to pathogen attacks [21,22,23,24]. Furthermore, the BTB-containing protein ARIA also affected the abscisic acid (ABA)-mediated signaling pathway through interactions with a transcription factor ABF2 [25]. The BTB domain-containing protein gene family of tomato was also well-known to be related closely to abiotic stresses [26].
Many BTB/POZ proteins contain a secondary protein domain, such as a meprin and TRAF (tumor necrosis factor receptor associated factor) homology (MATH) domain, a tumor necrosis factor receptor-associated element [27]. A previous study suggested that MATH domain-containing proteins positively regulate defense responses to abiotic stresses in Arabidopsis and rice [28]. Different functional roles have been identified for BTB proteins, including transcription repression, cytoskeleton regulation, and protein degradation, in which the amino-terminal MATH domain is responsible for substrate specificity [21]. In the model plant Arabidopsis, six genes (AtBPM1–AtBPM6) encoding BTB-MATH proteins were characterized [29], and these BPMs can interact with the homeodomain-leucine zipper transcription factors ATHB6 to regulate phytohormone ABA responses and responses to abiotic stresses [17]. Morimoto et al. (2017) [30] revealed that BPM proteins modulate plant thermotolerance by negatively regulating the degradation of DREB2A. BPM also can participate in other biological processes of plant through interacting with ERFs and MYBs [31,32]. Furthermore, 33 BTB-MATH proteins have been identified in maize and one member ZmMAB1 was studied playing key roles during meiosis II and the first mitotic division [33,34]. Overexpression of a wheat BTB-MATH gene TaMAB2 on Arabidopsis plants showed an effect on the plant’s growth and development [35]. Thus, BPMs play different roles during the plant growth and development processes.
Pepper (Capsicum annuum L.) is an economically important vegetable crop worldwide. However, environmental stresses are significant constraint in pepper production, which can remarkably affect pepper growth and development. Here, we have isolated and characterized the gene CaBPM4, which belongs to the BPM subfamily, and further analyzed its contribution to abiotic or biotic stress response in pepper plant using qRT-PCR and virus-induced gene silencing (VIGS). The study provides insights into the mechanisms by which CaBPM4 regulates pepper development under environmental stress and provides a foundation for future selective breeding efforts.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
Pepper (Capsicum annuum L.) plant cultivar ‘A3’ which is susceptible to HX-9 and resistant to PC strain of P. capsici were provided from the Capsicum Research Group, College of Horticulture, Northwest A&F University, China. Seeds were soaked in warm water (55 °C) for 20 min and dipped in the normal temperature water for 4–6 h. Seedlings were grown under the conditions described by Wang et al. (2013) [36].
2.2. Pathogen Preparation and Inoculation Procedures
The avirulent PC strain (incompatible with the ‘A3’ pepper cultivar) and the virulent HX-9 strain (compatible with ‘A3’) of Phytophthora capsici were used in this experiment. A P. capsici zoospore suspension was prepared according to the protocol used by Wang et al. (2013) [36] and Ali et al. (2019) [37]. The virulent HX-9 and avirulent PC strains of P. capsici were grown on potato dextrose agar (PDA) medium for seven days in the darkness at 28 °C. Cut-up portions of the media were placed into sterile distilled water under fluorescent light for four days. Zoospore release was induced by chilling cultures to 4 °C for 1 h and then incubation at room temperature for 1 h. The zoospore concentration was counted using a hemocytometer, and the concentration was adjusted to 1 × 104 zoospores per millimeter. Experimentation with P. capsici infection was conducted as follows: The drench-method was used to inoculate the zoospores into the pepper seedlings at the six true-leaf stage, and distilled water was used as a control. Leaf and root samples were collected at 0, 4, 12, 24, and 48 h after inoculation.
For virus-induced gene silencing (VIGS) of CaBPM4 in pepper plants, leaves and roots from control plants (TRV2:00) and silenced pepper plants (TRV2:CaBPM4) were collected after infection with P. capsici.
2.3. Stress Treatments
Pepper plants at six true-leaf stage were used for abiotic stresses and signaling molecule treatments. In case of abiotic stress, cold, drought, and salt stress were applied according to the protocol used by He et al., 2018 [38]. Control plants were treated with sterile water. Leaves from treated plants were collected at 0, 4, 8, 12, and 24 h after treatment. For each plant signaling molecule treatment, each signaling molecule (namely 5 mM salicylic acid (SA), 50 μM methyl jasmonate (MeJA), 10 mM H2O2, 100 μM abscisic acid (ABA), and 10 mM ethephon (ETH)) was supplemented with 0.5% Tween-20 and sprayed onto pepper seedlings (SA, MeJA, ABA was first solubilized in 0.1% v pure alcohol, respectively, and then sterile water was used to adjust the solution to working concentration, while ETH and H2O2 was solubilized with sterile water). Control plants (for SA, MeJA, ABA treatment) were sprayed with sterile water with a mixture of 0.5% Tween-20 and 0.1% alcohol. And plants sprayed with sterile water with a mixture of 0.5% Tween-20 were also treated as a control (for H2O2 and ETH treatment). Leaves from treated plants were collected at 0, 4, 8, 12, 24, and 48 h after treatment. All collected samples were instantly frozen in liquid nitrogen and stored at −80 °C. All treatments were performed and analyzed in triplicate across separate experiments.
2.4. RNA Extraction and Quantitative Real-Time PCR Analysis
To evaluate the tissue-specific expression of CaBPM4, samples from roots, stems, leaves, flowers, green fruits, and red fruits were collected from ‘A3’ cultivar pepper plants. RNA was extracted from pepper samples at different time points using Trizol reagent (Invitrogen, Carlsbad, CA, USA). Then, 0.5 µg of total RNA was used for first-strand cDNA synthesis using a PrimeScript™ Kit (TaKaRa Bio Inc., Dalian, China). Real-time quantitative PCR (qRT-PCR) was conducted using SYBR® Premix Ex Taq™ II (TaKaRa) as described by Guo et al. (2016) [39]. Actin (GenBank ID, AY572427.1) was used as an internal control (i.e., reference gene) [40]. The primer sequences are shown in Table 1. Relative CaBPM4 expression levels were determined using the comparative 2−∆∆CT method [41].

Table 1.
Primer sequences used in the experiment.
2.5. Cloning and Sequence Analysis of CaBPM4
The rapid amplification of cDNA ends (RACE) technique was used to obtain the full-length cDNA designated CaBPM4. According to the partial cDNA fragment of the reported sequence (accession number, CK901556.1), a set of gene-specific primers (CaBPM4F and CaBPM4R) for 5′ and 3′ RACE was designed. Contig Express software and BLAST were used to assemble the full-length cDNA sequences. The full-length cDNA sequence was obtained from the leaves of the pepper cultivar ‘A3’ using the primer pair CaBPM4QF and CaBPM4QR. The full-length protein sequences of CaBPM4 and homologs in other plant species were used for multiple sequence alignments with DNAMAN software. MEGA 6.0 was used to construct a phylogenetic tree with the neighbor-joining method.
