Genome-Wide Identification and Expression Analysis of the Dof Transcription Factor Gene Family in Gossypium hirsutum L.
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification and Annotation of Dof Gene Family Members in G. hirsutum
2.2. GhDof Protein Alignment and Phylogenetic Analysis
2.3. Duplication and Synteny Analysis of GhDof Genes
2.4. Plant Materials, Treatments, and Collections
2.5. RNA Isolation and qRT-PCR Experiments
3. Results
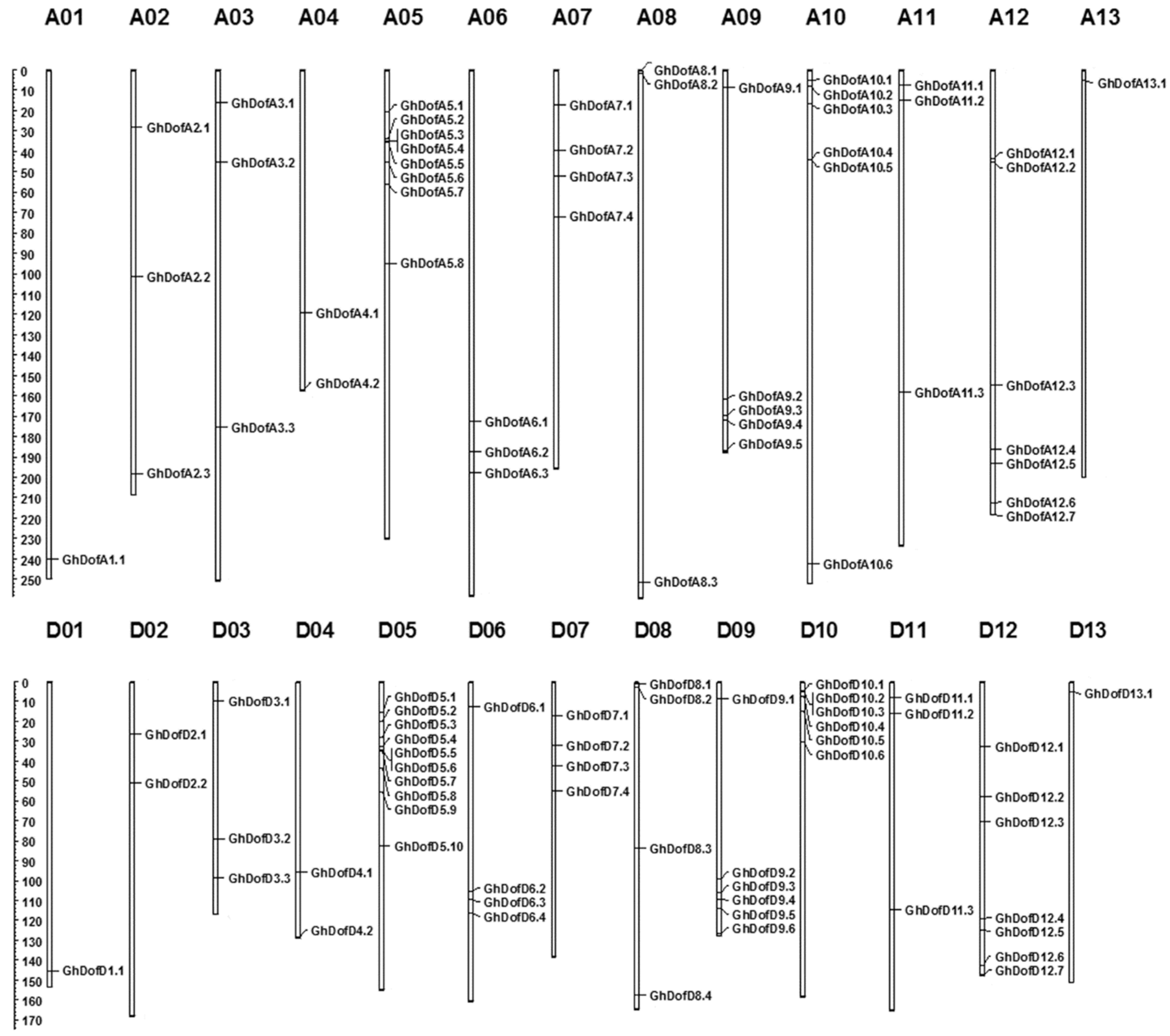
3.1. Identification of GhDof Transcription Factor Gene Family Members in G. hirsutum
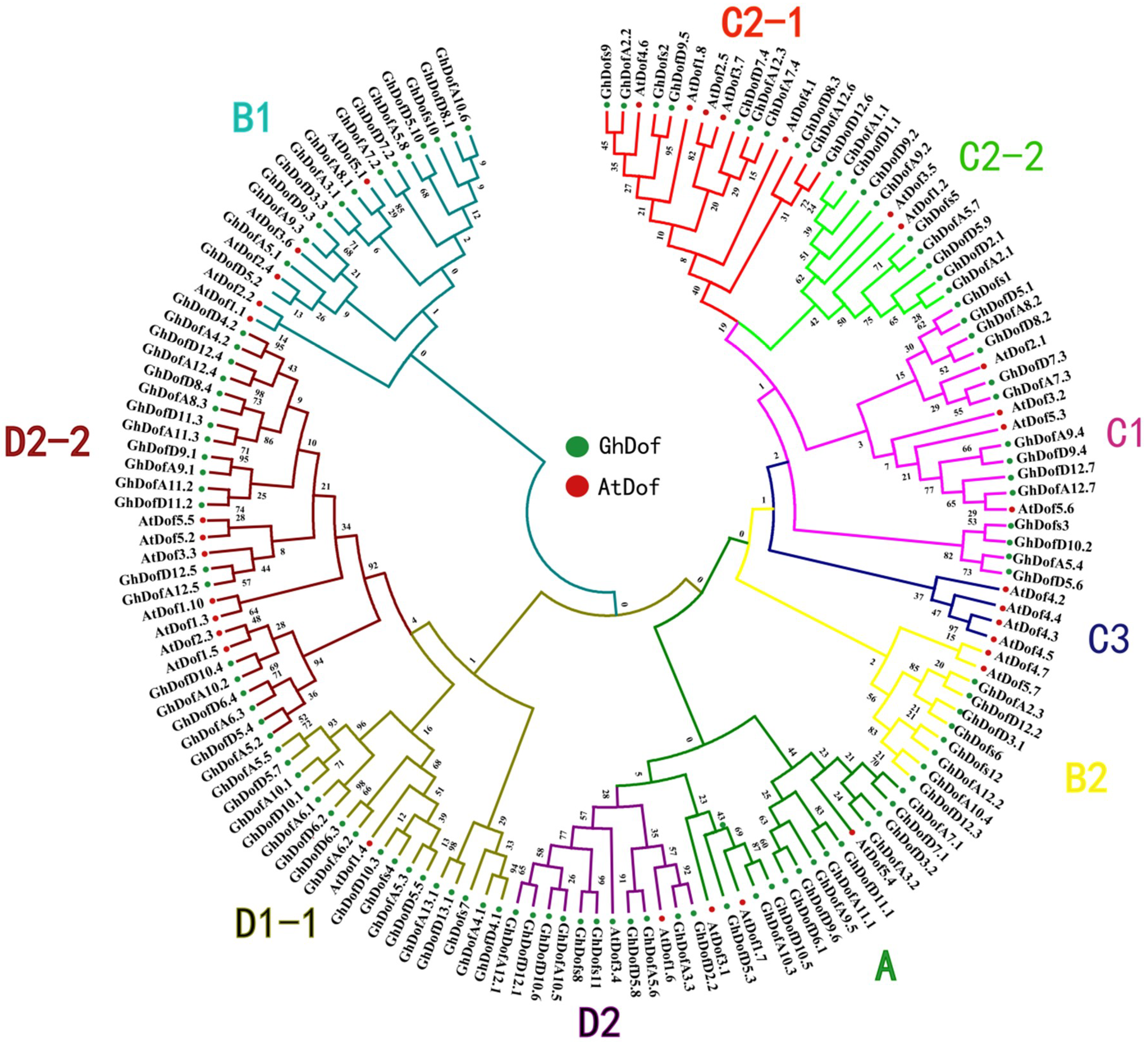
3.2. Phylogenetic Analysis of GhDof TFs
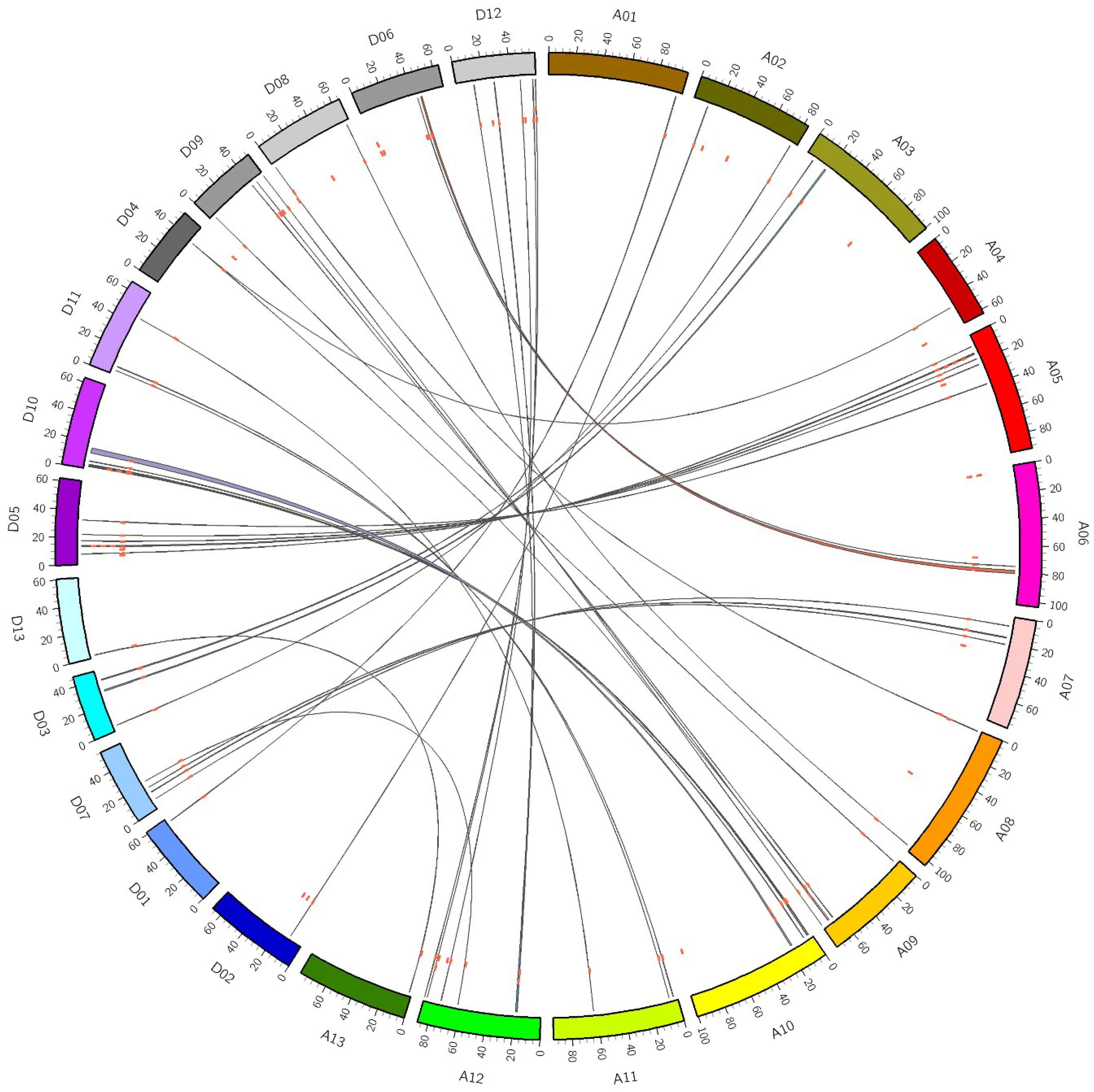
3.3. Gene Duplication and Synteny Analysis of GhDofs
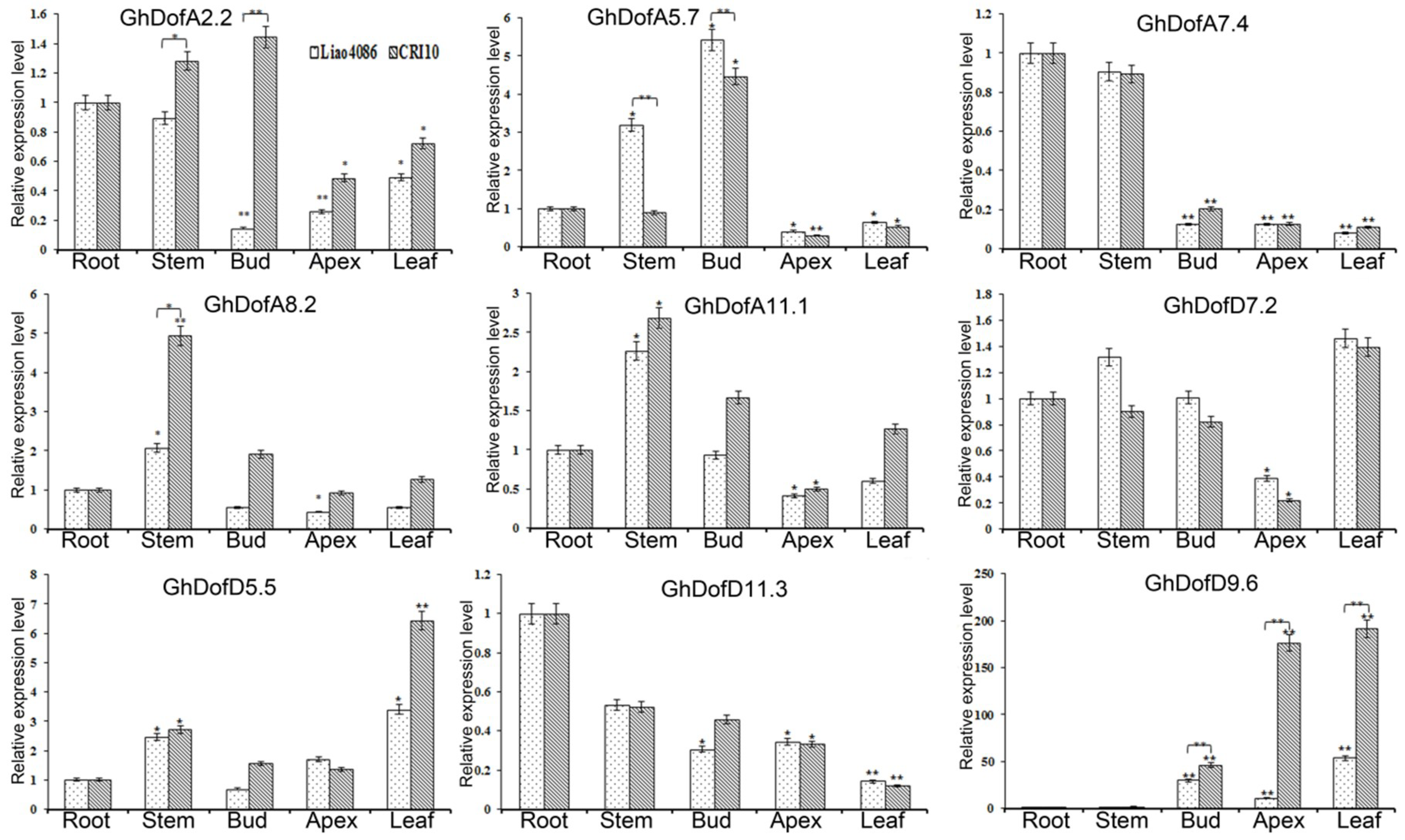
3.4. Gene Expression Analysis
3.4.1. Tissue-Specific Expression
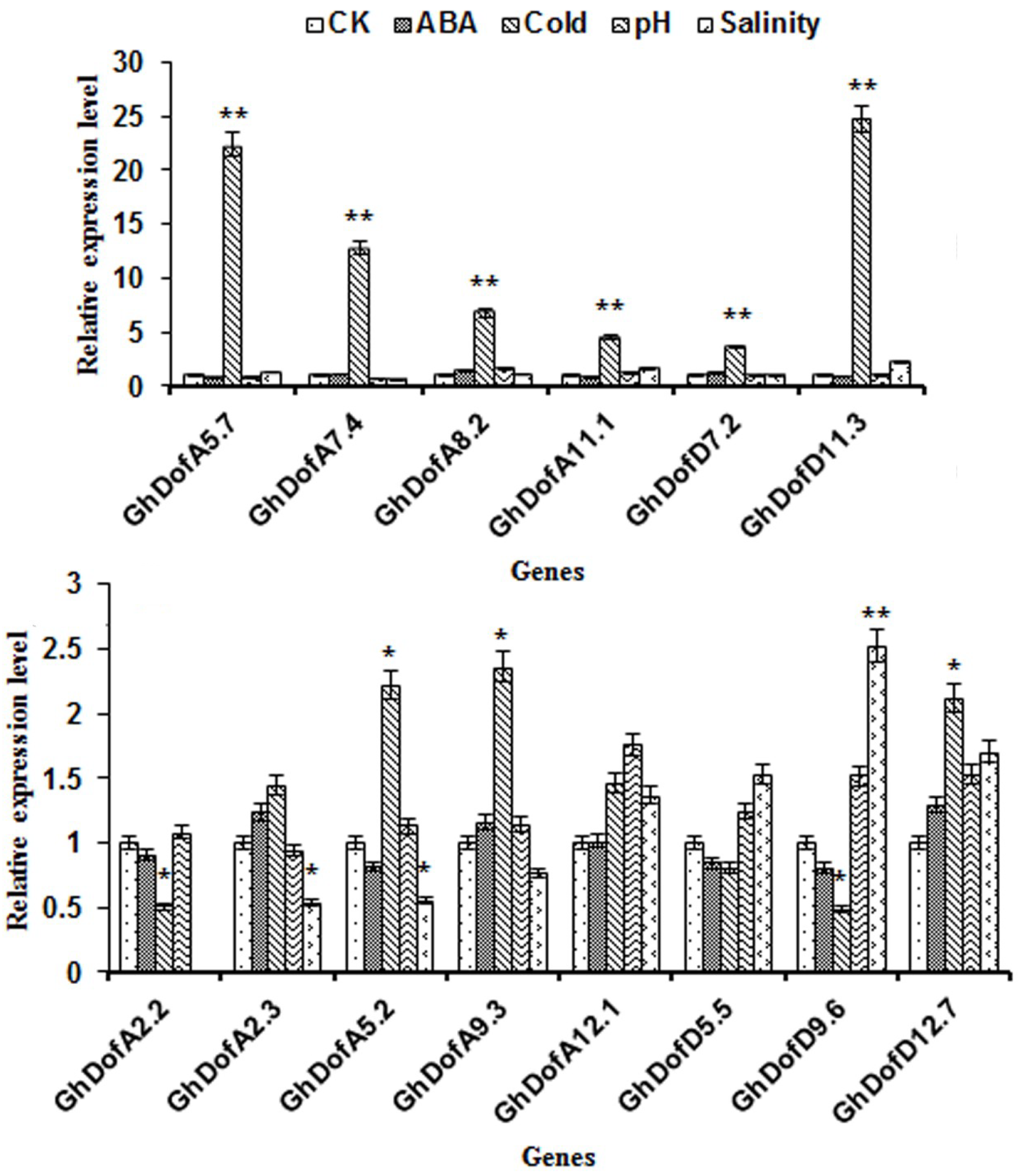
3.4.2. Analysis of Gene Expression under Abiotic Stresses
4. Discussion
4.1. GhDof Genes Are Amplified by Segmental Duplication
4.2. GhDof Genes Might Play Functions in G. hirsutum L. Leaf Senescence
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yanagisawa, S. The Dof Family of Plant Transcription Factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Umemura, Y.; Ishiduka, T.; Yamamoto, R.; Esaka, M. The Dof domain, a zinc finger DNA-binding domain conserved only in higher plants, truly functions as a Cys2/Cys2 Zn finger domain. Plant J. 2004, 37, 741–749. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S.; Schmidt, R.J. Diversity and similarity among recognition sequences of Dof transcription factors. Plant J. 1999, 17, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. A Novel DNA-binding domain that may form a single zinc finger motif. Nucleic Acids Res. 1995, 23, 3403–3410. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Liu, X.; Yin, D.; Yuan, H.; Xie, Q.; Zhao, X.; Li, X.; Zhu, L.; Li, S.; Li, D. Constitutive expression of OsDof4, encoding a C2-C2 zinc finger transcription factor, confesses its distinct flowering effects under long- and short-day photoperiods in rice (Oryza Sativa L.). BMC Plant Biol. 2017, 17, 166. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; Panigrahi, K.C.; Gissot, L.; Sauerbrunn, N.; Ruhl, M.; Jarillo, J.A.; Coupland, G. Arabidopsis Dof transcription factors act redundantly to reduce constans expression and are essential for a photoperiodic flowering response. Dev. Cell 2009, 17, 75–86. [Google Scholar] [CrossRef] [PubMed]
- Molina-Hidalgo, F.J.; Medina-Puche, L.; Canete-Gomez, C.; Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Solano, R.; Caballero, J.L.; Rodriguez-Franco, A.; Blanco-Portales, R.; Munoz-Blanco, J.; et al. The fruit-specific transcription factor FadoF2 regulates the production of eugenol in ripe fruit receptacles. J. Exp. Bot. 2017, 68, 4529–4543. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Li, S.; Zhu, Y.; Zhao, Q.; Zhu, D.; Yu, J. ZmDof3, a maize endosperm-specific Dof protein gene, regulates starch accumulation and aleurone development in maize endosperm. Plant Mol. Biol. 2017, 93, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Yang, W.; Wei, J.; Yoon, H.; An, G. Transcription factor OsDOF18 controls ammonium uptake by inducing ammonium transporters in rice roots. Mol. Cells 2017, 40, 178–185. [Google Scholar] [PubMed]
- Rueda-Lopez, M.; Pascual, M.B.; Pallero, M.; Henao, L.M.; Lasa, B.; Jauregui, I.; Aparicio-Tejo, P.M.; Canovas, F.M.; Avila, C. Overexpression of a pine Dof transcription factor in hybrid poplars: A comparative study in trees growing under controlled and natural conditions. PLoS ONE 2017, 12, e0174748. [Google Scholar] [CrossRef] [PubMed]
- Rymen, B.; Kawamura, A.; Schafer, S.; Breuer, C.; Iwase, A.; Shibata, M.; Ikeda, M.; Mitsuda, N.; Koncz, C.; Ohme-Takagi, M.; et al. ABA suppresses root hair growth via OBP4 transcriptional-regulator of the RSL2 promoter. Plant Physiol. 2017, 173, 1750–1762. [Google Scholar] [CrossRef] [PubMed]
- Castorina, G.; Fox, S.; Tonelli, C.; Galbiati, M.; Conti, L. A novel role for stomatal carpenter 1 in stomata patterning. BMC Plant Biol. 2016, 16, 172. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.-Q.; Wei, P.-C.; Tan, F.; Yu, M.; Zhang, X.-Y.; Chen, Q.-J.; Wang, X.-C. The transcription Factor AtDOF4.7 is involved in ethylene- and IDA-mediated organ abscission in Arabidopsis. Front. Plant Sci. 2016, 7, 863. [Google Scholar] [CrossRef] [PubMed]
