Transcriptomic Analysis of Banana in Response to Phosphorus Starvation Stress
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Growth Conditions
2.2. Measurement of Plant Weight and Total P Concentration
2.3. Sample Preparation and RNA-Seq
2.4. Identification of Differentially Expressed Genes (DEGs)
2.5. Quantitative Rreverse-Transcription PCR (qRT-PCR)
3. Results
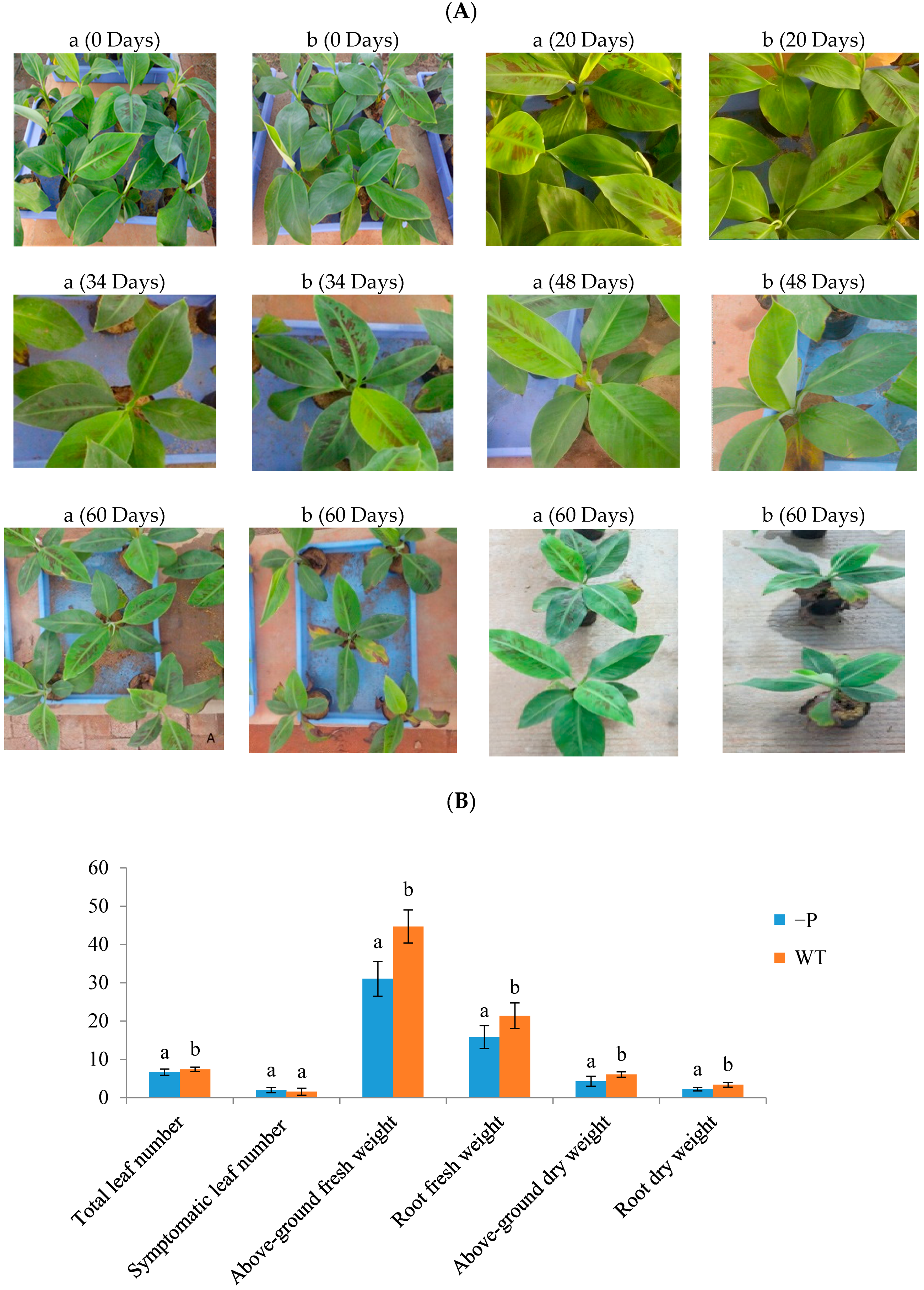
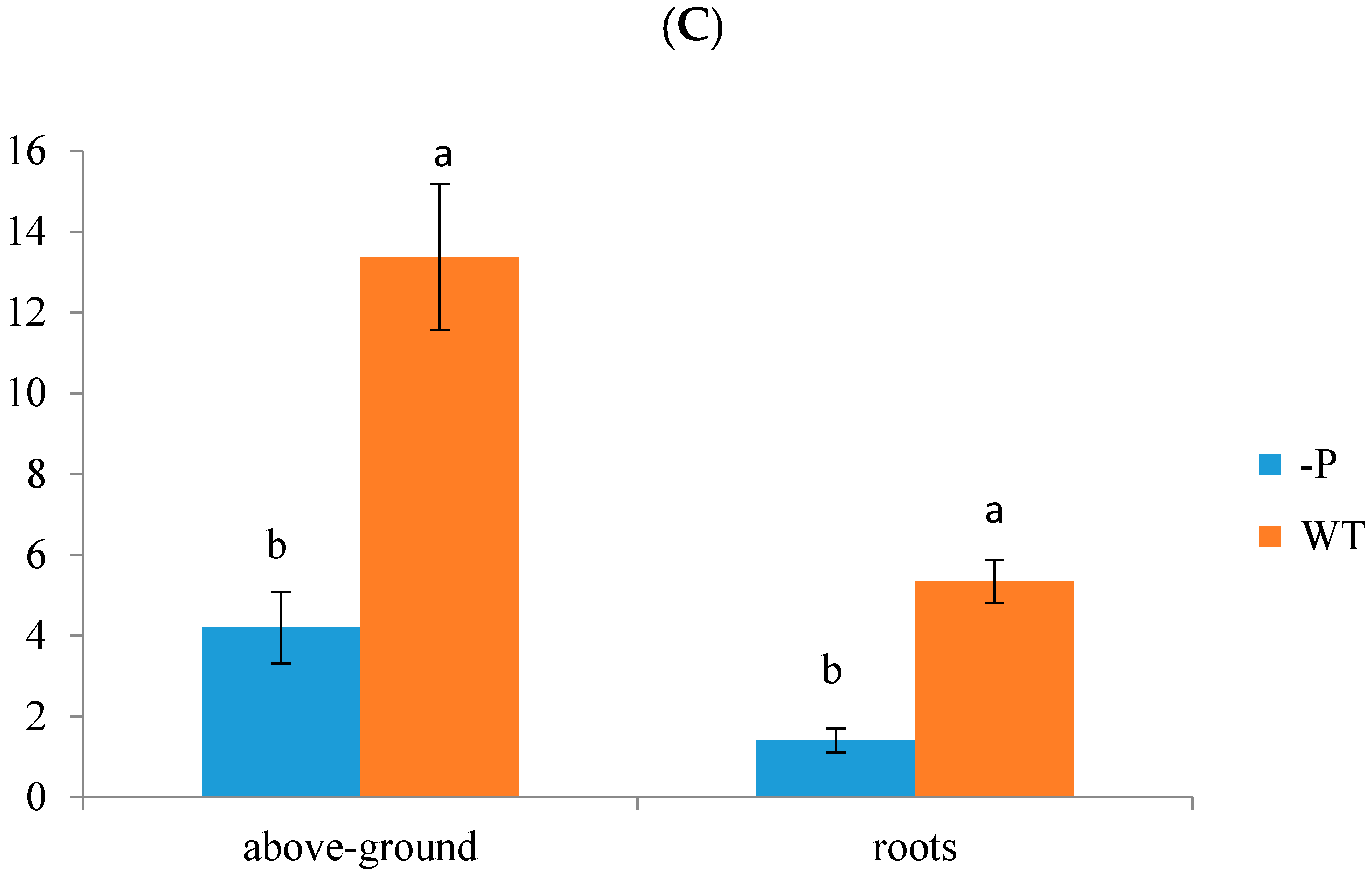
3.1. Physiological Indicators of Banana Response to Deficient P Stress
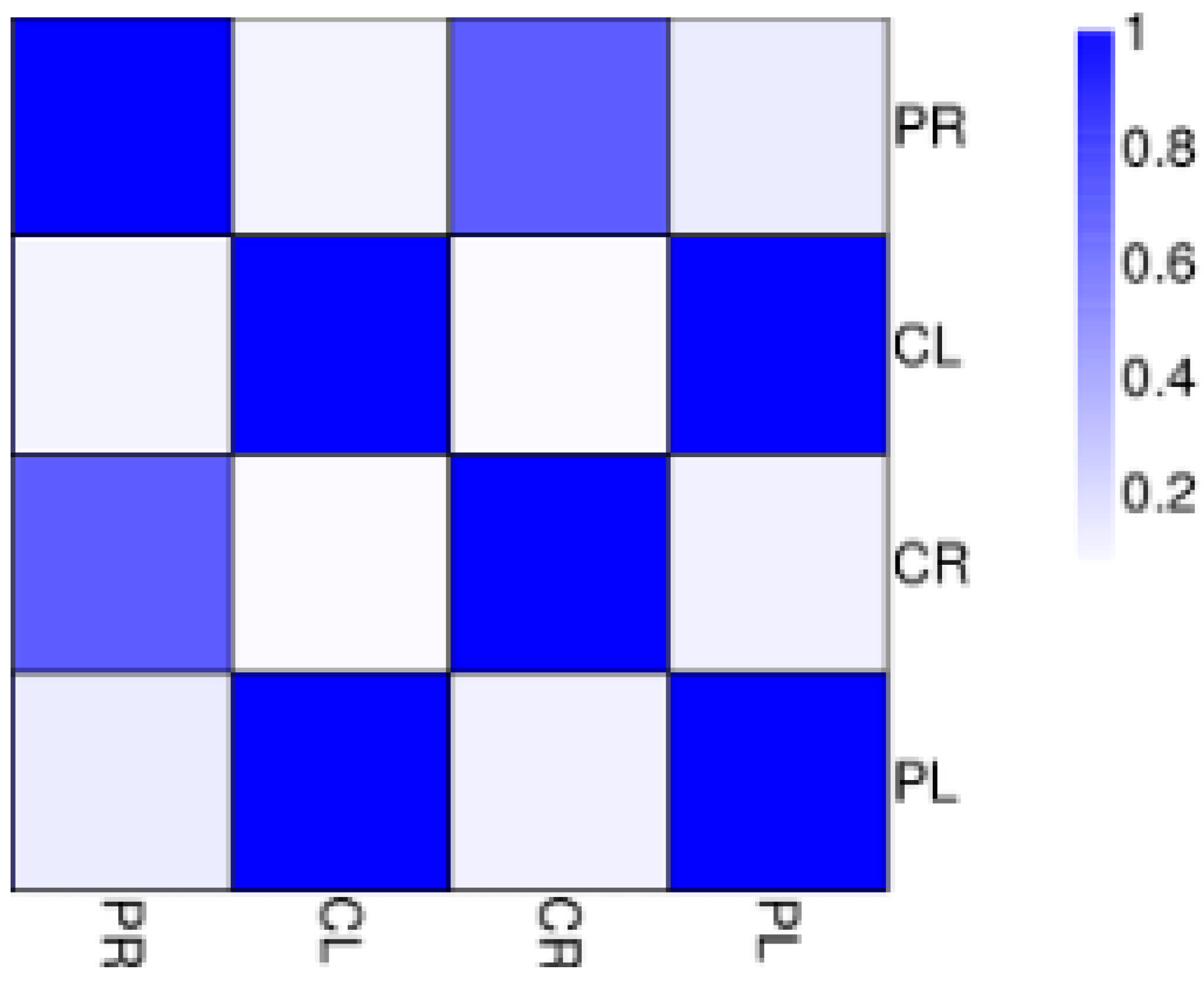
3.2. RNA Sequencing (RNA-Seq) and Alignment of Clean Reads to the Banana Reference Genome
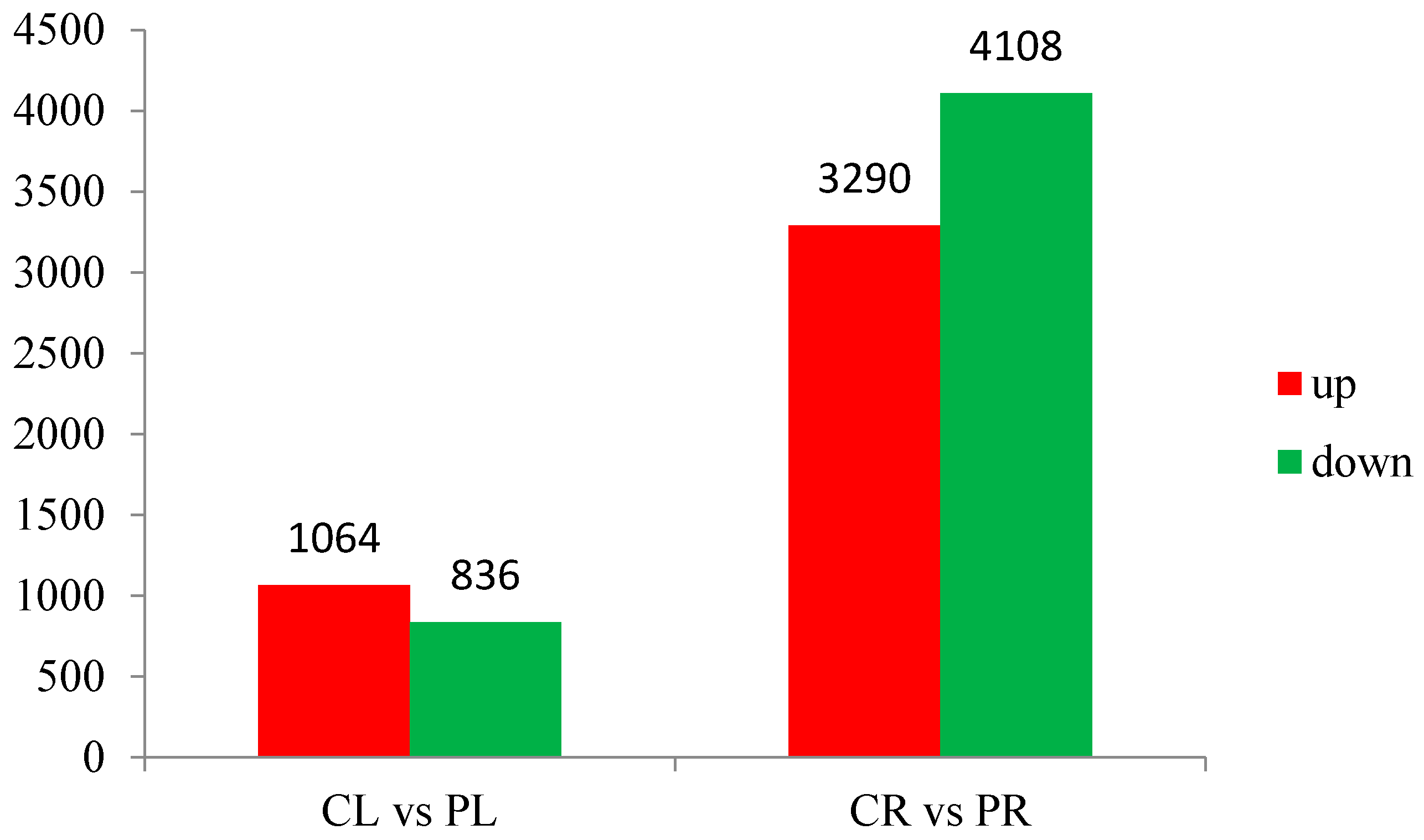
3.3. General Features of Differential Expression Profiling
3.4. Energy Metabolism-Related Genes during P Deficiency Stress
3.5. Signal Transduction-Related Genes under Low P Conditions
3.6. Control of Rhizosphere P Activation Genes
3.7. Genes Involved in Pi Mobilization
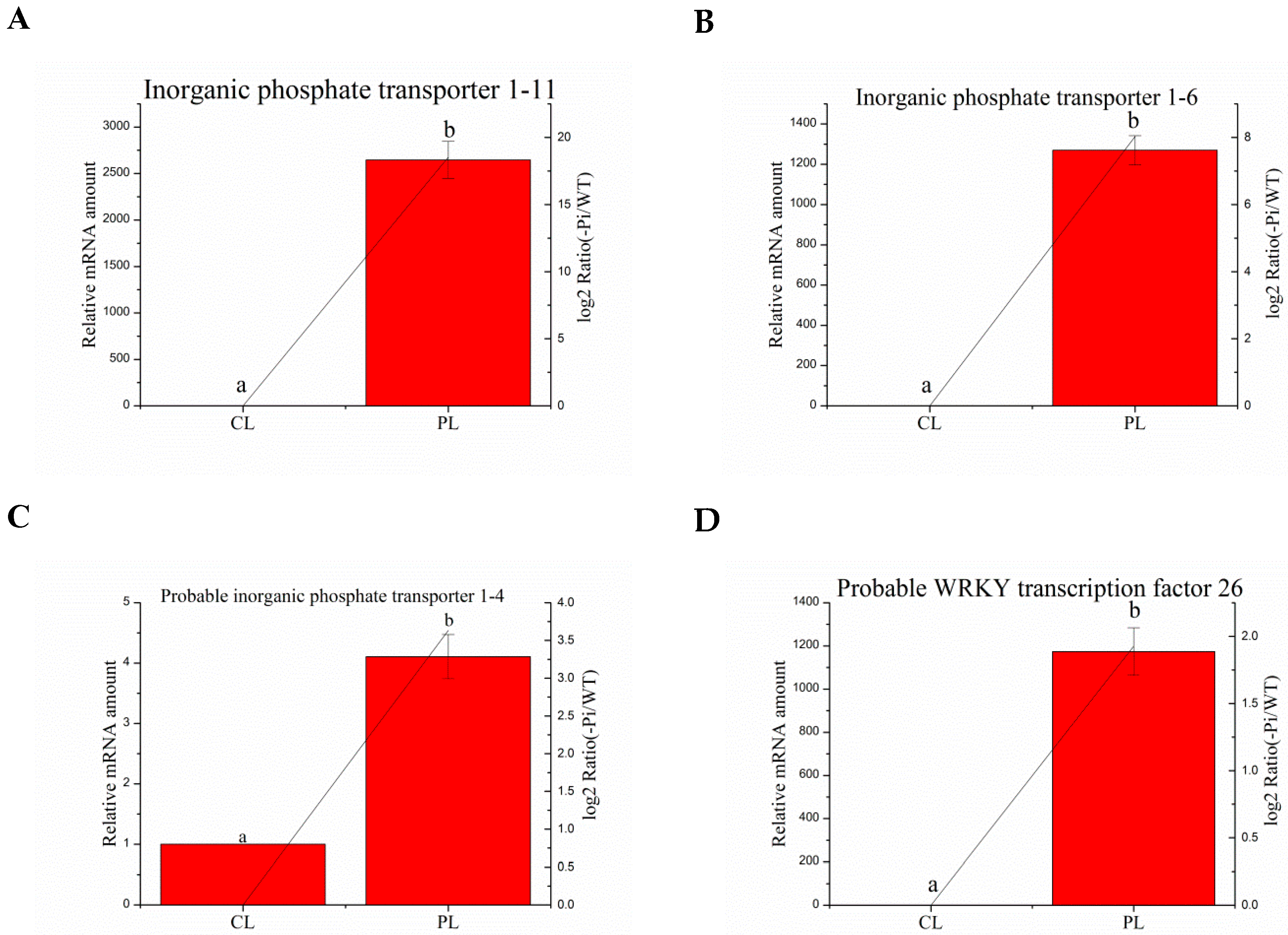
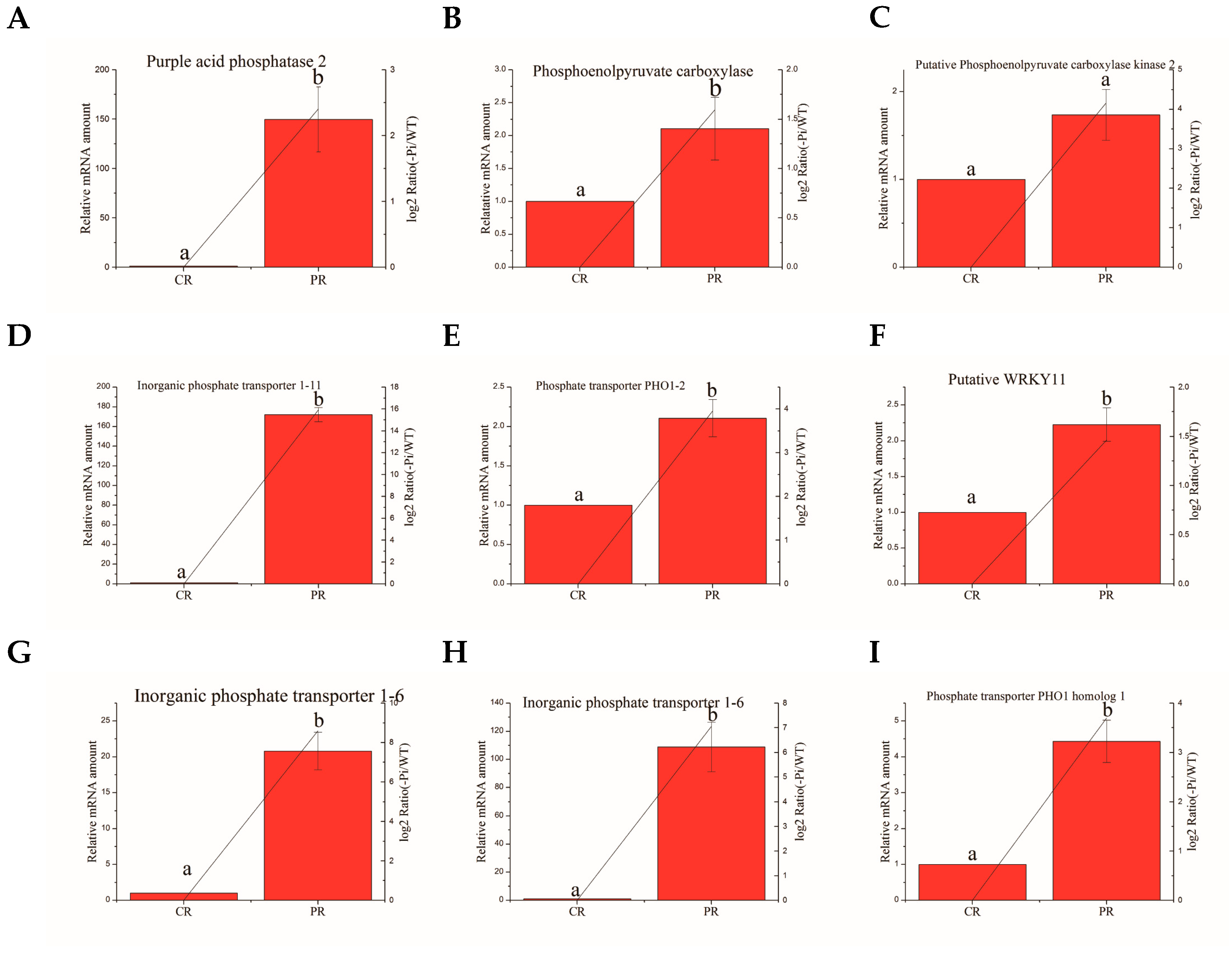
3.8. Validation of the Selected Degs by Quantitative RT-PCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tiessen, H. Phosphorus in the global environment. In The Ecophysiology of Plant-Phosphorus Interactions; White, P.J., Hammond, J.P., Eds.; Springer: Dordrecht, The Netherlands, 2008; pp. 1–7. [Google Scholar]
