Chromosomes I and X Harbor Consistent Genetic Factors Associated with the Anthocyanin Variation in Potato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Anthocyanin Extraction
2.3. Quantification of Anthocyanins in Cooked Tubers
2.4. QTL Identification and Candidate Gene for Anthocyanin Concentration
3. Results
3.1. Quantification of Anthocyanidins in Cooked Tubers
3.2. Genome-Wide Association Study for Anthocyanidins Concentration Regulation
4. Discussion
5. Conclusions
Supplementary Materials
Data Availability Statement
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization. 2018. Available online: http://www.fao.org/faostat/en/#home (accessed on 19 March 2019).
- Chun, O.K.; Kim, D.O.; Smith, N.; Schroeder, D.; Han, J.T.; Chang, Y.L. Daily consumption of phenolics and total antioxidant capacity from fruit and vegetables in the American diet. J. Sci. Food Agric. 2005, 85, 1715–1724. [Google Scholar] [CrossRef]
- Thompson, M.D.; Thompson, H.J.; McGinley, J.N.; Neil, E.S.; Rush, D.K.; Holm, D.G.; Stushnoff, C. Functional food characteristics of potato cultivars (Solanum tuberosum L.): Phytochemical composition and inhibition of 1-methyl-1-nitrosourea induced breast cancer in rats. J. Food Compos. Anal. 2009, 22, 571–576. [Google Scholar] [CrossRef]
- Andre, C.; Ghislain, M.; Bertin, P.; Oufir, M.; Herrera, M.; Hoffmann, L. Andean potato cultivars (Solanum tuberosum L.) as a source of antioxidant and mineral micronutrients. J. Agric. Food Chem. 2007, 55, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Godfray, H.C.; Garnett, T. Food security and sustainable intensification. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20120273. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.J. Roots and Tubers Galore: The Story of CIP’s Global Research Program and the People Who Shaped It; CIP: Lima, Peru, 1993. [Google Scholar]
- Estrada, R.N. La Biodiversidad en el Mejoramiento Genético de la Papa; Hardy, B., Martinez, E., Eds.; Editorial del Centro de información para el desarrollo: La Paz, Bolivia, 2004. [Google Scholar]
- Ezekiel, R.; Singh, N.; Sharma, S.; Kaur, A. Beneficial phytochemicals in potato—A review. Food Res. Int. 2013, 50, 487–496. [Google Scholar] [CrossRef]
- Huamán, Z.; Spooner, D.M. Reclassification of landrace populations of cultivated potatoes (Solanum sect. Petota). Am. J. Bot. 2002, 89, 947–965. [Google Scholar] [CrossRef]
