Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Drought Treatment
2.2. Transcriptome Data and Transcriptome Assembly
2.3. Long Non-Coding RNA Identification
2.4. Prediction and Functional Annotation of Long Non-Coding RNA Targets
2.5. Quantitative Reverse Transcription–Polymerase Chain Reaction Analysis
2.6. Statistical Analysis
3. Results and Discussion
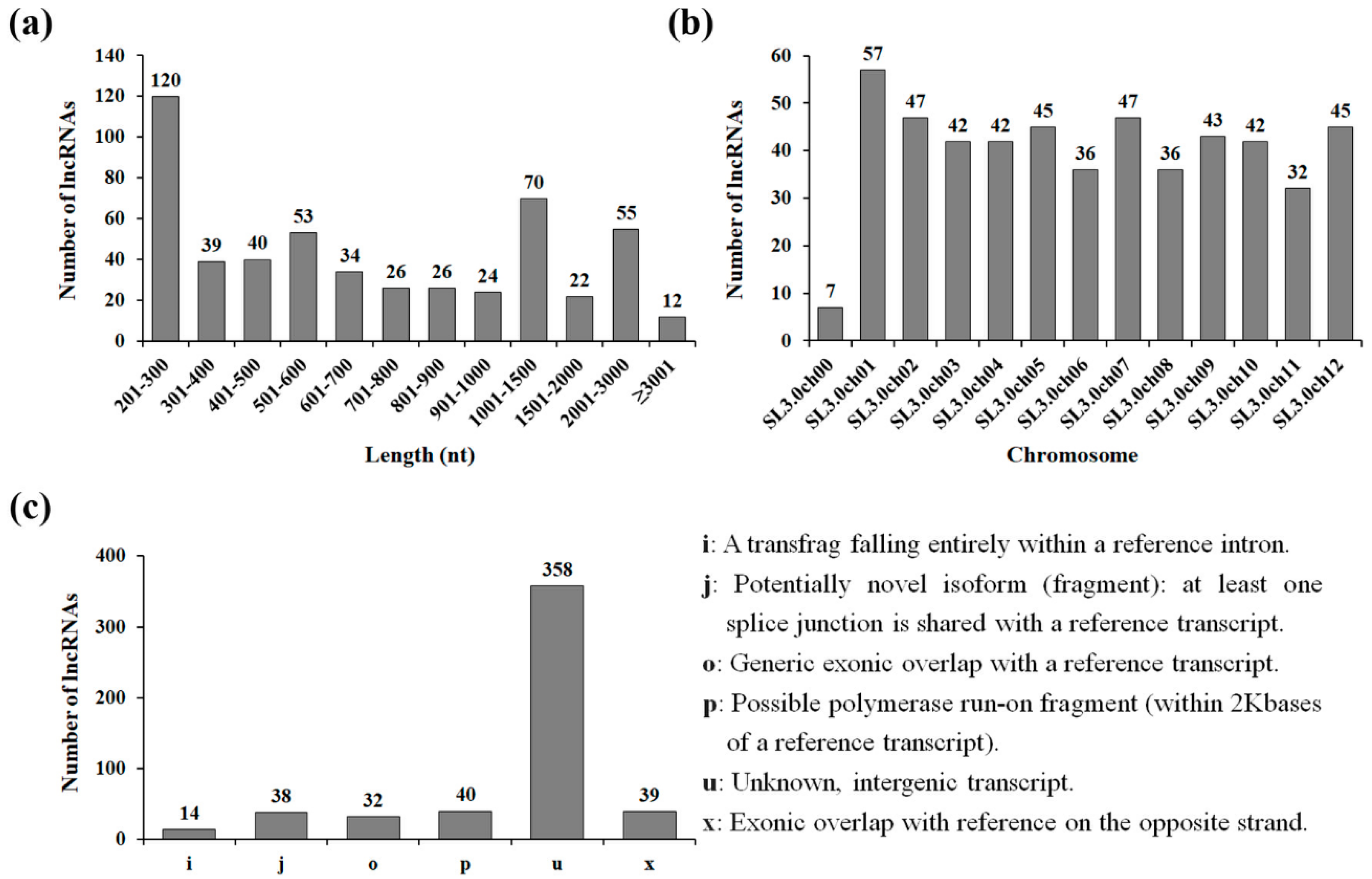
3.1. Identification and Characterization of Drought-Responsive Tomato Long Non-Coding RNAs
3.2. Drought-Responsive Tomato Long Non-Coding RNA Transcripts as Potential Targets of Tomato miRNAs
3.3. Functional Characterization of Drought-Responsive Tomato Long Non-Coding RNAs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Tilman, D.; Balzer, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef]
- Reichstein, M.; Bahn, M.; Ciais, P.; Frank, D.; Mahecha, M.D.; Seneviratne, S.I.; Zscheischler, J.; Beer, C.; Buchmann, N.; Frank, D.C.; et al. Climate extremes and the carbon cycle. Nature 2013, 500, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Zipper, S.C.; Qiu, J.; Kucharik, C.J. Drought effects on US maize and soybean production: Spatiotemporal patterns and historical changes. Environ. Res. Lett. 2016, 11, 094021. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Eom, S.H.; Baek, S.A.; Kim, J.K.; Hyun, T.K. Transcriptome analysis in Chinese cabbage (Brassica rapa ssp. pekinensis) provides the role of glucosinolate metabolism in response to drought stress. Molecules 2018, 23, E1186. [Google Scholar] [CrossRef] [PubMed]
- Chekanova, J.A. Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 2015, 27, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Rowley, M.J.; Böhmdorfer, G.; Wierzbicki, A.T. Analysis of long non-coding RNAs produced by a specialized RNA polymerase in Arabidopsis thaliana. Methods 2013, 63, 160–169. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Chang, H.Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 2012, 81, 145–166. [Google Scholar] [CrossRef]
- Liu, J.; Jung, C.; Xu, J.; Wang, H.; Deng, S.; Bernad, L.; Arenas-Huertero, C.; Chua, N.H. Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 2012, 24, 4333–4345. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zheng, H.; Sui, N. Regulation mechanism of long non-coding RNA in plant response to stress. Biochem. Biophys. Res. Commun. 2018, 503, 402–407. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, J.; Lian, B.; Gu, H.; Li, Y.; Qi, Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat. Commun. 2018, 9, 5056. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.H.; Quan, M.Y.; Zhang, D.Q. Genome-wide identification of novel long non-coding RNAs in Populus tomentosa tension wood, opposite wood and normal wood xylem by RNA-seq. Planta 2014, 241, 125–143. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.Y.; Liu, Z.C. Global identification and analysis of long non-coding RNAs in diploid strawberry Fragaria vesca, during flower and fruit development. BMC Genom. 2015, 16, 815. [Google Scholar] [CrossRef] [PubMed]
- Khemka, N.; Singh, V.K.; Garg, R.; Jain, M. Genome-wide analysis of long intergenic non-coding RNAs in chickpea and their potential role in flower development. Sci. Rep. 2016, 6, 33297. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xia, X.; Jiang, H.; Lu, Z.; Cui, J.; Cao, F.; Jin, B. Genome-wide identification and characterization of novel lncRNAs in Ginkgo biloba. Trees 2018, 32, 1429–1442. [Google Scholar] [CrossRef]
