Transcriptome Profiles of Contrasting Potato (Solanum tuberosum L.) Genotypes Under Water Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Biological Material
2.2. Greenhouse Stress Assays
2.3. Experimental Design and Statistical Analysis
2.4. RNA Extraction
2.5. Library Preparation and Sequencing
2.6. nCounter mRNA Quantification
3. Results
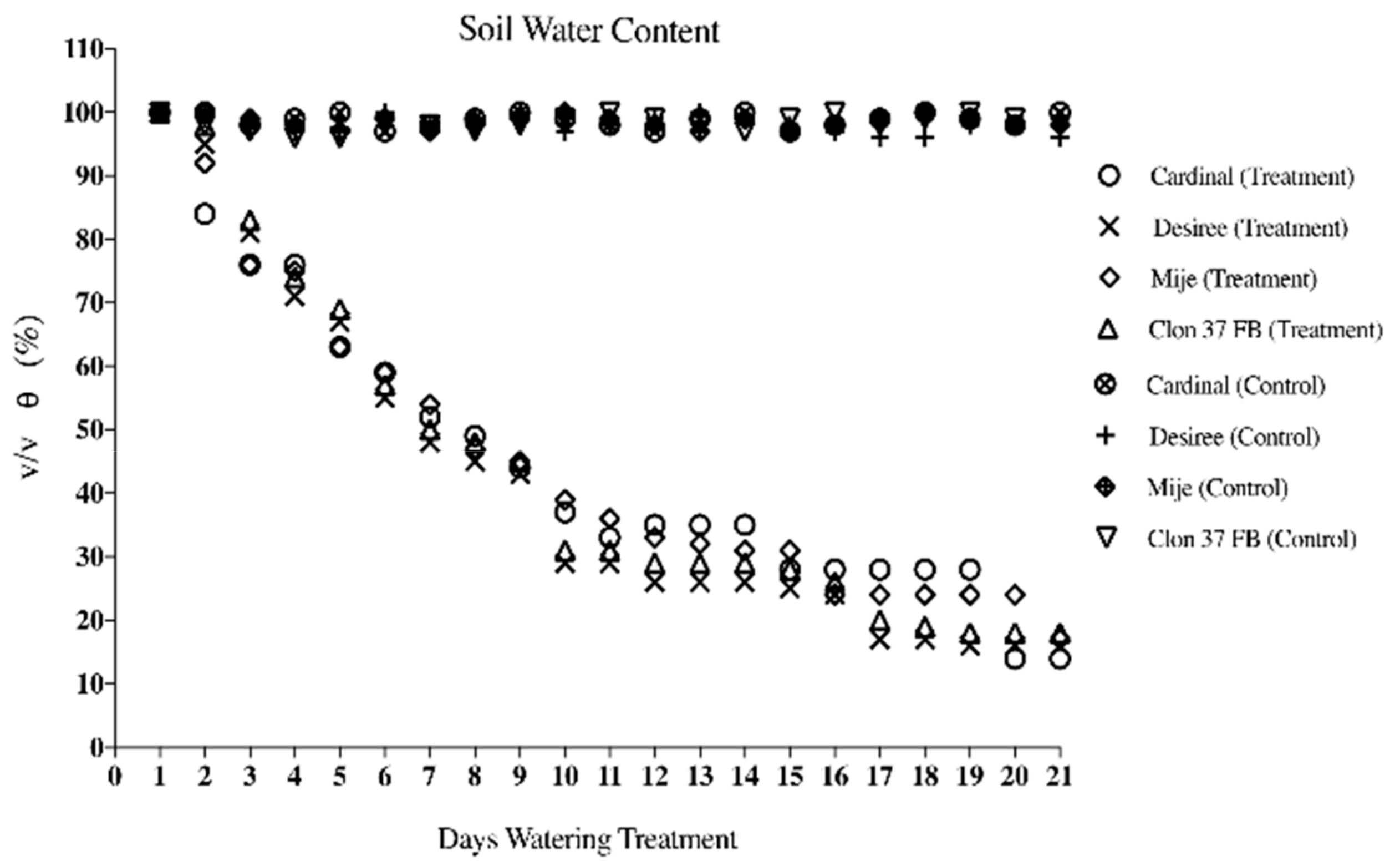
3.1. Soil Water Content
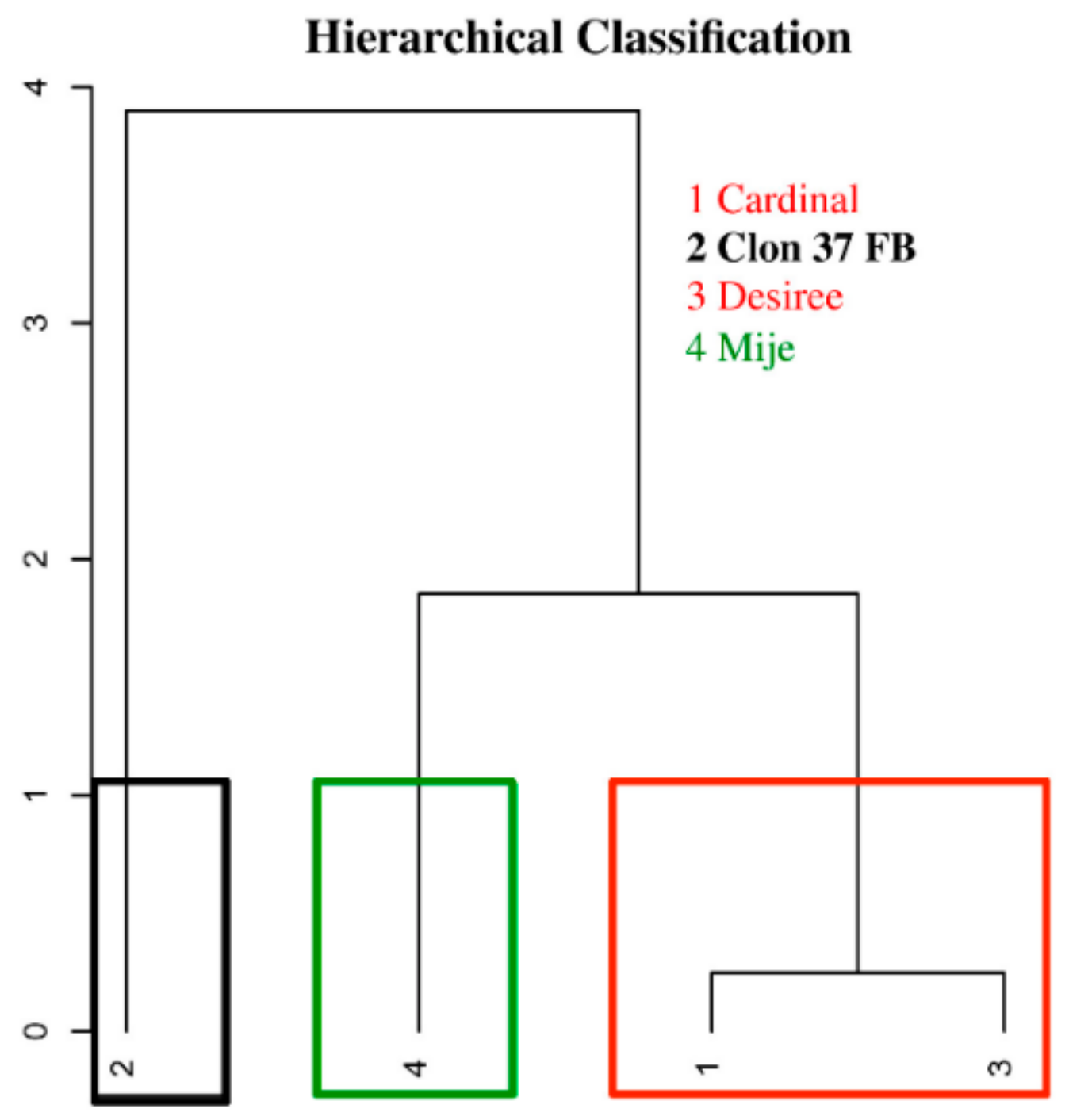
3.2. Relationship Between Performance Under Water Stress by Plant and Selection of Water Stress Tolerance Indices
3.3. Agronomic Characteristic Changes in Potato Plants Under Water Stress
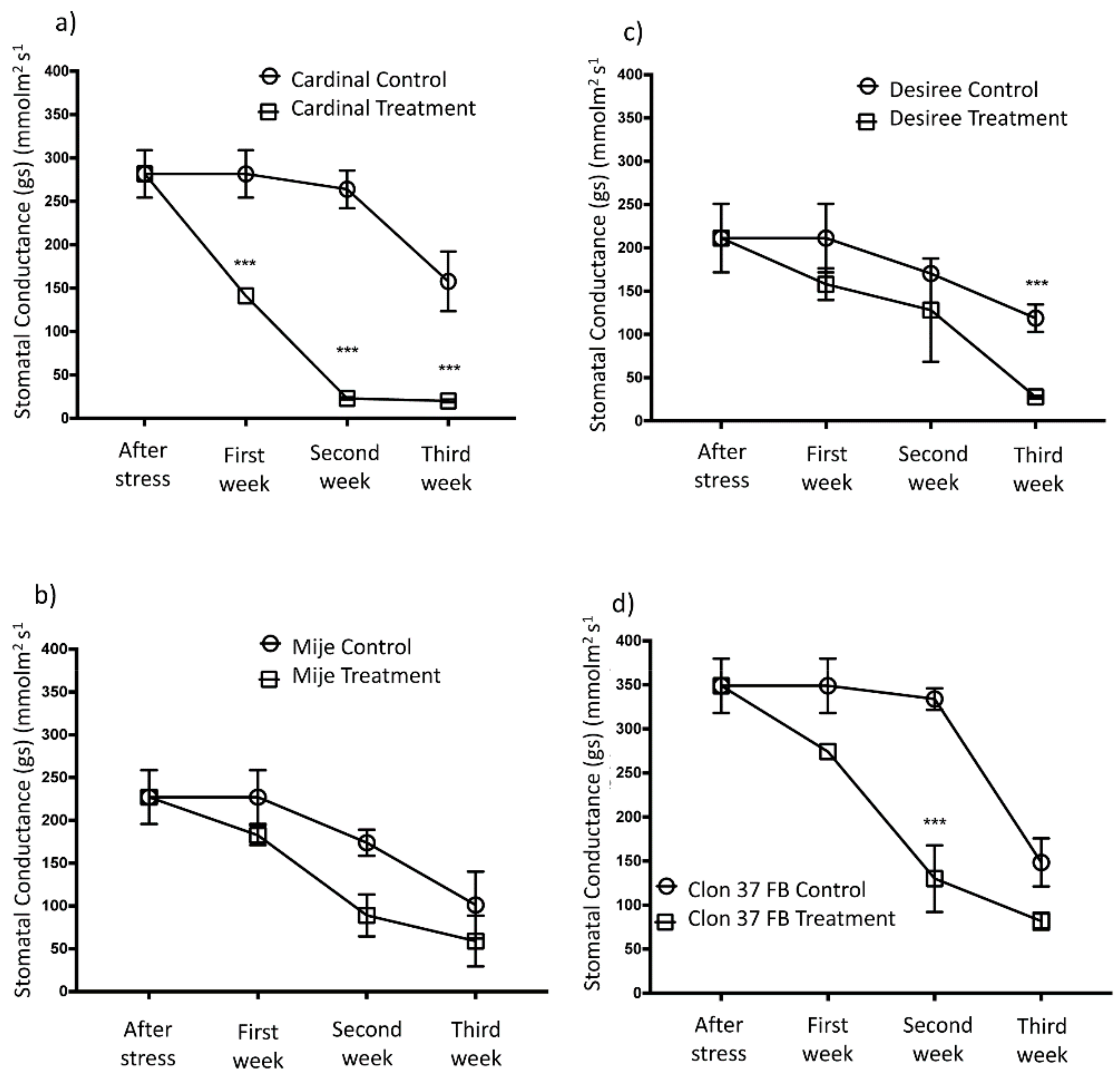
3.4. Gas Exchange in Potato Plants Under Water Stress
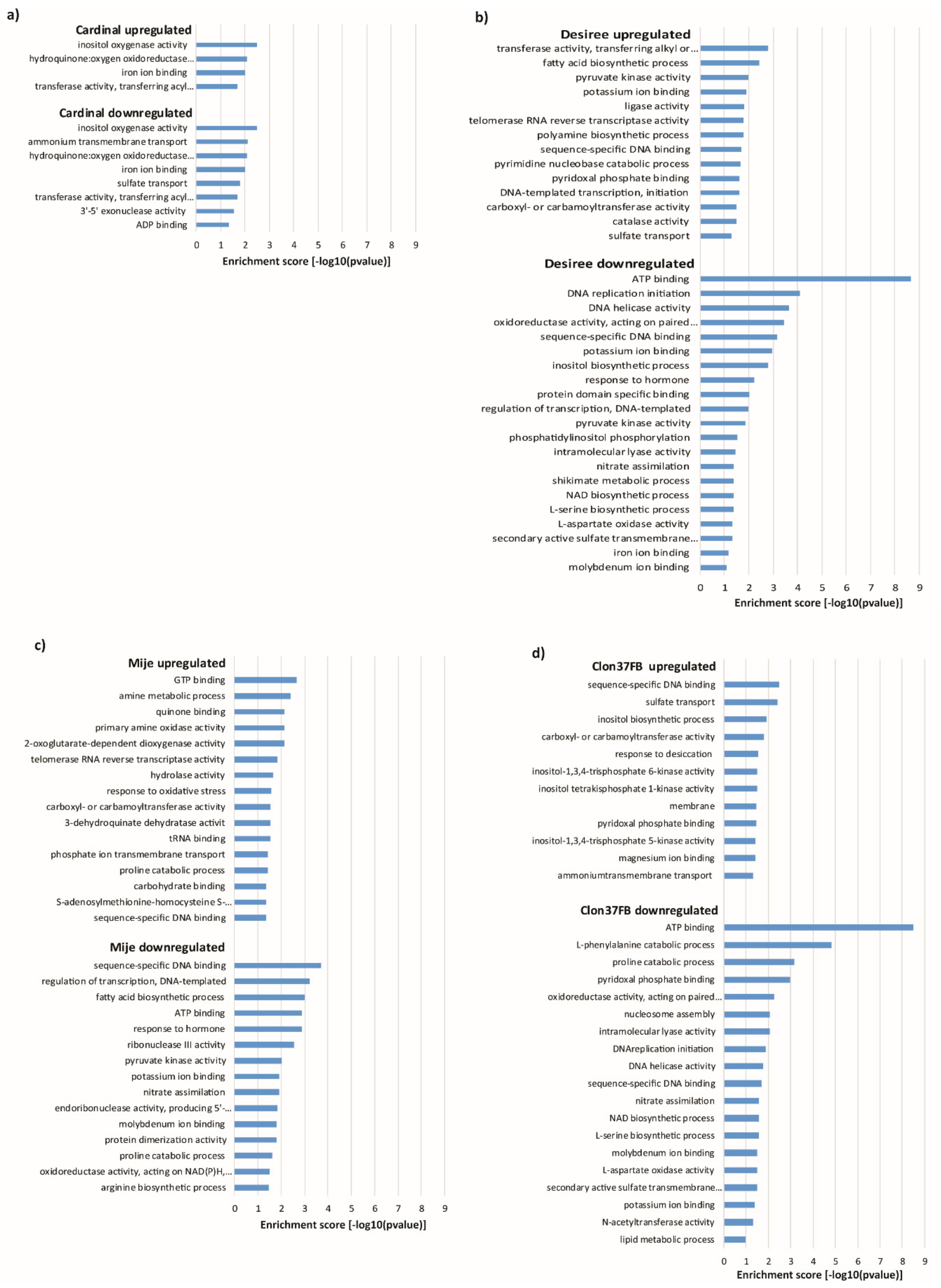
3.5. Transcriptome Sequencing Reveals Water Stress Responsive Genes
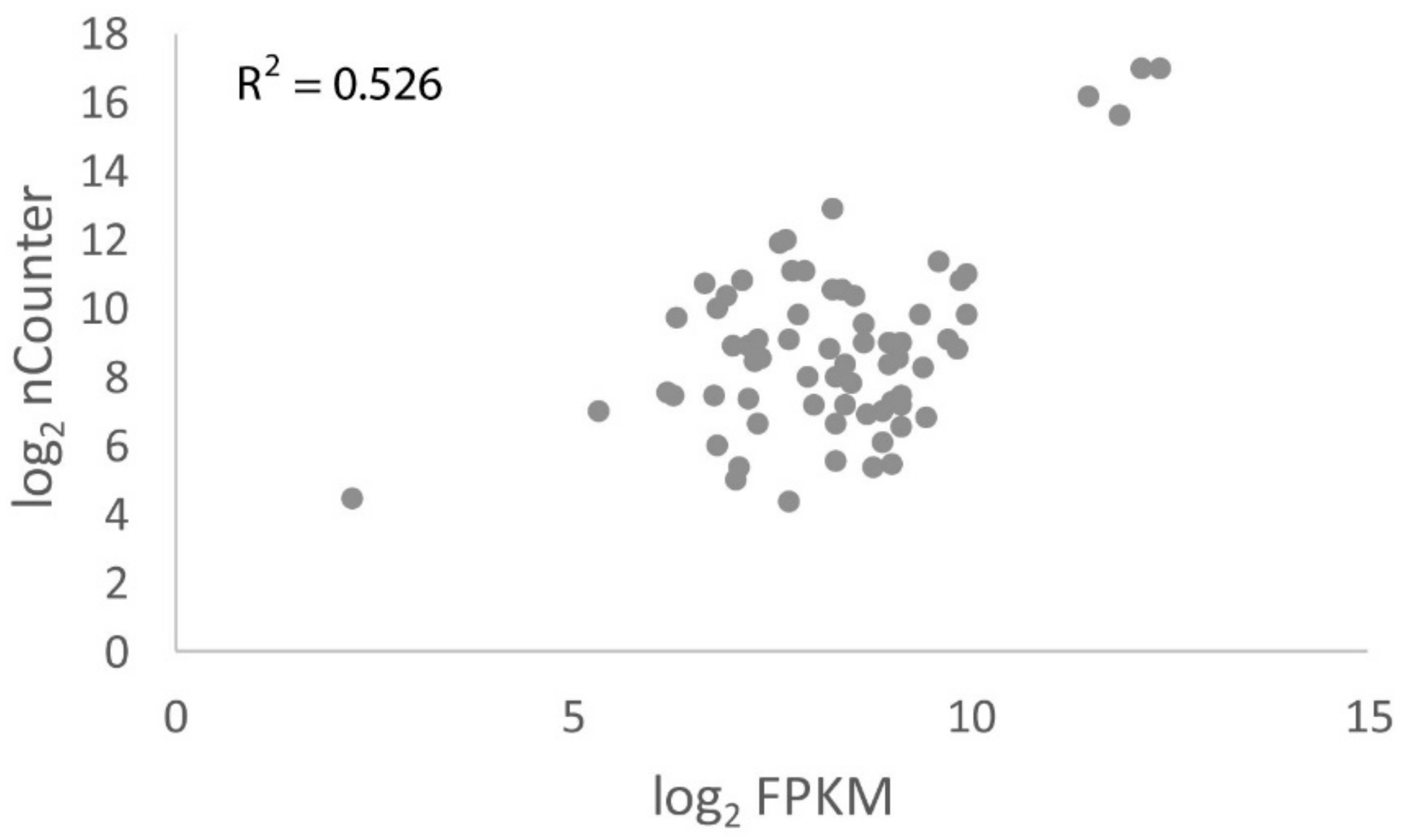
3.6. nCounter Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Haan, S.; Rodriguez, F. Potato Origin and Production. In Advances in Potato Chemistry and Technology, 2nd ed.; Singh, J., Kaur, L., Eds.; Academic Press: London, UK, 2016; pp. 1–32. [Google Scholar]
- Deblonde, P.M.K.; Ledent, J.F. Effects of moderate drought conditions on green leaf number, stem height, leaf length and tuber yield of potato cultivars. Eur. J. Agron. 2001, 14, 31–41. [Google Scholar] [CrossRef]