The isoelectric point (pI) and molecular weight (MW) of CaBPM4 were analyzed using the online tool (https://www.expasy.org/), and conserved domains were identified using an NCBI tool (available online: http://www. ncbi.nlm.nih.gov/cdd/) according to the method described by He et al. [38].
2.6. Virus-Induced Gene Silencing Analysis of CaBPM4 in Pepper Plants
The tobacco rattle virus (TRV)-based VIGS system was used for gene silencing in pepper plants as described by Choi et al. [42]. A 256-bp fragment harboring a conserved and a non-conserved region of CaBPM4 with primers CaBPM4VF and CaBPM4VR containing restriction sites for EcoRI and BamHI enzymes was cloned into the TRV2 vector. The VIGS in the pepper plants were applied according to the protocol used by Wang et al. (2013) [36]. Five weeks after infiltration, leaves and roots from control plants (TRV2:00) and silenced pepper plants (TRV2:CaBPM4) were collected after infection with P. capsici. Additionally, control (TRV2:00) and silenced (TRV2: CaBPM4) plants were subjected to 0.4 M sodium chloride (NaCl) and mannitol for salt and drought stress treatments, respectively. Leaf samples were collected for the measurement of peroxidase (POD) activity and Malondialdehyde (MDA) concentration. Peroxidase enzyme activity was quantified using the technique described by Beffa et al. [43]. MDA concentration was measured as described by Buege [44]. Triphenyltetrazolium chloride (TTC) was used to estimate root activity according to the methods described by Jin et al. [45]. Root samples collected from TRV2:00 and CaBPM4-silenced plants were incubated with 10 mL 1:1 (v/v) mixture of 1% TTC solution and 0.l M phosphate buffer (pH 7.0) for 1 h in the dark at 37 °C, followed by adding 2 mL of 1 M sulphuric acid to inhibit the reaction. Then rinsed root samples with distilled water and dry with absorbent papers, ground in mortar with 3 mL ethyl acetate and transferred to a 10 mL volumetric flask. The residues were rinsed with ethyl acetate and mixed with the earlier extracts and the final volume was adjusted to 10 mL. Measured the absorption of the extraction with a spectrophotometer at 485 nm. The reduced TTC amount was obtained from the standard curve and its intensity in the roots was calculated TTC reduction intensity (mg g−1 h−1) = reduced TTC amount/FW h (FW-fresh root mass; h-the incubation time).
2.7. Statistical Analysis
Data were evaluated by an analysis of variance (ANOVA) using SPSS (version 16.0, SPSS Inc., Chicago, IL, USA). Data are expressed as the means ± standard deviation (SD) of three replicates for all parameters. A least significant difference (p ≤ 0.05) test was used to identify significant differences among the treatments.
3. Results
3.1. Cloning and Sequence Analysis of Pepper CaBPM4
The full-length 1589-bp cDNA of CaBPM4 (Gene Bank accession number FJ617520) was obtained using RACE. The cDNA included a 41-bp 5′-untranslated region (UTR), a 1212-bp open reading frame (ORF), and a 336-bp 3′-UTR with an 18-bp poly (A+) tail. The ORF encoded a deduced protein consisting of 403 amino acids with a theoretical MW of 44.574 kDa and pI of 5.680. Motif scan indicated that the protein contained a MATH domain (32–144) at its N-terminus and a BTB domain (197–308) at its C-terminus (Figure 1). Additionally, high amino acid sequence homology was observed between CaBPM4 (ACM67640.1) and other BPM proteins based on multiple alignments obtained using ClustalW. The inferred phylogenetic tree revealed the amino acid sequence of CaBPM4 was closely related to BPM from potato (Solanum tuberosum) (Figure 2), indicating high conservation of the BPM sequence and structure throughout evolution.
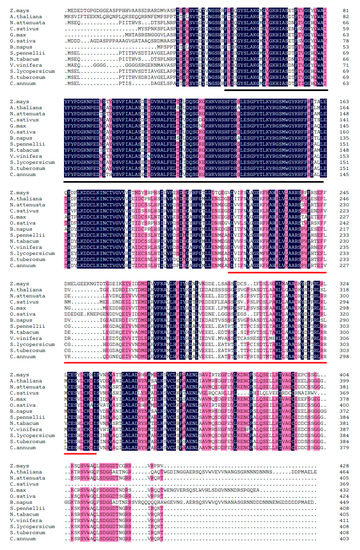
Figure 1.
Multiple sequence alignment of the ZmBPM4/mab17 (Zea mays, ACG37150.1), AtBPM4 (Arabidopsis thaliana, NP_566212.2), NaBPM4 (Nicotiana attenuate, XP_019251977.1), CsBPM4 (Cucumis sativus, XP_011653454.1), GmBPM4 (Glycine max, XP_006604260.1), OsBPM4 (Oryza sativa, XP_015646533.1), BnBPM4 (Brassica napus, XP_013729393.1), SpBPM4 (Solanum pennellii, XP_015056619.1), NtBPM4 (Nicotiana tabacum, XP_016504598.1), VvBPM4 (Vitis vinifera, XP_002277148.1), SlBPM4 (Solanum lycopersicum, XP_004251235.1), StBPM4 (Solanum tuberosum, XP_006340346.1) and CaBPM4 (ACM67640.1) proteins. Dark-blue and gray-pink shading reflects 100% and 75% percent amino acid residues conservation, respectively. Gaps (-) were introduced to maximize alignment. A red solid line shows the Broad-complex Tramtrack and Bric-a-brac (BTB) domain, and a dark solid line shows the meprin and TRAF homology (MATH) domain.
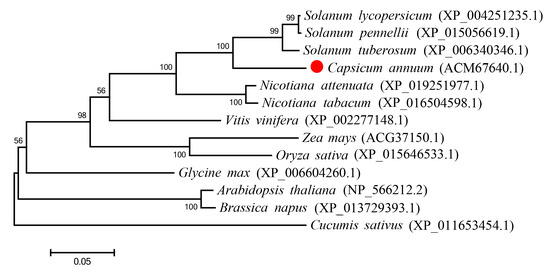
Figure 2.
Phylogenetic tree of CaBPM4 proteins and their homologues in other species. The rooted gene tree (majority-rule consensus from 1000 bootstrap replicates) resulted from was obtained by a heuristic searching in using MEGA6.0. Bootstrap values are indicated at each branch node. GenBank accession numbers are in parentheses after each species and gene name. The red point represents CaBPM4 (ACM67640.1). Scale bar indicates the similarity coefficient.