- Li, L.; Eichten, S.R.; Shimizu, R.; Petsch, K.; Yeh, C.T.; Wu, W.; Chettoor, A.M.; Givan, S.A.; Cole, R.A.; Fowler, J.E.; et al. Genome-wide discovery and characterization of maize long non-coding RNAs. Genome Biol. 2014, 15, R40. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.C.; Liao, J.Y.; Li, Z.Y.; Yu, Y.; Zhang, J.P.; Li, Q.F.; Qu, L.H.; Shu, W.S.; Chen, Y.Q. Genome-wide screening and functional analysis identify a large number of long noncoding RNAs involved in the sexual reproduction of rice. Genome Biol. 2014, 15, 512. [Google Scholar] [CrossRef] [PubMed]
- Chung, P.J.; Jung, H.; Jeong, D.H.; Ha, S.H.; Choi, Y.D.; Kim, J.K. Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genom. 2016, 17, 563. [Google Scholar] [CrossRef]
- Qin, T.; Zhao, H.; Cui, P.; Albesher, N.; Xiong, L. A nucleus-localized long non-coding RNA enhances drought and salt stress tolerance. Plant Physiol. 2017, 175, 1321–1336. [Google Scholar] [CrossRef]
- Ben Amor, B.; Wirth, S.; Merchan, F.; Laporte, P.; d’Aubenton-Carafa, Y.; Hirsch, J.; Maizel, A.; Mallory, A.; Lucas, A.; Deragon, J.M.; et al. Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 2009, 19, 57–69. [Google Scholar] [CrossRef]
- Lee, H.J.; Eom, S.H.; Lee, J.H.; Wi, S.H.; Kim, S.K.; Hyun, T.H. Genome-wide analysis of alternative splicing events during response to drought stress in tomato (Solanum lycopersicum L.). J. Hortic. Sci. Biotechnol. 2019. [Google Scholar] [CrossRef]
- Sun, L.; Zhang, Z.; Bailey, T.L.; Perkins, A.C.; Tallack, M.R.; Xu, Z.; Liu, H. Prediction of novel long non-coding RNAs based on RNA-Seq data of mouse Klf1 knockout study. BMC Bioinform. 2012, 13, 331. [Google Scholar] [CrossRef]
- Hou, X.; Du, Y.; Liu, X.; Zhang, H.; Liu, Y.; Yan, N.; Zhang, Z. Genome-wide analysis of long non-coding RNAs in potato and their potential role in tuber sprouting process. Int. J. Mol. Sci. 2017, 19, E101. [Google Scholar] [PubMed]
- Yu, T.; Zhu, H. Long non-coding RNAs: Rising regulators of plant reproductive development. Agronomy 2019, 9, 53. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, W.; Wang, Y. Genome-wide identification of long non-coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae. Mol. Plant Pathol. 2018, 19, 2177–2186. [Google Scholar] [CrossRef] [PubMed]
- Ulitsky, I.; Bartel, D.P. lincRNAs: Genomics, evolution, and mechanisms. Cell 2013, 154, 26–46. [Google Scholar] [CrossRef] [PubMed]
- Yamada, M. Functions of long intergenic non-coding (linc) RNAs in plants. J. Plant Res. 2017, 130, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Shuai, P.; Liang, D.; Tang, S.; Zhang, Z.; Ye, C.Y.; Su, Y.; Xia, X.; Yin, W. Genome-wide identification and functional prediction of novel and drought-responsive lincRNAs in Populus trichocarpa. J. Exp. Bot. 2014, 65, 4975–4983. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.D.; Sung, S. Long noncoding RNA: Unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 2012, 17, 16–21. [Google Scholar] [CrossRef]