- Andre, C.M.; Ghislain, M.; Bertin, P.; Oufir, M.; del Rosario Herrera, M.; Hoffmann, L.; Hausman, J.-F.; Larondelle, Y.; Evers, D. Andean Potato Cultivars (Solanum tuberosum L.) as a Source of Antioxidant and Mineral Micronutrients. J. Agric. Food Chem. 2007, 55, 366–378. [Google Scholar] [CrossRef] [PubMed]
- Obidiegwu, J.E.; Bryan, G.J.; Jones, H.G.; Prashar, A. Coping with drought: Stress and adaptive responses in potato and perspectives for improvement. Front. Plant Sci. 2015, 6, 542. [Google Scholar] [CrossRef] [PubMed]
- Campos, D.; Noratto, G.; Chirinos, R.; Arbizu, C.; Roca, W.; Cisneros-Zevallos, L. Antioxidant capacity and secondary metabolites in four species of Andean tuber crops: Native potato (Solanum sp.), mashua (Tropaeolum tuberosum Ruiz & PavónPav´Pavón), Oca (Oxalis tuberosa Molina) and ulluco (Ullucus tuberosus Caldas). J. Sci. Food Agric. 2006, 86, 1481–1488. [Google Scholar]
- Ritter, E.; Barandalla, L.; López, R.; de Galarreta, J.I.R. Exploitation of Exotic, Cultivated Solanum Germplasm for Breeding and Commercial Purposes. Potato Res. 2008, 51, 301–311. [Google Scholar] [CrossRef]
- André, C.M.; Schafleitner, R.; Legay, S.; Lefèvre, I.; Aliaga, C.A.A.; Nomberto, G.; Hoffmann, L.; Hausman, J.-F.; Larondelle, Y.; Evers, D. Gene expression changes related to the production of phenolic compounds in potato tubers grown under drought stress. Phytochemistry 2009, 70, 1107–1116. [Google Scholar] [CrossRef]