3.2. Tissue-Specific Expression of CaBPM4 in Pepper
Real-time quantitative PCR was conducted to determine the expression of CaBPM4 in different tissues. CaBPM4 transcripts were detectable in all tissues, with leaf tissue having the highest expression level, which was about 10-fold higher than that of root tissue, followed by flower tissue, with the second highest level. Among other tissues, the expression levels were highest in roots followed by red fruit, stem, and green fruit tissues, respectively, but there was no significant difference between these tissues (Figure 3).
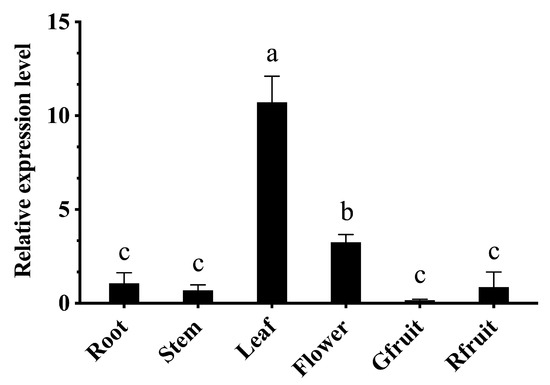
Figure 3.
Tissue-specific expression of CaBPM4 in different organs of pepper plants. The samples of root, stem and leaf are collected from A3 pepper plants at the six true-leaf stage, while the samples of flower, Gfruit (green fruit) and Rfruit (red fruit) are collected from mature plants. Relative expression levels of the CaBPM4 in different tissues were determined in comparison with that in roots. Results are presented as the means ± SD of three independent biological replicates. Small letters (a–c) represent significant differences (p < 0.05).
3.3. Expression of CaBPM4in Pepper Under P. capsici Infection
To investigate the role of CaBPM4 in pepper responses to biotic stress, we analyzed the temporal expression of CaBPM4 mRNA in leaves and roots after inoculation with the avirulent P. capsici strain PC (which is incompatible with ‘A3’ pepper plants) and the virulent P. capsici strain HX-9 (compatible with ‘A3’) using qRT-PCR. Transcript levels of CaBPM4 in leaf tissue samples were significantly downregulated at 4 h and reached their lowest level, 16-fold lower compared with the control, at 48; however, in the incompatible interaction, CaBPM4 transcript levels were downregulated significantly at 12 h and reached the lowest level of 16-fold less at 48 h (Figure 4A).
Figure 4.
Expression profiles of CaBPM4 in response to P. capsici infection and abiotic stresses. (A) The avirulent PC strain (incompatible with the ‘A3’ pepper cultivar) and the virulent HX-9 strain (compatible with ‘A3’) of P. capsici infection in roots; (B) the avirulent PC strain (incompatible with the ‘A3’ pepper cultivar) and the virulent HX-9 strain (compatible with ‘A3’) of P. capsici infection in leaves; (C) salt stress; (D) drought stress; (E) cold stress. Relative expression levels of the CaBPM4 at different time point were determined in comparison with that in the control plants at the corresponding same time point. Results are presented as mean means ± SD of three independent biological replicates. Small letters (a–d) represent significant differences (p < 0.05).
While in the root tissue, the expression level of CaBPM4 exhibited a trend similar to that in leaves during interactions (Figure 4B). CaBPM4 was downregulated after inoculation with P. capsici. At 4, 12, and 24 h the compatible virulent HX-9 and incompatible avirulent PC stain had significantly different effects on CaBPM4 expression.
3.4. CaBPM4 Expression in Response to Abiotic Stresses and Signaling Molecule Treatments in Pepper
CaBPM4 gene expression after abiotic stresses (i.e., cold, drought, and salt) was also observed. The expression level of CaBPM4 was increased up to three-fold at 8 h post-treatment with salt stress (Figure 4C), while exposure to drought stress significantly increased the expression level by up to 23-fold (23.71 ± 5.32) or more at 8 h (Figure 4D). CaBPM4 also responded to cold stress (4 °C), and its expression increased at 12 h post-treatment (Figure 4E).
We also studied the effects of SA, MeJA, ETH, ABA, and H2O2 on CaBPM4 expression in pepper plants. Plants treated with H2O2 showed a decrease in CaBPM4 transcript level, reaching a minimum at 48 h post-treatment, which is almost a 10-fold decrease in expression compared to control plants (Figure 5A). A similar trend in CaBPM4 expression under ABA treatment was detected as well, while transcript expression was decreased from 2 h post-treatment to 48 h post-treatment for ABA and H2O2 stress, reaching minimal expression compared with the control (Figure 5B). Likewise, plants treated with SA showed dramatically down-regulated CaBPM4 expression, and the differences in the transcript of CaBPM4 at post-treatment time points were significant compared with the control (Figure 5C). In response to the ETH treatment, the expression CaBPM4 increased at 4 and 8 h post-treatment compared to the control, continuing until 48 h (Figure 5D). Plants treated with MeJA showed initially upregulated expression, reaching the highest level at 8 h post-treatment, followed by downregulation, reaching a minimum at 12 h post-treatment, and the differences in CaBPM4 expression were significant (Figure 5E).
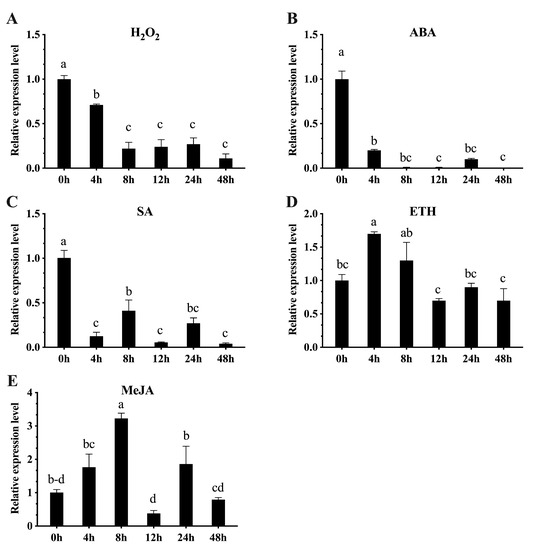
Figure 5.
Quantitative real-time PCR analysis of CaBPM4 gene after signaling molecule treatments in the leaves of pepper plant. (A) Hydrogen peroxide (H2O2) treatment; (B) abscisic acid (ABA) treatment; (C) salicylic acid (SA) treatment; (D) ethephon (ETH) treatment; (E) methyl jasmonate (MeJA) treatment. Relative expression levels of the CaBPM4 at different time point were determined in comparison with that in the control plants at the corresponding same time point. Results are presented as mean means ± SD of three independent biological replicates. Small letters (a–d) represent significant differences (p < 0.05).