- Nassar, A.; Sabally, K.; Kubow, S.; Leclerc, N.Y.; Donnelly, D.J. Some Canadian-grown potato cultivars contribute to a substantial content of essential dietary minerals. J. Agric. Food Chem. 2012, 60, 4688–4696. [Google Scholar] [CrossRef]
- André, C.M.; Schafleitner, R.; Legay, S.; Lefèvre, I.; Aliaga, C.A.; Nomberto, G.; Hoffmann, L.; Hausman, J.F.; Larondelle, Y.; Evers, D. Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 2009, 70, 1107–1116. [Google Scholar] [CrossRef]
- Hawkes, J.G. The Potato: Evolution, Biodiversity, and Genetic Resources; Belhaven Press: London, UK, 1990. [Google Scholar]
- Peña, C.; Restrepo-Sánchez, L.P.; Kushalappa, A.; Rodríguez-Molano, L.E.; Mosquera, T.; Narváez-Cuenca, C.E. Nutritional contents of advanced breeding clones of Solanum tuberosum group Phureja. LWT-Food Sci. Technol. 2015, 62, 76–82. [Google Scholar] [CrossRef]
- André, C.; Oufir, M.; Hoffmann, L.; Hausman, J.; Rogez, H.; Larondelle, Y. Influence of environment and genotype on polyphenol compounds and in vitro antioxidant capacity of native andean potatoes (Solanum tuberosum L.). J. Food Compos. Anal. 2009, 22, 517–524. [Google Scholar] [CrossRef]
- Ji, L.; Yogendra, K.N.; Mosa, K.A.; Kushalappa, A.C.; Piñeros-Niño, C.; Mosquera, T.; Narváez-Cuenca, C.E. Hydroxycinnamic acid functional ingredients and their biosynthetic genes in tubers of Solanum tuberosum Group Phureja. Cogent Food Agric. 2016, 2, 1138595. [Google Scholar] [CrossRef]
- Kubow, S.; Iskandar, M.M.; Sabally, K.; Azadi, B.; Ekbatan, S.S.; Kumarathasan, P.; Das, D.D.; Prakash, S.; Burgos, G.; zum Felde, T. Biotransformation of anthocyanins from two purple-fleshed sweet potato accessions in a dynamic gastrointestinal system. Food Chem. 2016, 192, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Piñeros-Niño, C.; Narváez-Cuenca, C.E.; Kushalappa, A.C.; Mosquera, T. Hydroxycinnamic acids in cooked potato tubers from Solanum tuberosum group Phureja. Food Sci. Nutr. 2017, 5, 380–389. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.R.; Wrolstad, R.; Durst, R.; Yang, C.P.; Clevidence, B. Breeding studies in potatoes containing high concentrations of anthocyanins. Am. J. Potato Res. 2003, 80, 241–249. [Google Scholar] [CrossRef]
- Brown, C.R.; Culley, D.; Bonierbale, M.; Amoros, W. Anthocyanin, carotenoid content, and antioxidant values in native South American potato cultivars. HortScience 2007, 42, 1733–1736. [Google Scholar] [CrossRef]
- Kaspar, K.L.; Park, J.S.; Brown, C.R.; Weller, K.; Ross, C.F.; Mathison, B.D.; Chew, B.P. Sensory Evaluation of Pigmented Flesh Potatoes (Solanum tuberosum L.). Food Nutr. Sci. 2013, 4, 77–81. [Google Scholar] [CrossRef][Green Version]