- Schafleitner, R.; Gutierrez Rosales, R.O.; Gaudin, A.; Alvarado Aliaga, C.A.; Martinez, G.N.; Tincopa Marca, L.R.; Bolivar, L.A.; Delgado, F.M.; Simon, R.; Bonierbale, M. Capturing candidate drought tolerance traits in two native Andean potato clones by transcription profiling of field grown plants under water stress. Plant Physiol. Biochem. 2007, 45, 673–690. [Google Scholar] [CrossRef]
- Vacher, J. Responses of two main Andean crops, quinoa (Chenopodium quinoa Willd) and papa amarga (Solanum juzepczukii Buk.) to drought on the Bolivian Altiplano: Significance of local adaptation. Agric. Ecosyst. Environ. 1998, 68, 99–108. [Google Scholar] [CrossRef]
- Chaves, M.M.; Pereira, J.S.; Maroco, J.; Rodrigues, M.L.; Ricardo, C.P.P.; Oso, M.L.; Rio, Â.; Carvalho, I.; Faria, T.; Pinheiro, C. How Plants Cope with Water Stress in the Field. Photosynthesis and Growth. Ann. Bot. 2002, 89, 907–916. [Google Scholar] [CrossRef]
- Moore, J.P.; Le, N.T.; Brandt, W.F.; Driouich, A.; Farrant, J.M. Towards a systems-based understanding of plant desiccation tolerance. Trends Plant Sci. 2009, 14, 110–117. [Google Scholar] [CrossRef]
- Zhu, J.-K. Cell signaling under salt, water and cold stresses. Curr. Opin. Plant Biol. 2001, 4, 401–406. [Google Scholar] [CrossRef]
- Cabello, R.; Monneveux, P.; Bonierbale, M.; Khan, M.A. Heritability of yield components under irrigated and drought conditions in andigenum potatoes. Am. J. Potato Res. 2014, 91, 492–499. [Google Scholar] [CrossRef]