- Xu, P.; Chen, H.; Ying, L.; Cai, W. AtDOF5.4/OBP4, a DOF transcription factor gene that negatively regulates cell cycle progression and cell expansion in Arabidopsis thaliana. Sci. Rep. 2016, 6, 27705. [Google Scholar] [CrossRef] [PubMed]
- Konishi, M.; Yanagisawa, S. Transcriptional repression caused by Dof5.8 is involved in proper vein network formation in Arabidopsis thaliana Leaves. J. Plant Res. 2015, 128, 643–652. [Google Scholar] [CrossRef] [PubMed]
- Feng, B.H.; Han, Y.C.; Xiao, Y.Y.; Kuang, J.F.; Fan, Z.Q.; Chen, J.Y.; Lu, W.J. The banana fruit Dof transcription factor Madof23 acts as a repressor and interacts with Maerf9 in regulating ripening-related genes. J. Exp. Bot. 2016, 67, 2263–2275. [Google Scholar] [CrossRef] [PubMed]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling Dof factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.; Liang, W.; Liu, Z.; Wang, Y.; Zhao, Y.; Ijaz, B.; Hua, J. Overexpression of GhDOF1 improved salt and cold tolerance and seed oil content in Gossypium Hirsutum. J. Plant Physiol. 2017, 218, 222–234. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, C.; Shu, W.; Ye, Z.; Li, H.; Zhang, Y. The transcription factor Sldof22 involved in ascorbate accumulation and salinity stress in tomato. Biochem. Biophys. Res. Commun. 2016, 474, 736–741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Fan, G.; Lu, C.; Xiao, G.; Zou, C.; Kohel, R.J.; Ma, Z.; Shang, H.; Ma, X.; Wu, J.; et al. Genome sequence of cultivated upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 