3.5. CaBPM4 Silencing Decreased the Defense Response of Pepper Against P. capsici
The expression of CaBPM4 was upregulated under abiotic stresses and MeJA and ETH treatments, while ABA, SA and H2O2 treatments as well as infection with the virulent (HX-9) and avirulent (PC) strains of P. capsici downregulated its expression. To confirm this finding, a silencing vector was constructed and transformed into the Agrobacterium strain GV3101. Visible photobleaching symptoms were observed in the leaves of CaPDS-silenced pepper seedlings 35 days after injection (Figure 6A). The CaBPM4 expression was analyzed by qRT-PCR and significant decrease was found in silenced lines (Figure 6B), confirming the reliability of VIGS in silencing the target gene in pepper. The CaBPM4-silenced seedlings with silencing efficiency over 50% were used for the subsequent experiment.
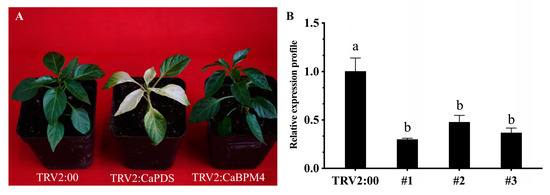
Figure 6.
Effect of CaBPM4-silencing in pepper plant. (A) Phenotypes of CaBPM4-silenced, TRV2:00 (negative control) and TRV2:CaPDS (positive control, in which the endogenous phytoene desaturase (PDS) gene was silenced to cause photobleaching), and images were obtained after 35-days inoculation. (B) Real-Time RT-PCR analysis of CaBPM4 expression levels in TRV2:00 and CaBPM4-silenced line (#1, #2, and #3 indicate three biological levels of efficiency of CaBPM4 gene silencing) 35 days after inoculation. Relative expression levels of the CaBPM4 were determined in comparison with that in TRV2:00. Small letters (a, b) represent significant differences (p < 0.05).
After inoculation with the PC strain of P. capsici, the expression levels of CaPR1, CaDEF1, CaPO1, and CaSAR82 were elevated. At 0 h, the expression of the pathogen-related genes CaPR1, CaDEF1, CaPO1, and CaSAR82 exhibited no significant difference between non-silenced plants and CaBPM4-silenced plants. Significant increases in the transcript abundances of CaPR1, CaDEF1, and CaSAR82 were detected at 24 h in the non-silenced plants, while the increase in the expression of defense-related genes was lower in the CaBPM4-silenced plants compared with control plants (Figure 7). However, CaPO1 expression was downregulated during P. capsici infection of both CaBPM4-silenced and control plants. At 2 day post-PC inoculation, CaDEF1 and CaSAR82 showed downregulation compared with that at 1 day, and significant difference were detected in the expression level of CaSAR82 between the control plants and CaBPM4-silenced plants. The expression of CaPO1 increased at 2 day, with a significant high level in the CaBPM4-silenced plants. Similarly, after inoculation with the HX-9 strain, the expression levels of the defense-related genes CaPR1, CaDEF1, and CaSAR82 were also upregulated 1 day post-inoculation. The up-regulation of CaDEF1 and CaSAR82 was significantly higher in non-silenced plants than in CaBPM4-silenced plants, while CaPO1 was downregulated at 1 day, followed by an increase at 2 day post-HX-9 inoculation. Furthermore, after P. capsici inoculation, we measured root activity in the silenced and control plants using the TTC method. Root activity in the silenced plants was lower than that in control plants, and in the case of the HX-9 strain, a significant difference was observed at 7 day post-inoculation between silenced and control plants (Figure 8).
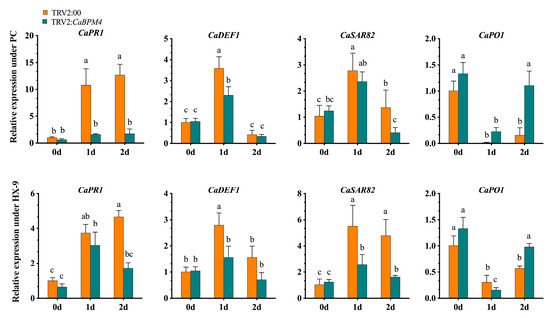
Figure 7.
Silencing of CaBPM4 reduced the expression of defense defense-related genes in pepper at 1 day post-inoculation with PC and HX-9 strains of P. capsici. Relative expression levels of the defense defense-related genes at different time point under P. capsici infection were determined in comparison with that in TRV2:00. Values are the means ± SD from three independent experiments. Small letters (a–c) represent significant differences (p < 0.05).
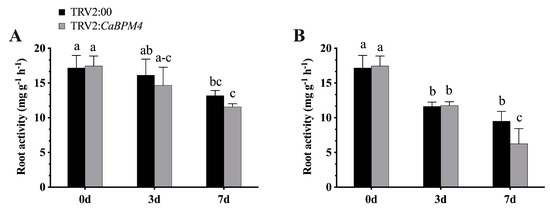
Figure 8.
Silencing of CaBPM4 reduced the root activity of pepper post P. capsici inoculation. The root activity parameter showed that the activeness and function of roots. (A) Root activity in CaBPM4-silenced and control plants after inoculation with the PC strain; (B) root activity in CaBPM4-silenced and control plants after inoculation with the HX-9 strain. Values are the means ± SD from three independent experiments. Small letters (a–c) represent significant differences (p < 0.05).
3.6. CaBPM4 Silencing Enhanced the Defense Response of Pepper against Abiotic Stresses
The above results suggested that the CaBPM4-silenced pepper plants showed reduced tolerance to biotic stress, while the effect of osmotic stress on CaBPM4 silenced pepper plants was unknown. Accordingly, we studied the effect of salt and drought on CaBPM4-silenced pepper plants and explored the possible physiological mechanism by which CaBPM4 contributes to osmotic stress responses in pepper. At 24 h post-treatment with 0.4 M NaCl (salt treatment), the control (TRV2:00) plants exhibited more severe wilting compared with the CaBPM4-silenced plants (Figure 9A). Under salt stress, CaBPM4-silenced plants showed a higher increase in POD activity at 6 and 24 h post-treatment compared to the control plants (Figure 9C). Likewise, MDA levels were also measured, with MDA concentrations increasing in both CaBPM4-silenced plants as well as control plants increased under salt stress, reaching a maximum at 24 h post-treatment. A lower increase was detected in the silenced plants, with levels significantly differing from the control plants (Figure 9E).