- Liu, X.; Hao, L.; Li, D.; Zhu, L.; Hu, S. Long non-coding RNAs and their biological roles in plants. Genom. Proteom. Bioinform. 2015, 13, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.J.; Wang, Z.M.; Wang, M.; Wang, X.J. Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 2013, 161, 1875–1884. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Zhuang, Z.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018, 46, W49–W54. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Shen, J.; Hou, X.; Yao, J.; Li, X.; Xiao, J.; Xiong, L. Gradual increase of miR156 regulates temporal expression changes of numerous genes during leaf development in rice. Plant Physiol. 2012, 158, 1382–1394. [Google Scholar] [CrossRef] [PubMed]
- Fu, C.; Sunkar, R.; Zhou, C.; Shen, H.; Zhang, J.Y.; Matts, J.; Wolf, J.; Mann, D.G.; Stewart, C.N., Jr.; Tang, Y.; et al. Overexpression of miR156 in switchgrass (Panicum virgatum L.) results in various morphological alterations and leads to improved biomass production. Plant Biotechnol. J. 2012, 10, 443–452. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, Z.; Amyot, L.; Tian, L.; Xu, Z.; Gruber, M.Y.; Hannoufa, A. Ectopic expression of miR156 represses nodulation and causes morphological and developmental changes in Lotus japonicus. Mol. Genet. Genom. 2015, 290, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Arshad, M.; Feyissa, B.A.; Amyot, L.; Aung, B.; Hannoufa, A. MicroRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) by silencing SPL13. Plant Sci. 2017, 258, 122–136. [Google Scholar] [CrossRef] [PubMed]
- Stief, A.; Altmann, S.; Hoffmann, K.; Pant, B.D.; Scheible, W.R.; Bäurle, I. Arabidopsis miR156 regulates tolerance to recurring environmental stress through spl transcription factors. Plant Cell 2014, 26, 1792–1807. [Google Scholar] [CrossRef] [PubMed]
- Macovei, A.; Gill, S.S.; Tuteja, N. MicroRNAs as promising tools for improving stress tolerance in rice. Plant Signal. Behav. 2012, 7, 1296–1301. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Liang, R.; Ge, L.; Li, W.; Xiao, H.; Lin, H.; Ruan, K.; Jin, Y. Identification of drought-induced microRNAs in rice. Biochem. Biophys. Res. Commun. 2007, 354, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.X.; Yu, D.Q. Overexpression of Arabidopsis miR396 enhances drought tolerance in transgenic tobacco plants. Acta Bot. Yunnan. 2010, 31, 421–426. [Google Scholar] [CrossRef]
- Huang, Z.A.; Huang, Y.A.; You, Z.H.; Zhu, Z.; Sun, Y. Novel link prediction for large-scale miRNA-lncRNA interaction network in a bipartite graph. BMC Med. Genom. 2018, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Zhang, X.; Wang, W.; Yuan, R.; Shen, F. Identification of Gossypium hirsutum long non-coding RNAs (lncRNAs) under salt stress. BMC Plant Biol. 2018, 18, 23. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Liu, L.; Li, J.; Le, T.D. LncmiRSRN: Identification and analysis of long non-coding RNA related miRNA sponge regulatory network in human cancer. Bioinformatics 2018, 34, 4232–4240. [Google Scholar] [CrossRef] [PubMed]
- Schor, I.E.; Bussotti, G.; Maleš, M.; Forneris, M.; Viales, R.R.; Enright, A.J.; Furlong, E.E.M. Non-coding RNA expression, function, and variation during Drosophila embryogenesis. Curr. Biol. 2018, 28, 3547–3561. [Google Scholar] [CrossRef] [PubMed]
- Scruggs, B.S.; Gilchrist, D.A.; Nechaev, S.; Muse, G.W.; Burkholder, A.; Fargo, D.C.; Adelman, K. Bidirectional transcription arises from two distinct hubs of transcription factor binding and active chromatin. Mol. Cell 2015, 58, 1101–1112. [Google Scholar] [CrossRef] [PubMed]