- Ramirez-Vallejo, P.; Kelly, J.D. Traits related to drought resistance in common bean. Euphytica 1998, 99, 127–136. [Google Scholar] [CrossRef]
- Seki, M.; Narusaka, M.; Ishida, J.; Nanjo, T.; Fujita, M.; Oono, Y.; Kamiya, A.; Nakajima, M.; Enju, A.; Sakurai, T.; et al. Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 2002, 31, 279–292. [Google Scholar] [CrossRef] [PubMed]
- Massa, A.N.; Childs, K.L.; Lin, H.; Bryan, G.J.; Giuliano, G.; Buell, C.R. The Transcriptome of the Reference Potato Genome Solanum tuberosum Group Phureja Clone DM1-3 516R44. PLoS ONE 2011, 6, e26801. [Google Scholar] [CrossRef] [PubMed]
- Evers, D.; Lefèvre, I.; Legay, S.; Lamoureux, D.; Hausman, J.-F.; Rosales, R.O.G.; Marca, L.R.T.; Hoffmann, L.; Bonierbale, M.; Schafleitner, R. Identification of drought-responsive compounds in potato through a combined transcriptomic and targeted metabolite approach. J. Exp. Bot. 2010, 61, 2327–2343. [Google Scholar] [CrossRef]
- Gong, L.; Zhang, H.; Gan, X.; Zhang, L.; Chen, Y.; Nie, F.; Shi, L.; Li, M.; Guo, Z.; Zhang, G.; et al. Transcriptome profiling of the potato (Solanum tuberosum L.) plant under drought stress and water-stimulus conditions. PLoS ONE 2015, 10, e0128041. [Google Scholar] [CrossRef]