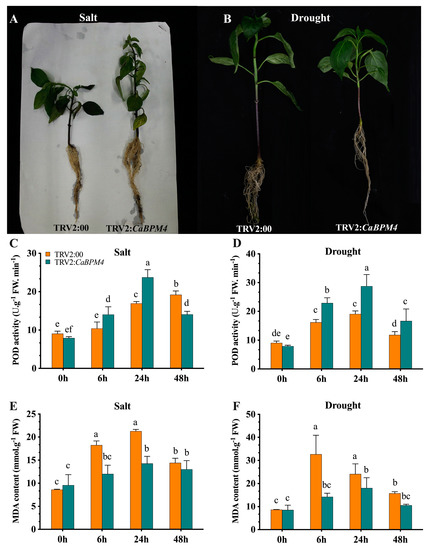
Figure 9.
Physiological attributes of CaBPM4-silenced and control pepper plants under salt and drought stress. (A) Phenotypes of the gene-silenced plants in response to salt stress; (B) phenotypes of the gene-silenced plants in response to drought stress; (C) the activity of POD after salt stress; (D) the activity of POD after drought stress; (E) MDA concentration after salt stress; (F) MDA concentration after drought stress. Values are the means ± SD from three independent experiments. Small letters (a–e) represent significant differences (p < 0.05).
Similarity, at 6 h post-treatment with 0.4 M mannitol (i.e., drought treatment), the control plants (TRV2:00) also exhibited more serious wilting compared the CaBPM4-silenced plants (Figure 9B). Exposure of the TRV2:00 and TRV2:CaBPM4 lines to drought stress resulted in increased POD activity at 6 and 24 h post-treatment compared with that at 0 h (Figure 9D), and the increases were more substantial for TRV2:CaBPM4 plants than TRV2:00 plants. The opposite trend was observed for MDA concentration, and MDA levels were higher in TRV2:00 plants. Afterwards, a slight decrease was detected at 48 h post-treatment in both lines (Figure 9F).
4. Discussion
Plants have evolved intricate regulatory and defense systems that resist biotic and abiotic stresses [46], and the BTB domain has been implicated in plant defense and environmental stress responses. Many BTB-containing proteins participate in biological regulatory networks, including transcriptional regulation, protein degradation, chromatin remodeling, and cytoskeletal regulation networks, under adverse environmental conditions [15,47]. Utilizing available whole genome sequences, 80 and 149 members of the BTB gene superfamily have been identified in Arabidopsis thaliana and rice, respectively [44,45]. Moreover, 38 BTB members were found in the tomato plant [26]. Furthermore, six BPMs (BTB-MATH) in Arabidopsis have been observed to assemble with cullins to form complexes [29]. ZmMAB1, a BPM protein in maize can interact with Cullin 3 proteins [33]. Furthermore, wheat BPM protein TaMAB2 functions as part of a CUL3 based ligase complex and involved in CUL3-based degradation [35]. However, very little is known about the effect of genes with BTB domains on the development of pepper. The current study described the characteristic of the newly discovered gene CaBPM4 from the BPM family in the pepper plant. CaBPM4 was inferred to encode a protein containing an N-terminal MATH domain and a C-terminal BTB domain. This novel pepper gene may play an important role in revealing the BPM signaling transduction mechanisms in pepper.
The tissue-specific expression patterns of BPM genes have been reported in various species, such as Arabidopsis [48], tomato [26], and rice [49]. However, there is no uniform pattern of gene expression for these BPM genes [17,31]. The qRT-PCR result from the present study showed that CaBPM4 has a high level of expression in leaf and flower tissues. In Arabidopsis, AtBPMs were expressed at a significantly higher level in floral buds and open flowers, and BPM-silenced plants exhibited altered flower development [17]. In tomato, SlBOPs (Solanum lycopersicum BLADE-ON-PETIOLE), which are BTB transcriptional regulators, can interact with the transcription factor TERMINATING FLOWER (TMF) to control inflorescence architecture [50]. This suggests that BTB-containing proteins may participate in flower development processes.
P. capsici was also revealed to downregulate CaBPM4 expression, which was contrary to the results of previous research that found rice genes including OsMB9, OsMB10, and OsMB11 show upregulated expression under infection [28]. Few plant species, including Arabidopsis, have shown trends similar to our result. Therefore, the underlying mechanisms of BPM proteins remain elusive and require further investigation.
Under abiotic stress, CaBPM4 expression was strongly induced, which is mostly consistent with previous research. In Arabidopsis, BPM1, BPM4, and BPM5 are reported to be upregulated under salt and drought stresses [31]. Kushwaha et al. (2016) [28] also noted that OsMB10 and OsMB11, which encode BPM proteins in rice, showed apparent increases in expression level under salinity and drought stress. Inhibitory effects of exogenous substances, especially ABA, on CaBPM4 expression were observed in the current study, which is unsurprising. In the tomato plant, SlBTB12 was inhibited by ABA across all time points [26]. GMPOZ, a nuclear factor found in soybean, also functions as a repressor of ABA-regulated genes [51]. Treatment with ABA lowered AtBT2 mRNA levels, while MeJA induced AtBT2 expression [18], indicating that BPM proteins can mediate the ABA pathway. AtBPM proteins were also linked to ethylene response. Specifically, AtBPMs can interact with members of the ethylene response factor/Apetala2 (ERF/AP2) transcription factor family [31]. Additionally, the BTB domain is associated with ethylene biosynthesis [52] and the salicylic acid signaling pathway [22,53].
To understand the role of CaBPM4 in the response of pepper plants to stress conditions, VIGS was conducted to silence CaBPM4 expression. In the current study, photobleaching symptoms were observed in the leaves of CaPDS-silenced plants, indicating the reliability of the VIGS assay. After inoculation with P. capsici, the transcript levels of defense-related genes in CaBPM4-silenced pepper plants were lower than those of control plants, and root activity in the silenced plants was also lower than that of control plants. This indicated that CaBPM4 silencing aggravated P. capsici infection in pepper by diminishing the defense system of pepper plants. A similar result was also observed in CabZIP63-silenced plants, where the transcript levels of the defense-related genes CaPR1 and CaDEF1 were significantly lower than those in the control pepper plants under R. solanacearum infection [54]. Similarly, CaPTI1 and CanTF silencing also altered the expression levels of CaPR1 [55], CaDEF1 [56], and CaSAR82 [57] as well as the root activity of silenced plants relative to those in control plants [38,45]. Zhang reported that overexpression of the gene GmBTB increased the transcript levels of GmNPR1 in GmBTB/POZ-OE plants [19]. Another study supporting our results showed that CaPAL1 silencing suppresses CaPR1 expression compared to control plants during Xcv infection [58]. Under infection with Xcv (especially with a virulent strain), CaPO2-silenced plants exhibited significantly lower induction of CaDEF1 and CaSAR82 [42].