2015, 33, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Fan, S.; Wei, H.; Tao, C.; Ma, Q.; Zhang, S.; Li, H.; Pang, C.; Yu, S. Itraq-based quantitative proteomic analysis reveals cold responsive proteins involved in leaf senescence in upland cotton (Gossypium hirsutum L.). Int. J. Mol. Sci. 2017, 18, 1984. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Wei, H.; Song, M.; Pang, C.; Liu, J.; Wang, L.; Zhang, J.; Fan, S.; Yu, S. Transcriptome profiling analysis reveals that flavonoid and ascorbate-glutathione cycle are important during another development in upland cotton. PLoS ONE 2012, 7, e49244. [Google Scholar] [CrossRef] [PubMed]
- Dou, L.; Zhang, X.; Pang, C.; Song, M.; Wei, H.; Fan, S.; Yu, S. Genome-wide analysis of the Wrky gene family in cotton. Mol. Genet. Genom. 2014, 289, 1103–1121. [Google Scholar] [CrossRef] [PubMed]
- Hir, R.L.; Bellini, C. The plant-specific Dof transcription factors family: New players involved in vascular system development and functioning in Arabidopsis. Front. Plant Sci. 2013, 4, 164. [Google Scholar] [PubMed]
- Shigyo, M.; Tabei, N.; Yoneyama, T.; Yanagisawa, S. Evolutionary processes during the formation of the plant-specific Dof transcription factor family. Plant Cell Physiol. 2007, 48, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wen, C.L.; Cheng, Q.; Zhao, L.; Mao, A.; Yang, J.; Yu, S.; Weng, Y.; Xu, Y. Identification and characterisation of Dof transcription factors in the cucumber genome. Sci. Rep. 2016, 6, 23072. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wang, C.; Pang, C.; Wei, H.; Wang, H.; Song, M.; Fan, S.; Yu, S. Characterization and functional analysis of PEBP family genes in upland cotton (Gossypium hirsutum L.). PLoS ONE 2016, 11, e0161080. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Fan, S.; Pang, C.; Wei, H.; Yu, S. Genetic analysis of the antioxidant enzymes, methane dicarboxylic aldehyde (MDA) and chlorophyll content in leaves of the short season cotton (Gossypium hirsutum L.). Euphytica 2014, 198, 153–162. [Google Scholar] [CrossRef]
- Zhu, Y.N.; Shi, D.Q.; Ruan, M.B.; Zhang, L.L.; Meng, Z.H.; Liu, J.; Yang, W.C. Transcriptome analysis reveals crosstalk of responsive genes to multiple abiotic stresses in cotton (Gossypium hirsutum L.). PLoS ONE 2013, 8, e80218. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Han, X.; Lv, J.; Zhao, L.; Xu, X.; Zhang, T.; Guo, W. Structure, expression differentiation and evolution of duplicated fiber developmental genes in Gossypium barbadense and G. Hirsutum. BMC Plant Biol. 2011, 11, 40. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.; Schmittgen, T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Cheng, J.; Cui, J.; Xu, X.; Liang, G.; Luo, X.; Chen, X.; Tang, X.; Hu, K.; Qin, C. Genome-wide identification and expression profile of Dof transcription factor gene family in pepper (Capsicum annuum L.). Front. Plant Sci. 2016, 7, 574. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Risueno, M.A.; Martinez, M.; Vicente-Carbajosa, J.; Carbonero, P. The family of DOF transcription factors: From green unicellular algae to vascular plants. Mol. Genet. Genom. 2007, 277, 379–390. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.; Pang, C.; Fan, S.; Song, M.; Wei, H.; Yu, S. Global analysis of the Gossypium Hirsutum L. Transcriptome during leaf senescence by RNA-Seq. BMC Plant Biol. 2015, 15, 43. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, H.; Gupta, S.; Singh, V.; Rastogi, S.; Yadav, D. Genome wide identification of dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis. Mol. Biol. Rep. 2011, 38, 5037–5053. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Qiu, L.-J. Retraction: Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS ONE 2016, 11, e0167019. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zhang, Y.; Zhang, C.; Zhang, T.; Hu, T.; Ye, J.; Zhang, J.; Wang, T.; Li, H.; Ye, Z. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. J. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Li, M.Y.; Wang, F.; Tang, J.; Xiong, A.S. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genom. 2015, 16, 33. [Google Scholar] [CrossRef] [PubMed]
- Venkatesh, J.; Park, S.; Se, W. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiol. Biochem. 2015, 94, 73–86. [Google Scholar] [CrossRef] [PubMed]
- Shu, Y.; Song, L.L.; Zhang, J.; Liu, Y.; Guo, C.H. Genome-wide identification and characterization of the Dof gene family in Medicago truncatula. Genet. Mol. Res. 2015, 14, 10645–10657. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Huang, Y.; Li, M.Y.; Wang, F.; Xu, Z.S.; Xiong, A.S. Dof transcription factors in carrot: Genome-wide analysis and their response to abiotic stress. Biotechnol. Lett. 2016, 38, 145–155. [Google Scholar] [CrossRef] [PubMed]

© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, H.; Dou, L.; Li, W.; Wang, P.; Zhao, Q.; Xi, R.; Pei, X.; Liu, Y.; Ren, Z. Genome-Wide Identification and Expression Analysis of the Dof Transcription Factor Gene Family in Gossypium hirsutum L. Agronomy 2018, 8, 186. https://doi.org/10.3390/agronomy8090186
Li H, Dou L, Li W, Wang P, Zhao Q, Xi R, Pei X, Liu Y, Ren Z. Genome-Wide Identification and Expression Analysis of the Dof Transcription Factor Gene Family in Gossypium hirsutum L. Agronomy. 2018; 8(9):186. https://doi.org/10.3390/agronomy8090186
Chicago/Turabian StyleLi, Huaizhu, Lingling Dou, Wei Li, Ping Wang, Qin Zhao, Ruimin Xi, Xiaoyu Pei, Yangai Liu, and Zhongying Ren. 2018. "Genome-Wide Identification and Expression Analysis of the Dof Transcription Factor Gene Family in Gossypium hirsutum L." Agronomy 8, no. 9: 186. https://doi.org/10.3390/agronomy8090186
APA StyleLi, H., Dou, L., Li, W., Wang, P., Zhao, Q., Xi, R., Pei, X., Liu, Y., & Ren, Z. (2018). Genome-Wide Identification and Expression Analysis of the Dof Transcription Factor Gene Family in Gossypium hirsutum L. Agronomy, 8(9), 186. https://doi.org/10.3390/agronomy8090186