- Padam, B.S.; Tin, H.S.; Chye, F.Y.; Abdullah, M.I. Banana by-products: An under-utilized renewable food biomass with great potential. J. Food Sci. Technol. 2014, 51, 3527–3545. [Google Scholar] [CrossRef] [PubMed]
- Pant, B.D.; Burgos, A.; Pant, P.; Cuadros-Inostroza, A.; Willmitzer, L.; Scheible, W.R. The transcription factor PHR1 regulates lipid remodeling and triacylglycerol accumulation in Arabidopsis thaliana during phosphorus starvation. J. Exp. Bot. 2015, 66, 1907–1918. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The water culture method of growing plants without soil. Calif. Agric. Exp. Stn. Circ. 1950, 347, 357–359. [Google Scholar]
- Tiong, S.M.I. The incorporation of potassium antimony tartrate in molybdenum blue method for the automated colorimetric determination of available phosphorus in soils. Pertanika 1987, 10, 27–29. [Google Scholar]
- Li, H.; Durbin, R. Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 2010, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- De, S.N. The encode project. Nat. Methods 2012, 9, 1046. [Google Scholar]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar]
- Du, H.; Liu, H.B.; Xiong, L.Z. Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 2013, 4, 397. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Hata, S.; Kyozuka, J.; Shimamoto, K.; Izui, K. Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. Plant J. 2000, 23, 319–327. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Wang, J.; Zhao, J.; Tian, J.; Liao, H. Control of phosphate homeostasis through gene regulation in crops. Curr. Opin. Plant Biol. 2014, 21, 59–66. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Tran, H.T. Metabolic adaptations of phosphate-starved plants. Plant Physiol. 2011, 156, 1006–1015. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Wang, X.; Tong, Y.; Chen, X.; Liao, H. Bioengineering and management for efficient phosphorus utilization in crops and pastures. Curr. Opin. Biotechnol. 2012, 23, 866–871. [Google Scholar] [CrossRef] [PubMed]
- Vance, C.P.; Uhdestone, C.; Allan, D.L. Phosphorus acquistion and use: Critical adaptations by plants for securing a nonrenewable resource. New Phytol. 2003, 157, 423–447. [Google Scholar] [CrossRef]
- Fang, Z.Y.; Shao, C.; Meng, Y.; Wu, P.; Chen, M. Phosphate signaling in Arabidopsis and Oryza sativa. Plant Sci. 2009, 176, 170–180. [Google Scholar] [CrossRef]
- Chiou, T.J.; Lin, S.I. Signaling network in sensing phosphate availability in plants. Annu. Rev. Plant Biol. 2011, 62, 185–206. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Boil. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Plaxton, W.C.; Podestá, F.E. The functional organization and control of plant respiration. Crit. Rev. Plant Sci. 2006, 25, 159–198. [Google Scholar] [CrossRef]
- Fita, A.; Bowen, H.C.; Hayden, R.M.; Nuez, F.; Picó, B.; Hammond, J.P. Diversity in expression of phosphorus (p) responsive genes in Cucumis melo L. PLoS ONE 2012, 7, e35387. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-T.; Jiang, H.