- Tsuda, T.; Horio, F.; Osawa, T. The role of anthocyanins as an antioxidant under oxidative stress in rats. Biofactors 2000, 13, 133–139. [Google Scholar] [CrossRef]
- Eichhorn, S.; Winterhalter, P. Anthocyanins from pigmented potato (Solanum tuberosum L.) varieties. Food Res. Int. 2005, 38, 943–948. [Google Scholar] [CrossRef]
- Lachman, J.; Hamouz, K.; Šulc, M.; Orsák, M.; Pivec, V.; Hejtmánková, A.; Dvořák, P.; Čepl, J. Cultivar differences of total anthocyanins and anthocyanidins in red and purple-fleshed potatoes and their relation to antioxidant activity. Food Chem. 2009, 114, 836–843. [Google Scholar] [CrossRef]
- Lewis, C. Anthocyanins and Related Compounds in Potatoes (Solanum tuberosum L.). Ph.D. Thesis, University of Canterbury, Christchurch, New Zealand, 1996. [Google Scholar]
- Zhang, Y.; Jung, C.S.; De Jong, W.S. Genetic analysis of pigmented tuber flesh in potato. Theor. Appl. Genet. 2009, 119, 143–150. [Google Scholar] [CrossRef]
- He, F.; Mu, L.; Yan, G.L.; Liang, N.N.; Pan, Q.H.; Wang, J.; Reeves, M.J.; Duan, C.Q. Biosynthesis of anthocyanins and their regulation in colored grapes. Molecules 2010, 15, 9057–9091. [Google Scholar] [CrossRef] [PubMed]
- Champagne, A.; Hilbert, G.; Legendre, L.; Lebot, V. Diversity of anthocyanins and other phenolic compounds among tropical root crops from Vanuatu, South Pacific. J. Food Compos. Anal. 2011, 24, 315–325. [Google Scholar] [CrossRef]
- Jiao, Y.; Jiang, Y.; Zhai, W.; Yang, Z. Studies on antioxidant capacity of anthocyanin extract from purple sweet potato (Ipomoea batatas L.). Afr. J. Biotechnol. 2012, 11, 7046–7054. [Google Scholar] [CrossRef]
- Hamouz, K.; Lachman, J.; Pazderů, K.; Tomášek, J.; Hejtmánková, K.; Pivec, V. Differences in anthocyanin content and antioxidant activity of potato tubers with different flesh colour. Plant Soil Environ. 2011, 57, 478–485. [Google Scholar] [CrossRef]
- Han, K.H.; Sekikawa, M.; Shimada, K.I.; Hashimoto, M.; Hashimoto, N.; Noda, T.; Tanaka, H.; Fukushima, M. Anthocyanin-rich purple potato flake extract has antioxidant capacity and improves antioxidant potential in rats. Br. J. Nutr. 2006, 96, 1125–1134. [Google Scholar] [CrossRef] [PubMed]
- Pillai, S.S.; Navarre, D.A.; Bamberg, J. Analysis of Polyphenols, Anthocyanins and Carotenoids in Tubers from Solanum tuberosum Group Phureja, Stenotomum and Andigena. Am. J. Potato Res. 2013, 90, 440–450. [Google Scholar] [CrossRef]