- Watkinson, J.I.; Hendricks, L.; Sioson, A.A.; Vasquez-Robinet, C.; Stromberg, V.; Heath, L.S.; Schuler, M.; Bohnert, H.J.; Bonierbale, M.; Grene, R. Accessions of Solanum tuberosum ssp. andigena show differences in photosynthetic recovery after drought stress as reflected in gene expression profiles. Plant Sci. 2006, 171, 745–758. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2006, 58, 221–227. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, B.; Ma, C.; Zhang, G.; Chang, J.; Si, H.; Wang, D. Transcriptome characterization and sequencing-based identification of drought-responsive genes in potato. Mol. Biol. Rep. 2014, 41, 505–517. [Google Scholar] [CrossRef]
- Barra, M.; Correa, J.; Salazar, E.; Sagredo, B. Response of Potato (Solanum tuberosum L.) Germplasm to Water Stress Under In Vitro Conditions. Am. J. Potato Res. 2013, 90, 591–606. [Google Scholar] [CrossRef]
- Murashige, T.; Skoog, F. A Revised Medium for Rapid Growth and Bio Assays with Tobacco Tissue Cultures. Physiol. Plant. 1962, 15, 473–497. [Google Scholar] [CrossRef]
- Toledo, J.; Espinoza, N.; Golmirzaie, A. Cultivo de Tejidos: Manejo de Plantulas In Vitro en la Produccion de Semilla de Papa; Martínez, E., Ed.; Centro internacional de la Papa: Lima, Peru, 1998; pp. 1–13. [Google Scholar]
- Rosielle, A.A.; Hamblin, J. Theoretical Aspects of Selection for Yield in Stress and Non-Stress Environment1. Crop Sci. 1981, 21, 943–946. [Google Scholar] [CrossRef]