- Ørom, U.A.; Derrien, T.; Beringer, M.; Gumireddy, K.; Gardini, A.; Bussotti, G.; Lai, F.; Zytnicki, M.; Notredame, C.; Huang, Q.; et al. Long noncoding RNAs with enhancer-like function in human cells. Cell 2010, 143, 46–58. [Google Scholar] [CrossRef]
- Camblong, J.; Iglesias, N.; Fickentscher, C.; Dieppois, G.; Stutz, F. Antisense RNA stabilization induces transcriptional gene silencing via histone deacetylation in S. cerevisiae. Cell 2007, 131, 706–717. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, Y.; Liu, X.; Jiang, J. Comparative transcriptome profiling of two tomato genotypes in response to potassium-deficiency stress. Int. J. Mol. Sci. 2018, 19, E2402. [Google Scholar] [CrossRef]
- Shabala, S. Regulation of potassium transport in leaves: From molecular to tissue level. Ann. Bot. 2003, 92, 627–634. [Google Scholar] [CrossRef]
- Tanaka, Y.; Sano, T.; Tamaoki, M.; Nakajima, N.; Kondo, N.; Hasezawa, S. Cytokinin and auxin inhibit abscisic acid-induced stomatal closure by enhancing ethylene production in Arabidopsis. J. Exp. Bot. 2006, 57, 2259–2266. [Google Scholar] [CrossRef] [PubMed]
- Veach, Y.K.; Martin, R.C.; Mok, D.W.; Malbeck, J.; Vankova, R.; Mok, M.C. O-glucosylation of cis-zeatin in maize. Characterization of genes, enzymes, and endogenous cytokinins. Plant Physiol. 2003, 131, 1374–1380. [Google Scholar] [CrossRef] [PubMed]
- Havlová, M.; Dobrev, P.I.; Motyka, V.; Storchová, H.; Libus, J.; Dobrá, J.; Malbeck, J.; Gaudinová, A.; Vanková, R. The role of cytokinins in responses to water deficit in tobacco plants over-expressing trans-zeatin O-glucosyltransferase gene under 35S or SAG12 promoters. Plant Cell Environ. 2008, 31, 341–353. [Google Scholar] [CrossRef]
- Keshishian, E.A.; Hallmark, H.T.; Ramaraj, T.; Plačková, L.; Sundararajan, A.; Schilkey, F.; Novák, O.; Rashotte, A.M. Salt and oxidative stresses uniquely regulate tomato cytokinin levels and transcriptomic response. Plant Direct 2018, 2, e00071. [Google Scholar] [CrossRef] [PubMed]
- Hoshiyasu, S.; Kohzuma, K.; Yoshida, K.; Fujiwara, M.; Fukao, Y.; Yokota, A.; Akashi, K. Potential involvement of N-terminal acetylation in the quantitative regulation of the ε subunit of chloroplast ATP synthase under drought stress. Biosci. Biotechnol. Biochem. 2013, 77, 998–1007. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eom, S.H.; Lee, H.J.; Lee, J.H.; Wi, S.H.; Kim, S.K.; Hyun, T.K. Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato. Agronomy 2019, 9, 629. https://doi.org/10.3390/agronomy9100629
Eom SH, Lee HJ, Lee JH, Wi SH, Kim SK, Hyun TK. Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato. Agronomy. 2019; 9(10):629. https://doi.org/10.3390/agronomy9100629
Chicago/Turabian StyleEom, Seung Hee, Hee Ju Lee, Jin Hyoung Lee, Seung Hwan Wi, Sung Kyeom Kim, and Tae Kyung Hyun. 2019. "Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato" Agronomy 9, no. 10: 629. https://doi.org/10.3390/agronomy9100629
APA StyleEom, S. H., Lee, H. J., Lee, J. H., Wi, S. H., Kim, S. K., & Hyun, T. K. (2019). Identification and Functional Prediction of Drought-Responsive Long Non-Coding RNA in Tomato. Agronomy, 9(10), 629. https://doi.org/10.3390/agronomy9100629