-X.; Qi, Y.-P.; Chen, L.-S. Differential expression of genes involved in alternative glycolytic pathways, phosphorus scavenging and recycling in response to aluminum and phosphorus interactions in Citrus roots. Mol. Biol. Rep. 2012, 39, 6353–6366. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Hurley, B.A.; Plaxton, W.C. Feeding hungry plants: The role of purple acid phosphatases in phosphate nutrition. Plant Sci. 2010, 179, 14–27. [Google Scholar] [CrossRef]
- Wang, L.; Lu, S.; Zhang, Y.; Li, Z.; Du, X.; Liu, D. Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J. Integr. Plant Biol. 2014, 56, 299–314. [Google Scholar] [CrossRef] [PubMed]
- Tran, H.T.; Plaxton, W.C. Proteomic analysis of alterations in the secretome of Arabidopsis thaliana suspension cells subjected to nutritional phosphate deficiency. Proteomics 2010, 8, 4317–4326. [Google Scholar] [CrossRef] [PubMed]
- Veljanovski, V.; Vanderbeld, B.; Knowles, V.L.; Snedden, W.A.; Plaxton, W.C. Biochemical and molecular characterization of AtPAP26, a vacuolar purple acid phosphatase up-regulated in phosphate-deprived Arabidopsis suspension cells and seedlings. Plant Physiol. 2006, 142, 1282–1293. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Shi, W.; Jia, L.; Liang, J.; Zhang, J. TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ. 2012, 35, 1393–1406. [Google Scholar] [CrossRef] [PubMed]