- Fernandez, G.C. Effective selection criteria for assessing plant stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and other Food Crops in Temperature and Water Stress, Shanhua, Taiwan, 13–16 August 1992; pp. 257–270. [Google Scholar]
- Hossain, A.B.S.; Sears, R.G.; Cox, T.S.; Paulsen, G.M. Desiccation Tolerance and Its Relationship to Assimilate Partitioning in Winter Wheat. Crop Sci. 1990, 30, 622–627. [Google Scholar] [CrossRef]
- Fischer, R.; Maurer, R. Drought resistance in spring wheat cultivars. I. Grain yield responses. Aust. J. Agric. Res. 1978, 29, 897–912. [Google Scholar] [CrossRef]
- Bouslama, M.; Schapaugh, W.T. Stress Tolerance in Soybeans. I. Evaluation of Three Screening Techniques for Heat and Drought Tolerance. Crop Sci. 1984, 24, 933–937. [Google Scholar] [CrossRef]
- Levy, D.; Coleman, W.K.; Veilleux, R.E. Adaptation of Potato to Water Shortage: Irrigation Management and Enhancement of Tolerance to Drought and Salinity. Am. J. Potato Res. 2013, 90, 186–206. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S.L. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)—From genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef] [PubMed]
- Tomato Genome Consortium The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [CrossRef] [PubMed]
- Alexa, A.; Rahnenfuhrer, J. topGO: Enrichment Analysis for Gene Ontology. R Package Version 2. 2010. Available online: https://bioconductor.org/packages/release/bioc/html/topGO.html (accessed on 15 July 2019).