In contrast, the CaBPM4-silenced plants exposed to drought and salt stress showed enhanced tolerance compared with control plants. Less wilting, higher POD activity, and lower MDA concentration in CaBPM4-silenced plants suggested that CaBPM4 silencing can improve plant tolerance to abiotic stress. In Arabidopsis, BPM-silenced plants showed an improved thermotolerance under high humidity, as shown by a higher survival rate and lower ion leakage after heat shock. AtBPMs can interact with DREB2A, a key transcription factor in both drought and heat stress tolerance [59], and are responsible for DREB2A degradation, thus affecting plant responses to abiotic stress [30]. BPM proteins can also interact with ATHB6, a negative regulator of ABA responses [60], resulting in modulations of ABA signaling and plant responses to cold and salt stresses [17]. BPM proteins can also interact with some other factors that respond to abiotic stress [61]. Collectively, these data suggest a putative BPM function of CaBPM4 in pepper responses to stresses.
5. Conclusions
In brief, CaBPM4 encodes a protein containing an N-terminal MATH domain and a C-terminal BTB domain and the amino acid sequence of CaBPM4 was closely related to BPM from potato (Solanum tuberosum). CaBPM4 gene has a higher expression in leaf and flower tissue other than in root, stem, fruit tissue and it was induced by both biotic and abiotic stresses. P. capsici infection resulted in the downregulation of CaBPM4 expression, while salt and drought stress enhanced CaBPM4 expression. CaBPM4 silencing in pepper significantly weakened the defense response by downregulating the expression of defense-related genes and inhibiting root activity under P. capsici infection, while enhancing tolerance to salt and drought stress in CaBPM4-silenced plants. These results suggest that CaBPM4 can respond to different stresses by regulating plant defense systems. Additional research is needed to characterize the detailed mechanism by which CaBPM4 regulates the defense response to biotic and abiotic stress in pepper plants.
Author Contributions
Conceptualization, Y.-M.H. and Z.-H.G.; methodology, Y.-M.H., K.-K.L. and Z.-H.G.; software, Y.-M.H. and G.-X.C.; investigation, Y.-M.H., K.-K.L. and H.-X.Z.; resources, A.-M.W. and Z.-H.G.; data curation, Y.-M.H., M.A.; writing—original draft preparation, Y.-M.H.; writing—review and editing, Y.-M.H. M.A., S.U.H. and Z.-H.G.; funding acquisition, A.-M.W. and Z.-H.G. All authors read and approved the final manuscript.
Acknowledgments
This work was supported through funding from the National Key R&D Program of China (No. 2016YFD0101900), the National Natural Science Foundation of China (No. U1603102).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Nguyen, H.T.; Leipner, J.; Stamp, P.; Guerra-Peraza, O. Low temperature stress in maize (Zea mays L.) induces genes involved in photosynthesis and signal transduction as studied by suppression subtractive hybridization. Plant Physiol. Biochem. 2009, 47, 116–122. [Google Scholar] [CrossRef]
- Chaves, M.M.; Flexas, J.; Pinheiro, C. Photosynthesis under drought and salt stress: Regulation mechanisms from whole plant to cell. Ann. Bot. 2009, 103, 551–560. [Google Scholar] [CrossRef]
- Barber, V.A.; Juday, G.P.; Finney, B.P. Reduced growth of Alaskan white spruce in the twentieth century from temperature-induced drought stress. Nature 2000, 405, 668–673. [Google Scholar] [CrossRef]
- Lamour, K.H.; Stam, R.; Jupe, J.; Huitema, E. The oomycete broad-host-range pathogen Phytophthora capsici. Mol. Plant Pathol. 2012, 13, 329–337. [Google Scholar] [CrossRef]
- Kim, T.H.; Ok, S.H.; Kim, D.; Suh, S.C.; Ok Byun, M.; Shin, J.S. Molecular characterization of a biotic and abiotic stress resistance-related gene RelA/SpoT homologue (PepRSH) from pepper. Plant Sci. 2009, 176, 635–642. [Google Scholar] [CrossRef]
- Petrov, V.; Hille, J.; Mueller-Roeber, B.; Gechev, T.S. ROS-mediated abiotic stress-induced programmed cell death in plants. Front. Plant Sci. 2015, 6, 69. [Google Scholar] [CrossRef]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef]
- Ülker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef]
- Dang, F.; Wang, Y.; Yu, L.U.; Eulgem, T.; Lai, Y.A.N.; Liu, Z.; Wang, X.U.; Qiu, A.; Zhang, T.; Lin, J.; et al. CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection ABSTRACT. Plant. Cell Environ. 2013, 36, 757–774. [Google Scholar] [CrossRef]
- Huh, S.U.; Choi, L.M.; Lee, G.; Kim, Y.J.; Paek, K. Plant Science Capsicum annuum WRKY transcription factor d (CaWRKYd) regulates hypersensitive response and defense response upon Tobacco mosaic virus infection. Plant Sci. 2012, 197, 50–58. [Google Scholar] [CrossRef]
- Abe, H. Role of Arabidopsis MYC and MYB Homologs in Drought-and Abscisic Acid-Regulated Gene Expression. Plant Cell Online 1997, 9, 1859–1868. [Google Scholar]
- Sun, X.; Li, Y.; Cai, H.; Bai, X.; Ji, W.; Ding, X.; Zhu, Y. The Arabidopsis AtbZIP1 transcription factor is a positive regulator of plant tolerance to salt, osmotic and drought stresses. J. Plant Res. 2012, 125, 429–438. [Google Scholar] [CrossRef]
- Zhou, X.T.; Jia, L.J.; Wang, H.Y.; Zhao, P.; Wang, W.Y.; Liu, N.; Song, S.W.; Wu, Y.; Su, L.; Zhang, J.; et al. The potato transcription factor StbZIP61 regulates dynamic biosynthesis of salicylic acid in defense against Phytophthora infestans infection. Plant J. 2018, 95, 1055–1068. [Google Scholar] [CrossRef]