- Fossen, T.; Øvstedal, D.O.; Slimestad, R.; Andersen, Ø.M. Anthocyanins from a Norwegian potato cultivar. Food Chem. 2003, 81, 433–437. [Google Scholar] [CrossRef]
- Burmeister, A.; Bondiek, S.; Apel, L.; Kühne, C.; Hillebrand, S.; Fleischmann, P. Comparison of carotenoid and anthocyanin profiles of raw and boiled Solanum tuberosum and Solanum phureja tubers. J. Food Compos. Anal. 2011, 24, 865–872. [Google Scholar] [CrossRef]
- Hejtmánková, K.; Kotíková, Z.; Hamouz, K.; Pivec, V.; Vacek, J.; Lachman, J. Influence of flesh colour, year and growing area on carotenoid and anthocyanin content in potato tubers. J. Food Compos. Anal. 2013, 32, 20–27. [Google Scholar] [CrossRef]
- Grotewold, E. The genetics and biochemistry of floral pigments. Rev. Plant Biol. 2006, 57, 761–780. [Google Scholar] [CrossRef]
- Holton, T.A.; Cornish, E.C. Genetics and biochemistry of anthocyanin biosynthesis. Plant Cell 1995, 7, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Martin, C.; Prescott, A.; Mackay, S.; Bartlett, J.; Vrijlandt, E. Control of anthocyanin biosynthesis in flowers of Antirrhinum majus. Plant J. 1991, 1, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Van Eck, H.J.; Jacobs, J.M.E.; Van Den Berg, P.M.M.M.; Stiekema, W.J.; Jacobsen, E. The inheritance of anthocyanin pigmentation in potato (Solatium tuberosum L.) and mapping of tuber skin colour loci using RFLPs. Heredity (Edinb.) 1994, 73, 410–421. [Google Scholar] [CrossRef]
- De Jong, W.S.; Eannetta, N.T.; De Jong, D.M.; Bodis, M. Candidate gene analysis of anthocyanin pigmentation loci in the Solanaceae. Theor. Appl. Genet. 2004, 108, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Dodds Ks Long, D.H. The inheritance of colour in diploid potatoes. J. Genet. 1955, 53, 136–149. [Google Scholar] [CrossRef]
- Salaman, R.N. The inheritance of colour and other characters in the potato. J. Genet. 1910, 1, 7–46. [Google Scholar] [CrossRef]
- Dodds, E.S.; Long, D.H. The inheritance oe colour in diploid potatoes II. A three-factor linkage group. J. Genet. 1956, 54, 27. [Google Scholar] [CrossRef]
- De Jong, H. Inheritance of anthocyanin pigmentation in the cultivated potato: A critical review. Am. Potato J. 1991, 68, 585–593. [Google Scholar] [CrossRef]
- De Jong, W.S.; De Jong, D.M.; De Jong, H.; Kalazich, J.; Bodis, M. An allele of dihydroflavonol 4-reductase associated with the ability to produce red anthocyanin pigments in potato (Solanum tuberosum L.). Theor. Appl. Genet. 2003, 107, 1375–1383. [Google Scholar] [CrossRef]