- Hirsch, C.D.; Hamilton, J.P.; Childs, K.L.; Cepela, J.; Crisovan, E.; Vaillancourt, B.; Hirsch, C.N.; Habermann, M.; Neal, B.; Buell, C.R. Spud DB: A Resource for Mining Sequences, Genotypes, and Phenotypes to Accelerate Potato Breeding. Plant Genome 2014, 7, 42. [Google Scholar] [CrossRef]
- Geiss, G.K.; Bumgarner, R.E.; Birditt, B.; Dahl, T.; Dowidar, N.; Dunaway, D.L.; Fell, H.P.; Ferree, S.; George, R.D.; Grogan, T.; et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat. Biotechnol. 2008, 26, 317–325. [Google Scholar] [CrossRef] [PubMed]
- Tai, H.H.; Zebarth, B.J. Effect of Time of Day of Sampling on Potato Foliar Gene Expression Used to Assess Crop Nitrogen Status. Am. J. Potato Res. 2015, 92, 284–293. [Google Scholar] [CrossRef]
- Gálvez, J.H.; Tai, H.H.; Lagüe, M.; Zebarth, B.J.; Strömvik, M.V. The nitrogen responsive transcriptome in potato (Solanum tuberosum L.) reveals significant gene regulatory motifs. Sci. Rep. 2016, 6, 26090. [Google Scholar] [CrossRef]
- Quiñones, V.; Zanlungo, S.; Holuigue, L.; Litvak, S.; Jordana, X. The cox1 initiation codon is created by RNA editing in potato mitochondria. Plant Physiol. 1995, 108, 1327–1328. [Google Scholar] [CrossRef]
- Nakane, E.; Kawakita, K.; Doke, N.; Yoshioka, H. Elicitation of primary and secondary metabolism during defense in the potato. J. Gen. Plant Pathol. 2003, 69, 378–384. [Google Scholar] [CrossRef]
- Drouin, G.; Dover, G.A. Independent gene evolution in the potato actin gene family demonstrated by phylogenetic procedures for resolving gene conversions and the phylogeny of angiosperm actin genes. J. Mol. Evol. 1990, 31, 132–150. [Google Scholar] [CrossRef]
- Romero, A.P.; Alarcón, A.; Valbuena, R.I.; Galeano, C.H. Physiological Assessment of Water Stress in Potato Using Spectral Information. Front. Plant Sci. 2017, 8, 1608. [Google Scholar] [CrossRef]
- Saravia, D.; Farfán-Vignolo, E.R.; Gutiérrez, R.; De Mendiburu, F.; Schafleitner, R.; Bonierbale, M.; Khan, M.A. Yield and Physiological Response of Potatoes Indicate Different Strategies to Cope with Drought Stress and Nitrogen Fertilization. Am. J. Potato Res. 2016, 93, 288–295. [Google Scholar] [CrossRef]
- Rudack, K.; Seddig, S.; Sprenger, H.; Köhl, K.; Uptmoor, R.; Ordon, F. Drought stress-induced changes in starch yield and physiological traits in potato. J. Agron. Crop Sci. 2017, 203, 494–505. [Google Scholar] [CrossRef]
- Pieczynski, M.; Wyrzykowska, A.; Milanowska, K.; Boguszewska-Mankowska, D.; Zagdanska, B.; Karlowski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. Genomewide identification of genes involved in the potato response to drought indicates functional evolutionary conservation with Arabidopsis plants. Plant Biotechnol. J. 2018, 16, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Pirasteh-Anosheh, H.; Saed-Moucheshi, A.; Pakniyat, H.; Pessarakli, M. Stomatal responses to drought stress. In Water Stress and Crop Plants; Ahmad, P., Ed.; John Wiley & Sons, Ltd.: Chichester, UK, 2016; pp. 24–40. [Google Scholar]
- Olvera-Carrillo, Y.; Luis Reyes, J.; Covarrubias, A.A. Late embryogenesis abundant proteins: Versatile players in the plant adaptation to water limiting environments. Plant Signal. Behav. 2011, 6, 586–589. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Mukherjee, S.; Basak, P.; Majumder, A.L. Significance of galactinol and raffinose family oligosaccharide synthesis in plants. Front. Plant Sci. 2015, 6, 656. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine Function in Plants: Metabolism, Regulation on Development, and Roles in Abiotic Stress Responses. Front. Plant Sci. 2019, 9, 1945. [Google Scholar] [CrossRef]
- He, M.; Dijkstra, F.A. Drought effect on plant nitrogen and phosphorus: A meta-analysis. New Phytol. 2014, 204, 924–931. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.E.; Jones, J.D.G. Plant Disease Resistance Genes. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1997, 48, 575–607. [Google Scholar] [CrossRef]