- Diatloff, E.; Roberts, M.; Sanders, D.; Roberts, S.K. Characterization of anion channels in the plasma membrane of Arabidopsis epidermal root cells and the identification of a citrate-permeable channel induced by phosphate starvation. Plant Physiol. 2004, 136, 4136–4149. [Google Scholar] [CrossRef] [PubMed]
- Ticconi, C.A.; Abel, S. Short on phosphate: Plant surveillance and countermeasures. Trends Plant Sci. 2004, 9, 548–555. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.E. Regulating the phosphorus nutrition of plants: Molecular biology meeting agronomic needs. Plant Soil 2009, 322, 17–24. [Google Scholar] [CrossRef]
- Secco, D.; Wang, C.; Shou, H.; Whelan, J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012, 586, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Hamburger, D.; Rezzonico, E.; Macdonaldcomber, P.J.; Somerville, C.; Poirier, Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 2002, 14, 889–902. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Yi, K.K.; Dang, L.; Huang, H.J.; Wu, W.; Wu, P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008, 54, 965–975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hürlimann, H.C.; Pinson, B.; Stadler-Waibel, M.; Zeeman, S.C.; Freimoser, F.M. The SPX domain of the yeast low-affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep. 2009, 10, 1003–1008. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, P.; Shou, H.X.; Xu, G.H.; Lian, X.M. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr. Opin. Plant Biol. 2013, 16, 205–212. [Google Scholar] [CrossRef] [PubMed]