- Jung, C.S.; Griffiths, H.M.; De Jong, D.M.; Cheng, S.; Bodis, M.; De Jong, W.S. The potato P locus codes for flavonoid 3′, 5′-hydroxylase. Theor. Appl. Genet. 2005, 110, 269–275. [Google Scholar] [CrossRef]
- Baudry, A.; Caboche, M.; Lepiniec, L. TT8 controls its own expression in a feedback regulation involving TTG1 and homologous MYB and bHLH factors, allowing a strong and cell-specific accumulation of flavonoids in Arabidopsis thaliana. Plant J. 2006, 46, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Hall, R.D.; Beekwilder, J. Control of anthocyanin and non-flavonoid compounds by anthocyanin-regulating MYB and bHLH transcription factors in Nicotiana benthamiana leaves. Front. Plant Sci. 2014, 5, 519. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, K.L.; Espley, R.V.; Wang, L.; Yang, H.; Yu, B.; Dare, A.; Varkonyi-Gasic, E.; Wang, J.; Zhang, J.; et al. Functional diversification of the potato R2R3 MYB anthocyanin activators AN1, MYBA1, and MYB113 and their interaction with basic helix-loop-helix cofactors. J. Exp. Bot. 2016, 67, 2159–2176. [Google Scholar] [CrossRef] [PubMed]
- Payyavula, R.; Singh, R.; Navarre, D. Transcription factors, sucrose and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp. Bot. 2013, 64, 5115–5131. [Google Scholar] [CrossRef] [PubMed]
- Rommens, C.M.; Richael, C.M.; Yan, H.; Navarre, D.A.; Ye, J.; Krucker, M.; Swords, K. Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol. J. 2008, 6, 870–886. [Google Scholar] [CrossRef] [PubMed]
- Springob, K.; Nakajima, J.; Yamazaki, M.; Saito, K. Recent advances in the biosynthesis and accumulation of anthocyanins. Nat. Prod. Rep. 2003, 20, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Mano, H.; Ogasawara, F.; Sato, K.; Higo, H.; Minobe, Y. Isolation of a Regulatory Gene of Anthocyanin Biosynthesis in Tuberous Roots of Purple-Fleshed Sweet Potato. Plant Physiol. 2007, 143, 1252–1268. [Google Scholar] [CrossRef]
- Zhang, Y.; Cheng, S.; De Jong, D.; Griffiths, H.; Halitschke, R.; De Jong, W. The potato R locus codes for dihydroflavonol 4-reductase. Theor. Appl. Genet. 2009, 119, 931–937. [Google Scholar] [CrossRef]
- D’hoop, B.B.; Keizer, P.L.C.; Paulo, M.J.; Visser, R.G.F.; van Eeuwijk, F.A.; van Eck, H.J. Identification of agronomically important QTL in tetraploid potato cultivars using a marker-trait association analysis. Theor. Appl. Genet. 2014, 127, 731–748. [Google Scholar] [CrossRef]
- Kumar, D.; Singh, B.P.; Kumar, P. An overview of the factors affecting sugar content of potatoes. Ann. Appl. Biol. 2004, 145, 247–256. [Google Scholar] [CrossRef]