- Breen, S.; Williams, S.J.; Outram, M.; Kobe, B.; Solomon, P.S. Emerging Insights into the Functions of Pathogenesis-Related Protein 1. Trends Plant Sci. 2017, 22, 871–879. [Google Scholar] [CrossRef]
- Sung, D.Y.; Vierling, E.; Guy, C.L. Comprehensive Expression Profile Analysis of the Arabidopsis Hsp70 Gene Family. Plant Physiol. 2001, 126, 789–800. [Google Scholar] [CrossRef]


| Total Fresh Weight of Tuber | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Control (g/plant) | Treatment (g/plant) | Response (treatment/control) × 100 (%) | |||||||||
| Cardinal | 35.1 | ± | 12.7 | Ba | 13.4 | ± | 1.2 | Bb | 42.24 | ± | 12.9 | A |
| Desiree | 32.5 | ± | 3.7 | Ba | 25.7 | ± | 2.4 | Cb | 83.15 | ± | 7.9 | A |
| Mije | 24.9 | ± | 2.1 | Ba | 24.7 | ± | 1.4 | Ca | 99.84 | ± | 7.4 | AB |
| Clone 37 FB | 5.3 | ± | 0.9 | Aa | 7.8 | ± | 1.2 | Ab | 151.7 | ± | 45.6 | B |
| Average Fresh Weight of Tubers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Control (g/tuber) | Treatment (g/tuber) | Response (treatment/control) × 100 (%) | |||||||||
| Cardinal | 7.0 | ± | 2.5 | Ba | 3.01 | ± | 0.4 | Ab | 44.65 | ± | 9.39 | A |
| Desiree | 9.9 | ± | 2.3 | Ca | 8.6 | ± | 0.8 | Ba | 92.43 | ± | 18.23 | A |
| Mije | 3.2 | ± | 0.3 | ABa | 2.8 | ± | 0.1 | Aa | 88.77 | ± | 13.5 | A |
| Clone 37 FB | 1.6 | ± | 0.3 | Aa | 2.6 | ± | 0.4 | Ab | 165.3 | ± | 38.2 | B |
| Average Dry Weight of Tubers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Control (g/tuber) | Treatment (g/tuber) | Response (treatment/control) × 100 (%) | |||||||||
| Cardinal | 1.5 | ± | 0.7 | ABa | 0.7 | ± | 0.1 | Aa | 53.61 | ± | 21.32 | A |
| Desiree | 2.1 | ± | 0.7 | Ba | 1.7 | ± | 0.4 | Bb | 93.60 | ± | 18 | A |
| Mije | 0.5 | ± | 0.1 | Aa | 0.6 | ± | 0.04 | Aa | 121.15 | ± | 26.74 | A |
| Clone 37 FB | 0.2 | ± | 0.1 | Aa | 0.3 | ± | 0.1 | Aa | 64.83 | ± | 75.02 | A |
| Average Water Content of Tubers | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Control (g/tuber) | Treatment (g/tuber) | Response (treatment /control) × 100 (%) | |||||||||
| Cardinal | 5.5 | ± | 2.4 | ABa | 2.3 | ± | 0.4 | Aa | 44.20 | ± | 10.10 | A |
| Desiree | 7.8 | ± | 2.0 | Ba | 6.8 | ± | 0.4 | Ba | 93.25 | ± | 21.1 | A |
| Mije | 2.7 | ± | 0.3 | Aa | 2.2 | ± | 0.1 | Aa | 82.98 | ± | 15.64 | A |
| Clone 37 FB | 1.4 | ± | 0.4 | Aa | 2.3 | ± | 0.5 | Aa | 179.65 | ± | 54.09 | B |
| Average Water Content of Shoot + Root | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genotype | Control (g/tuber) | Treatment (g/tuber) | Response (treatment /control) × 100 (%) | |||||||||
| Cardinal | 12.02 | ± | 2.9 | Aa | 8.5 | ± | 2.9 | ABa | 71.59 | ± | 22.67 | A |
| Desiree | 23.6 | ± | 6.1 | Ba | 14.7 | ± | 2.8 | Cb | 64.17 | ± | 12.66 | A |
| Mije | 6.3 | ± | 1.4 | Aa | 4.2 | ± | 1.9 | Ab | 57.13 | ± | 24.74 | A |
| Clone 37 FB | 6.1 | ± | 0.7 | Aa | 11.9 | ± | 1.5 | BCa | 196.81 | ± | 44.66 | B |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barra, M.; Meneses, C.; Riquelme, S.; Pinto, M.; Lagüe, M.; Davidson, C.; Tai, H.H. Transcriptome Profiles of Contrasting Potato (Solanum tuberosum L.) Genotypes Under Water Stress. Agronomy 2019, 9, 848. https://doi.org/10.3390/agronomy9120848
Barra M, Meneses C, Riquelme S, Pinto M, Lagüe M, Davidson C, Tai HH. Transcriptome Profiles of Contrasting Potato (Solanum tuberosum L.) Genotypes Under Water Stress. Agronomy. 2019; 9(12):848. https://doi.org/10.3390/agronomy9120848
Chicago/Turabian StyleBarra, Macarena, Claudio Meneses, Stephanie Riquelme, Manuel Pinto, Martin Lagüe, Charlotte Davidson, and Helen H. Tai. 2019. "Transcriptome Profiles of Contrasting Potato (Solanum tuberosum L.) Genotypes Under Water Stress" Agronomy 9, no. 12: 848. https://doi.org/10.3390/agronomy9120848
APA StyleBarra, M., Meneses, C., Riquelme, S., Pinto, M., Lagüe, M., Davidson, C., & Tai, H. H. (2019). Transcriptome Profiles of Contrasting Potato (Solanum tuberosum L.) Genotypes Under Water Stress. Agronomy, 9(12), 848. https://doi.org/10.3390/agronomy9120848