- Raghothama, K.G. Phosphate acquisition. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999, 50, 665–693. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, S.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mudge, S.R.; Rae, A.L.; Diatloff, E.; Smith, F.W. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002, 31, 341–353. [Google Scholar] [CrossRef] [PubMed]
- Ai, P.H.; Sun, S.B.; Zhao, J.N.; Fan, X.R.; Xin, W.J.; Guo, Q.; Yu, L.; Shen, Q.R.; Wu, P.; Miller, A.J.; et al. Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 2009, 57, 798–809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bustos, R.; Castrillo, G.; Linhares, F.; Puga, M.I.; Rubio, V.; Pérez-Pérez, J.; Solano, R.; Leyva, A.; Paz-Ares, J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010, 6, e1001102. [Google Scholar] [CrossRef] [PubMed]
- Rubio, V.; Linhares, F.; Solano, R.; Martín, A.C.; Iglesias, J.; Leyva, A.; Paz-Ares, J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001, 15, 2122–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Ying, S.H.; Li, K.; Wu, P.; Shou, H. Involvement of OsSPX1 in phosphate homeostasis in rice. Plant J. Cell Mol. Biol. 2010, 57, 895–904. [Google Scholar] [CrossRef] [PubMed]
- Franco-Zorrilla, J.M.; González, E.; Bustos, R.; Linhares, F.; Leyva, A.; Paz-Ares, J. The transcriptional control of plant responses to phosphate limitation. J. Exp. Bot. 2004, 55, 285–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, K.; Chai, Y.R.; Zhang, K.; Wang, R.; Chen, L.; Lei, B.; Lu, J.; Xu, X.F.; Li, J.N. Cloning and characterization of phosphorus starvation inducible Brassica napus PURPLE ACID PHOSPHATASE 12 gene family, and imprinting of a recently evolved MITE-minisatellite twin structure. Theor. Appl. Genet. 2008, 117, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Xu, Q.; Zhang, F.C.; Chen, Y.; Li, L.Q.; Wu, W.H.; Chen, Y.F. WRKY42 modulates phosphate homeostasis through regulating phosphate translocation and acquisition in Arabidopsis. Plant Physiol. 2015, 167, 1579–1591. [Google Scholar] [CrossRef] [PubMed]
- Devaiah, B.N.; Karthikeyan, A.S.; Raghothama, K.G. WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 2007, 143, 1789–1801. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
| Sample | CL | CR | PL | PR |
|---|---|---|---|---|
| Total reads | 12,121,980 (100.00%) | 12,121,219 (100.00%) | 12,071,536 (100.00%) | 12,002,078 (100.00%) |
| Total mapped reads | 10,134,956 (83.61%) | 9,963,385 (82.20%) | 10,059,569 (83.33%) | 10,002,358 (83.34%) |
| Perfect match | 6,594,550 (54.40%) | 6,336,833 (52.28%) | 6,601,731 (54.69%) | 6,357,459 (52.97%) |
| Mismatch | 3,540,406 (29.21%) | 3,626,552 (29.92%) | 3,457,838 (28.64%) | 3,644,899 (30.37%) |
| Unique match | 9,886,657 (81.56%) | 9,624,069 (79.40%) | 9,821,699 (81.36%) | 9,571,156 (79.75%) |
| Multi-position match | 248,299 (2.05%) | 339,316 (2.80%) | 237,870 (1.97%) | 431,202 (3.59%) |