- Duarte-Delgado, D.; Ñústez-López, C.E.; Narváez-Cuenca, C.E.; Restrepo-Sánchez, L.P.; Melo, S.E.; Sarmiento, F.; Kushalappa, A.C.; Mosquera-Vásquez, T. Natural variation of sucrose, glucose and fructose contents in Colombian genotypes of Solanum tuberosum Group Phureja at harvest. J. Sci. Food Agric. 2016, 96, 4288–4294. [Google Scholar] [CrossRef] [PubMed]
- Colmenares, G.A. Stratified/PCA: Un método de preprocesamiento de datos y variables para la construcción de modelos de redes neuronales. Rev. Econ. 2000, 16, 1–31. [Google Scholar]
- Ripley, B.D. The R project in statistical computing. MSOR Connect. Newsl. LTSN Maths Stats OR Netw. 2001, 1, 23–25. [Google Scholar] [CrossRef]
- Poland, J.A.; Rife, T.W. Genotyping-by-sequencing for plant breeding and genetics. Plant Genome 2012, 5, 92–102. [Google Scholar] [CrossRef]
- Sonah, H.; Bastien, M.; Iquira, E.; Tardivel, A.; Légaré, G.; Boyle, B.; Normandeau, É.; Laroche, J.; Larose, S.; Jean, M.; et al. An Improved Genotyping by Sequencing (GBS) Approach Offering Increased Versatility and Efficiency of SNP Discovery and Genotyping. PLoS ONE 2013, 8, e54603. [Google Scholar] [CrossRef]
- Duitama, J.; Quintero, J.C.; Cruz, D.F.; Quintero, C.; Hubmann, G.; Foulquie-Moreno, M.R.; Verstrepen, K.J.; Thevelein, J.M.; Tohme, J. An integrated framework for discovery and genotyping of genomic variants from high-throughput sequencing experiments. Nucleic Acids Res. 2014, 42, e44. [Google Scholar] [CrossRef] [PubMed]
- Potato Genome Sequencing Consortium. Genome sequence and analysis of the tuber crop potato. Nature 2011, 475, 189–195. [Google Scholar] [CrossRef]
- Marchini, J.; Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 2010, 11, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Juyó, D.; Sarmiento, F.; Álvarez, M.; Brochero, H.; Gebhardt, C.; Mosquera, T. Genetic diversity and population structure in diploid potatoes of Solanum tuberosum group phureja. Crop Sci. 2015, 55, 760–769. [Google Scholar] [CrossRef]
- Zhang, Z.; Ersoz, E.; Lai, C.Q.; Todhunter, R.J.; Tiwari, H.K.; Gore, M.A.; Bradbury, P.J.; Yu, J.; Arnett, D.K.; Ordovas, J.M.; et al. Mixed linear model approach adapted for genome-wide association studies. Nat. Genet. 2010, 42, 355–360. [Google Scholar] [CrossRef]
- Lipka, A.E.; Tian, F.; Wang, Q.; Peiffer, J.; Li, M.; Bradbury, P.J.; Gore, M.A.; Buckler, E.S.; Zhang, Z. GAPIT: Genome association and prediction integrated tool. Bioinformatics 2012, 28, 2397–2399. [Google Scholar] [CrossRef] [PubMed]