- Fedele, M.; Benvenuto, G.; Pero, R.; Majello, B.; Battista, S.; Lembo, F.; Vollono, E.; Day, P.M.; Santoro, M.; Lania, L.; et al. A novel member of the BTB/POZ family, PATZ, associates with the RNF4 RING finger protein and acts as a transcriptional repressor. J. Biol. Chem. 2000, 275, 7894–7901. [Google Scholar] [CrossRef]
- Melnick, A.; Ahmad, K.F.; Arai, S.; Polinger, A.; Ball, H.; Borden, K.L.; Carlile, G.W.; Prive, G.G.; Licht, J.D. In-depth mutational analysis of the promyelocytic leukemia zinc finger BTB/POZ domain reveals motifs and residues required for biological and transcriptional functions. Mol. Cell. Biol. 2000, 20, 6550–6567. [Google Scholar] [CrossRef]
- Furukawa, M.; Xiong, Y. BTB Protein Keap1 Targets Antioxidant Transcription Factor Nrf2 for Ubiquitination by the Cullin 3-Roc1 Ligase. Mol. Cell. Biol. 2005, 25, 162–171. [Google Scholar] [CrossRef]
- Lechner, E.; Leonhardt, N.; Eisler, H.; Parmentier, Y.; Alioua, M.; Jacquet, H.; Leung, J.; Genschik, P. MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signaling. Dev. Cell 2011, 21, 1116–1128. [Google Scholar] [CrossRef]
- Mandadi, K.K.; Misra, A.; Ren, S.; McKnight, T.D. BT2, a BTB Protein, Mediates Multiple Responses to Nutrients, Stresses, and Hormones in Arabidopsis. Plant Physiol. 2009, 150, 1930–1939. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, H.; Li, R.; Han, D.; Wang, L.; Wu, J.; Xu, P.; Zhang, S. GmBTB/POZ, a novel BTB/POZ domain-containing nuclear protein, positively regulates the response of soybean to Phytophthora sojae infection. Mol. Plant Pathol. 2019, 20, 78–91. [Google Scholar] [CrossRef]
- He, Q.; Naqvi, S.; McLellan, H.; Boevink, P.C.; Champouret, N.; Hein, I.; Birch, P.R.J. Plant pathogen effector utilizes host susceptibility factor NRL1 to degrade the immune regulator SWAP70. Proc. Natl. Acad. Sci. USA 2018, 115, 201808585. [Google Scholar] [CrossRef]
- Boyle, P.; Le Su, E.; Rochon, A.; Shearer, H.L.; Murmu, J.; Chu, J.Y.; Fobert, P.R.; Despres, C. The BTB/POZ Domain of the Arabidopsis Disease Resistance Protein NPR1 Interacts with the Repression Domain of TGA2 to Negate Its Function. Plant Cell 2009, 21, 3700–3713. [Google Scholar] [CrossRef]
- Chern, M.S.; Fitzgerald, H.A.; Yadav, R.C.; Canias, P.E.; Aarts, M.G.; Dong, X.; Ronald, P.C. Evidence for a resistance signalling pathway in rice similar to the NPR1-mediated, SA-dependent pathway in Arabidopsis. Plant J. 2001, 27, 101–113. [Google Scholar] [CrossRef]
- Makandar, R.; Essig, J.S.; Schapaugh, M.A.; Trick, H.N.; Shah, J. Genetically Engineered Resistance to Fusarium Head Blight in Wheat by Expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 2006, 19, 123–129. [Google Scholar] [CrossRef]
- Lin, W.; Lu, C.; Wu, J.; Cheng, M.; Lin, Y.; Black, L.; Green, S.K.; Wang, J.; Cheng, C. Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Transgenic Res. 2004, 13, 567–581. [Google Scholar] [CrossRef]
- Škrbić, B.; Godula, M.; Durišić-Mladenović, N.; Živančev, J. ARIA, an Arabidopsis Arm Repeat Protein Interacting with a Transcriptional Regulator of Abscisic Acid-Responsive Gene Expression, is a Novel Abscisic Acid Signaling Component. Agron. Res. 2011, 9, 461–468. [Google Scholar]
- Li, J.; Su, X.; Wang, Y.; Yang, W.; Pan, Y.; Su, C.; Zhang, X. Genome-wide identification and expression analysis of the BTB domain-containing protein gene family in tomato. Genes Genom. 2018, 40, 1–15. [Google Scholar] [CrossRef]
- Kuo, S.P.; Bradley, L.A.; Trussell, L.O. NIH Public Access. Hear. Res. 2010, 29, 9625–9634. [Google Scholar]
- Kushwaha, H.R.; Joshi, R.; Pareek, A.; Singla-pareek, S.L. MATH-Domain Family Shows Response toward Abiotic Stress in Arabidopsis and Rice. Front. Plant Sci. 2016, 7, 1–19. [Google Scholar] [CrossRef]
- Weber, H. Arabidopsis AtCUL3a and AtCUL3b Form Complexes with Members of the BTB/POZ-MATH Protein Family. Plant Physiol. 2005, 137, 83–93. [Google Scholar] [CrossRef]
- Morimoto, K.; Ohama, N.; Kidokoro, S.; Mizoi, J.; Takahashi, F.; Todaka, D.; Mogami, J.; Sato, H.; Qin, F.; Kim, J.S.; et al. BPM-CUL3 E3 ligase modulates thermotolerance by facilitating negative regulatory domain-mediated degradation of DREB2A in Arabidopsis. Proc. Natl. Acad. Sci. USA 2017, 114, E8528–E8536. [Google Scholar] [CrossRef]
- Weber, H.; Hellmann, H. Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 2009, 276, 6624–6635. [Google Scholar] [CrossRef]
- Chen, L.; Bernhardt, A.; Lee, J.; Hellmann, H. Identification of Arabidopsis MYB56 as a novel substrate for CRL3BPM E3 ligases. Mol. Plant 2015, 8, 242–250. [Google Scholar] [CrossRef]
- Juranic, M.; Srilunchang, K.O.; Krohn, N.G.; Leljak-Levanic, D.; Sprunck, S.; Dresselhaus, T. Germline-Specific MATH-BTB Substrate Adaptor MAB1 Regulates Spindle Length and Nuclei Identity in Maize. Plant Cell 2012, 24, 4974–4991. [Google Scholar] [CrossRef]
- Škiljaica, A. Wheat MATH-BTB Proteins and Their Role in Early Embryogenesis (Proteini MATH-BTB u Pšenice i Njihova Uloga u Ranoj EmbriogeneziI); University of Zagreb: Zagreb, Croatia, 2016. [Google Scholar]
- Juranic, M.; Sprunck, S.; Math-btb, T.P.D.Á. De novo zygotic transcription in wheat (Triticum aestivum L.) includes genes encoding small putative secreted peptides and a protein involved in proteasomal degradation. Plant Reprod. 2013, 26, 267–285. [Google Scholar]
- Wang, J.E.; Liu, K.K.; Li, D.W.; Zhang, Y.L.; Zhao, Q.; He, Y.M.; Gong, Z.H. A novel peroxidase CanPOD gene of pepper is involved in defense responses to Phytophtora capsici infection as well as abiotic stress tolerance. Int. J. Mol. Sci. 2013, 14, 3158–3177. [Google Scholar] [CrossRef]