- Sukumaran, S.; Xiang, W.; Bean, S.R.; Pedersen, J.F.; Kresovich, S.; Tuinstra, M.R.; Tesso, T.T.; Hamblin, M.T.; Yu, J. Association Mapping for Grain Quality in a Diverse Sorghum Collection. Plant Genome J. 2012, 5, 126–135. [Google Scholar] [CrossRef]
- Turner, S.D. qqman: An R package for visualizing GWAS results using QQ and manhattan plots. Biorxiv 2014, 005165. [Google Scholar] [CrossRef]
- Johnson, R.C.; Nelson, G.W.; Troyer, J.L.; Lautenberger, J.A.; Kessing, B.D.; Winkler, C.A.; O’Brien, S.J. Accounting for multiple comparisons in a genome-wide association study (GWAS). BMC Genom. 2010, 11, 724. [Google Scholar] [CrossRef] [PubMed]
- Jung, C.S.; Griffiths, H.M.; De Jong, D.M.; Cheng, S.; Bodis, M.; Kim, T.S.; De Jong, W.S. The potato developer (D) locus encodes an R2R3 MYB transcription factor that regulates expression of multiple anthocyanin structural genes in tuber skin. Theor. Appl. Genet. 2009, 120, 45–57. [Google Scholar] [CrossRef]
- Vos, P.G.; Paulo, M.J.; Voorrips, R.E.; Visser, R.G.F.; van Eck, H.J.; van Eeuwijk, F.A. Evaluation of LD decay and various LD-decay estimators in simulated and SNP-array data of tetraploid potato. Theor. Appl. Genet. 2017, 130, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Spelt, C.; Quattrocchio, F.; Mol, J.N.; Koes, R. Anthocyanin1 of Petunia Encodes a Basic Helix-Loop-Helix Protein That Directly Activates Transcription of Structural Anthocyanin Genes. Plant Cell 2000, 12, 1619–1631. [Google Scholar] [CrossRef]
- Allan, A.; Hellens, R.; Laing, W. MYB transcription factors that colour our fruit. Trends Plant Sci. 2008, 13, 99–102. [Google Scholar] [CrossRef]
- Gonzalez, A.; Zhao, M.; Leavitt, J.; Lloyd, A. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008, 53, 814–827. [Google Scholar] [CrossRef]
- Mathews, H.; Clendennen, S.K.; Caldwell, C.G.; Liu, X.L.; Connors, K.; Matheis, N.; Schuster, D.K.; Menasco, D.J.; Wagoner, W.; Lightner, J.; et al. Activation tagging in tomato identifies a transcriptional regulator of anthocyanin biosynthesis, modification, and transport. Plant Cell 2003, 15, 1689–1703. [Google Scholar] [CrossRef]
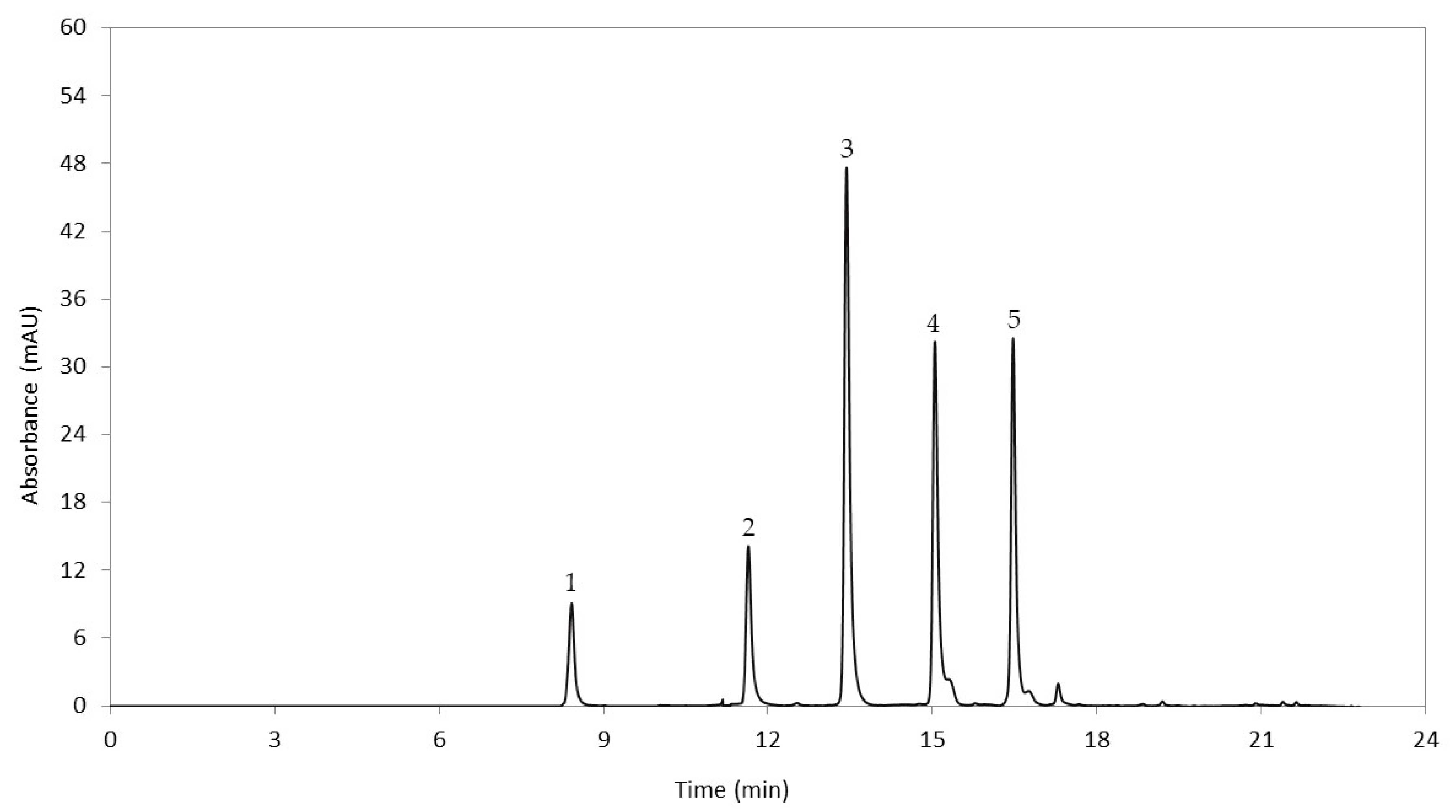
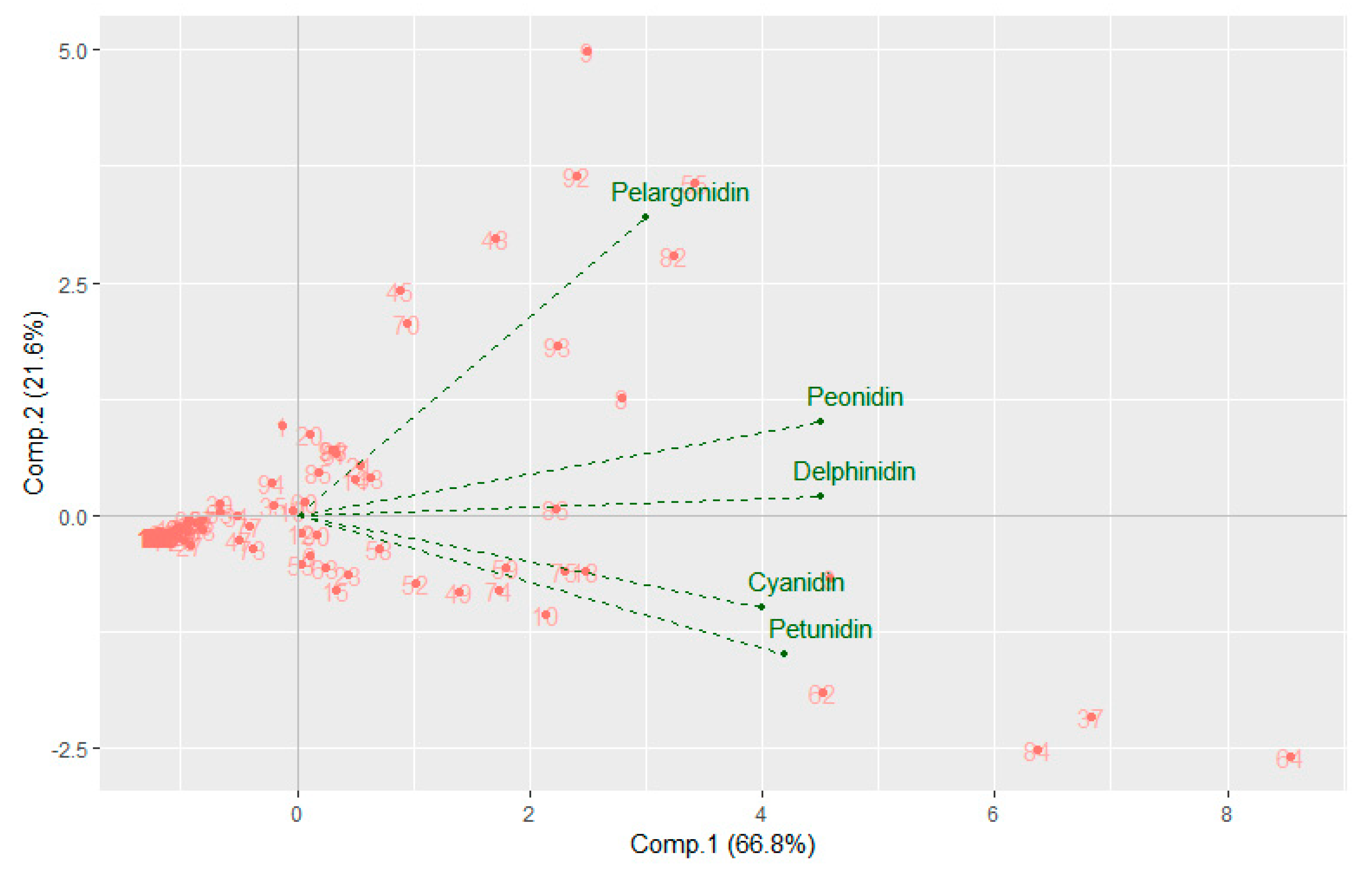
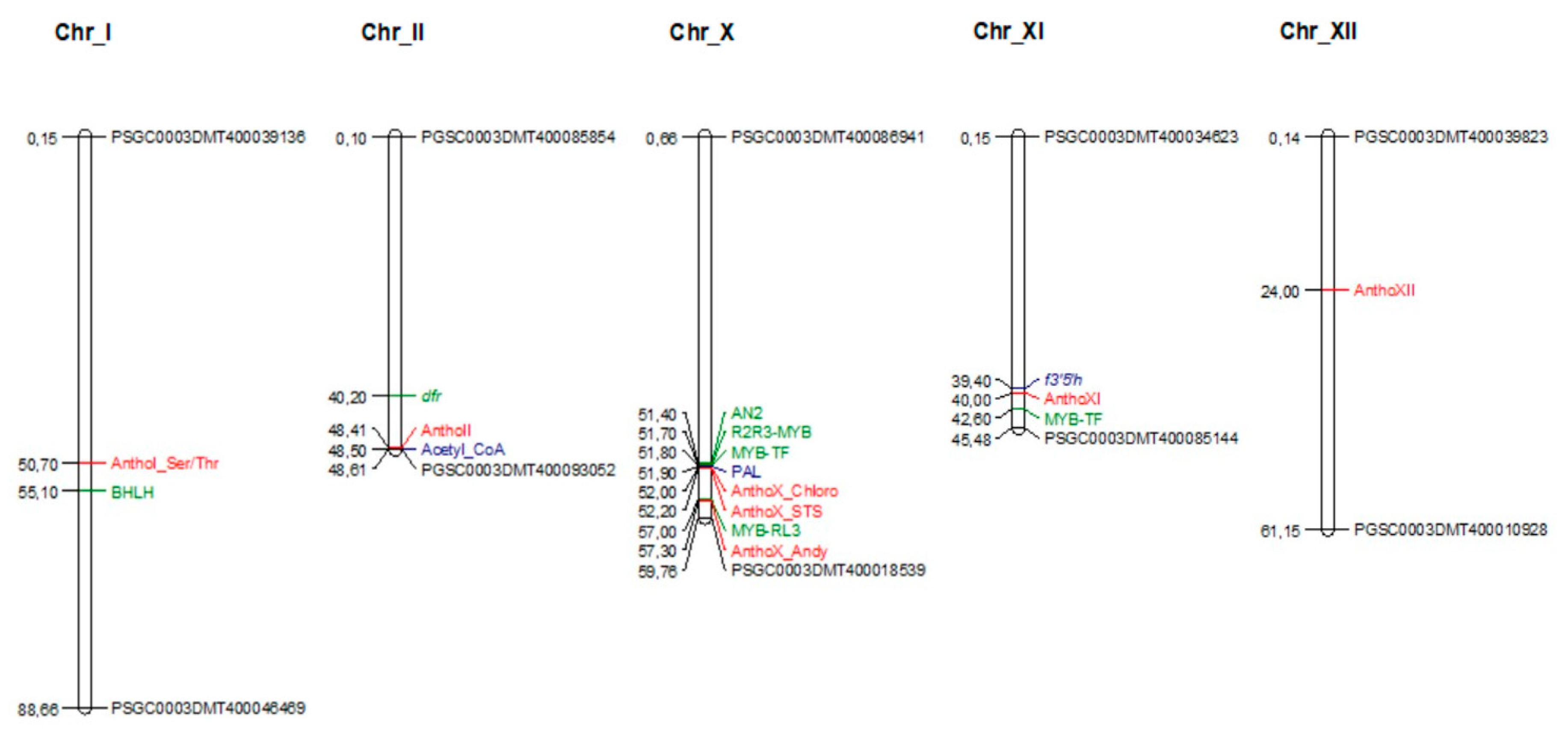
| Anthocyanidins | QTL Name | Chr | Position | p-Value | Model R2 | Effect | Gene Annotation |
|---|---|---|---|---|---|---|---|
| Cyanidin | AnthoX_Chloro | X | 52004868 | 1.23 × 10−6 | 0.400 | −0.222 | Chloroplast threonine deaminase 1 |
| AnthoX_STS | X | 52261553 | 4.75 × 10−6 | 0.374 | −0.024 | STS14 protein | |
| AnthoI_Ser/Thr | I | 50785405 | 1.51 × 10−4 | 0.313 | 0.313 | Serine/threonine-protein phosphatase | |
| Delphinidin | AnthoX_Chloro | X | 52004868 | 5.97 × 10−5 | 0.384 | 0.121 | Chloroplast threonine deaminase 1 |
| AnthoX_STS | X | 52261553 | 5.97 × 10−5 | 0.384 | 0.048 | STS14 protein | |
| AnthoXI | XI | 40001900 | 1.04 × 10−4 | 0.374 | −0.100 | No functional annotation | |
| Pelargonidin | AnthoX_Chloro | X | 52261553 | 6.06 × 10−7 | 0.439 | −0.670 | STS14 protein |
| AnthoX_STS | X | 52004868 | 8.58 × 10−7 | 0.433 | −0.308 | Chloroplast threonine deaminase 1 | |
| AnthoX_Adeny | X | 57301864 | 2.91 × 10−6 | 0.411 | −0.670 | Adenylyl-sulfate kinase | |
| AnthoX_ChloroI | X | 52004940 | 1.02 × 10−5 | 0.389 | −0.330 | Chloroplast threonine deaminase 1 | |
| Peonidin | AnthoX_Chloro | X | 52004868 | 2.87 × 10−5 | 0.444 | −0.071 | Chloroplast threonine deaminase 1 |
| AnthoX_STS | X | 52261553 | 2.87 × 10−5 | 0.444 | −0.052 | STS14 protein | |
| Petunidin | AnthoXII | XII | 24259182 | 1.99 × 10−5 | 0.382 | −0.483 | Hypothetical gene of unknown function |
| AnthoX_Chloro | X | 52004868 | 7.11 × 10−5 | 0.361 | −0.103 | Chloroplast threonine deaminase 1 | |
| AnthoII | II | 48517142 | 1.08 × 10−4 | 0.354 | −0.523 | No functional annotation |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parra-Galindo, M.-A.; Piñeros-Niño, C.; Soto-Sedano, J.C.; Mosquera-Vasquez, T. Chromosomes I and X Harbor Consistent Genetic Factors Associated with the Anthocyanin Variation in Potato. Agronomy 2019, 9, 366. https://doi.org/10.3390/agronomy9070366
Parra-Galindo M-A, Piñeros-Niño C, Soto-Sedano JC, Mosquera-Vasquez T. Chromosomes I and X Harbor Consistent Genetic Factors Associated with the Anthocyanin Variation in Potato. Agronomy. 2019; 9(7):366. https://doi.org/10.3390/agronomy9070366
Chicago/Turabian StyleParra-Galindo, María-Angélica, Clara Piñeros-Niño, Johana Carolina Soto-Sedano, and Teresa Mosquera-Vasquez. 2019. "Chromosomes I and X Harbor Consistent Genetic Factors Associated with the Anthocyanin Variation in Potato" Agronomy 9, no. 7: 366. https://doi.org/10.3390/agronomy9070366
APA StyleParra-Galindo, M.-A., Piñeros-Niño, C., Soto-Sedano, J. C., & Mosquera-Vasquez, T. (2019). Chromosomes I and X Harbor Consistent Genetic Factors Associated with the Anthocyanin Variation in Potato. Agronomy, 9(7), 366. https://doi.org/10.3390/agronomy9070366