- Ali, M.; Gai, W.X.; Khattak, A.M.; Khan, A.; Haq, S.U.; Ma, X.; Wei, A.M.; Muhammad, I.; Jan, I.; Gong, Z.H. Knockdown of the chitin-binding protein family gene CaChiIV1 increased sensitivity to Phytophthora capsici and drought stress in pepper plants. Mol. Genet. Genom. 2019, 1–16. [Google Scholar] [CrossRef]
- He, Y.; Luo, D.; Khan, A.; Liu, K.; Arisha, M.H.; Zhang, H. CanTF, a Novel Transcription Factor in Pepper, Is Involved in Resistance to Phytophthora capsici as well as Abiotic Stresses CanTF, a Novel Transcription Factor in Pepper, Is Involved in Resistance to Phytophthora capsici as well as Abiotic Stresses. Plant Mol. Biol. Rep. 2018, 36, 776–789. [Google Scholar] [CrossRef]
- Guo, M.; Liu, J.; Ma, X.; Zhai, Y.; Gong, Z.; Lu, M. Plant Science Genome-wide analysis of the Hsp70 family genes in pepper (Capsicum annuum L.) and functional identification of CaHsp70-2 involvement in heat stress. Plant Sci. 2016, 252, 246–256. [Google Scholar] [CrossRef]
- Palma, M.; Corpas, F.J.; Mateos, R.M.; Bonilla-valverde, D.; Rı, L.A. Physiologia Plantarum NADP-dehydrogenases from pepper fruits: Effect of maturation. Physiol. Plant. 2009, 135, 130–139. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Choi, H.W.; Kim, Y.J.; Lee, S.C.; Hong, J.K.; Hwang, B.K. Hydrogen peroxide generation by the pepper extracellular peroxidase CaPO2 activates local and systemic cell death and defense response to bacterial pathogens. Plant Physiol. 2007, 145, 890–904. [Google Scholar] [CrossRef]
- Beffa, R.; Martin, H.V.; Pilet, P. Oxidases and Peroxidases from Maize Roots. Plant Physiol. 1990, 94, 485–491. [Google Scholar] [CrossRef]
- Buege, J.A.; Aust, S.D. Microsomal lipid peroxidation. J. Phys. Conf. Ser. 1978, 52, 302–310. [Google Scholar]
- Jin, J.H.; Zhang, H.X.; Tan, J.Y.; Yan, M.J.; Li, D.W.; Khan, A.; Gong, Z.H. A New Ethylene-Responsive Factor CaPTI1 Gene of Pepper (Capsicum annuum L.) Involved in the Regulation of Defense Response to Phytophthora capsici. Front. Plant Sci. 2016, 6, 1–12. [Google Scholar] [CrossRef]
- Choi, H.W.; Hwang, B.K. The pepper extracellular peroxidase CaPO2 is required for salt, drought and oxidative stress tolerance as well as resistance to fungal pathogens. Planta 2012, 235, 1369–1382. [Google Scholar] [CrossRef]
- Lee, D.K.; Shic Kim, Y.; Kim, J.K. Determination of the optimal condition for ethylmethane sulfonate-mediated mutagenesis in a Korean commercial rice, Japonica cv. Dongjin. Appl. Biol. Chem. 2017, 60, 241–247. [Google Scholar] [CrossRef]
- Gingerich, D.J.; Gagne, J.M.; Salter, D.W.; Hellmann, H.; Estelle, M.; Ma, L.; Vierstra, R.D. Cullins 3a and 3b Assemble with Members of the Broad Complex/Tramtrack/Bric-a-Brac (BTB) Protein Family to Form Essential. J. Biol. Chem. 2005, 280, 18810–18821. [Google Scholar] [CrossRef]
- Gingerich, D.J.; Hanada, K.; Shiu, S.H.; Vierstra, R.D. Large-Scale, Lineage-Specific Expansion of a Bric-a-Brac/Tramtrack/Broad Complex Ubiquitin-Ligase Gene Family in Rice. Plant Cell Online 2007, 19, 2329–2348. [Google Scholar] [CrossRef]
- Xu, C.; Park, S.J.; Van Eck, J.; Lippman, Z.B. Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 2016, 30, 2048–2061. [Google Scholar] [CrossRef]
- Woodger, F.J.; Jacobsen, J.V.; Gubler, F. GMPOZ, a BTB/POZ domain nuclear protein, is a regulator of hormone responsive gene expression in barley aleurone. Plant Cell Physiol. 2004, 45, 945–950. [Google Scholar] [CrossRef]
- Levin, S.; Hartl, D.; Clark, A.; Petrov, D.; Bustamante, C.; Garwin, L.; Friedman, N. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 2004, 428, 945–950. [Google Scholar]
- Wu, Q.; Wang, X.Z.; Tang, Y.Y.; Yu, H.T.; Ding, Y.F.; De Yang, C.; Cui, F.G.; Zhang, J.C.; Wang, C.T. Molecular cloning and characterization of NPR1 gene from Arachis hypogaea. Mol. Biol. Rep. 2014, 41, 5247–5256. [Google Scholar] [CrossRef]
- Shen, L.; Liu, Z.; Yang, S.; Yang, T.; Liang, J.; Wen, J.; Liu, Y.; Li, J.; Shi, L.; Tang, Q.; et al. Pepper CabZIP63 acts as a positive regulator during Ralstonia solanacearum or high temperature-high humidity challenge in a positive feedback loop with CaWRKY40. J. Exp. Bot. 2016, 67, 2439–2451. [Google Scholar] [CrossRef]
- Kim, Y.J.; Hwang, B.K. Pepper gene encoding a basic pathogenesis-related 1 protein is pathogen and ethylene inducible. Physiol. Plant. 2000, 108, 51–60. [Google Scholar]
- Do, H.M.; Lee, S.C.; Jung, H.W.; Sohn, K.H.; Hwang, B.K. Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Plant Sci. 2004, 166, 1297–1305. [Google Scholar]
- Lee, S.C.; Hwang, B.K. Identification of the pepper SAR8. 2 gene as a molecular marker for pathogen infection, abiotic elicitors and environmental stresses in Capsicum annuum. Planta 2003, 216, 387–396. [Google Scholar]
- Kim, D.S.; Hwang, B.K. An important role of the pepper phenylalanine ammonia-lyase gene (PAL1) in salicylic acid-dependent signalling of the defence response to microbial pathogens. J. Exp. Bot. 2014, 65, 2295–2306. [Google Scholar] [CrossRef]
- Nakashima, K.; Ito, Y.; Yamaguchi-shinozaki, K. Transcriptional Regulatory Networks in Response to Abiotic Stresses in Transcriptional Regulatory Networks in Response to Abiotic Stresses in Arabidopsis and Grasses 1. Plant Physiol. 2009, 149, 88–95. [Google Scholar] [CrossRef]
- Himmelbach, A.; Hoffmann, T.; Leube, M.; Höhener, B.; Grill, E. Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO J. 2002, 21, 3029–3038. [Google Scholar] [CrossRef]
- Lin, R.; Park, H.; Wang, H. Role of Arabidopsis RAP2. 4 in Regulating Light- and Ethylene-Mediated Developmental Processes and Drought Stress Tolerance. Mol. Plant 2008, 1, 42–57. [Google Scholar] [CrossRef]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).