| Total unmapped reads | 1,987,024 (16.39%) | 2,157,834 (17.8%) | 2,011,967 (16.67%) | 1,999,720 (16.66%) |
| Glycolysis | Tricarboxylic Acid Cycle | ||||
|---|---|---|---|---|---|
| Gene ID | Gene ID | ||||
| Ma05_g07350 | Up | Ma03_g06530 | Up | Ma05_g07350 | Up |
| Ma10_g09450 | Up | Ma02_g23000 | Up | Ma10_g09450 | Up |
| Ma03_g09370 | Up | Ma05_g07150 | Up | Ma03_g09370 | Up |
| Ma02_g01890 | Up | Ma08_g16810 | Up | Ma02_g01890 | Up |
| Ma08_g33800 | Up | Ma10_g23690 | Down | Ma06_g27170 | Up |
| Ma07_g23360 | Up | Ma09_g09160 | Down | Ma06_g03480 | Up |
| Ma08_g19340 | Up | Ma06_g10020 | Down | Ma06_g18050 | Up |
| Ma08_g32440 | Up | Ma08_g26340 | Down | Ma05_g02120 | Up |
| Ma06_g27170 | Up | Ma02_g06250 | Down | Ma02_g23000 | Up |
| Ma06_g03480 | Up | Ma07_g19140 | Down | Ma05_g03680 | Up |
| Ma03_g04300 | Up | Ma09_g16720 | Down | Ma02_g22920 | Up |
| Ma03_g10380 | Up | Ma06_g33960 | Down | Ma04_g27480 | Down |
| Ma00_g00620 | Up | Ma01_g10270 | Down | Ma06_g33960 | Down |
| Ma06_g03680 | Up | Ma04_g27480 | Down | Ma10_g29220 | Down |
| Ma06_g18050 | Up | Ma10_g29220 | Down | ||
| Ma05_g21200 | Up | Ma06_g25360 | Down | ||
| Ma05_g02120 | Up | Ma08_g11500 | Down | ||
| Ma11_g17540 | Up | Ma04_g17780 | Down | ||
| Ma01_g13550 | Up | Ma03_g09070 | Down | ||
| Aboveground Part | ||||
| Calcium/Calmodulin-Dependent Protein Kinase | Ca2+-Transporting ATPase | Calcium-Binding Protein | ||
| Ma04_g28490 | Ma07_g27150 | Ma10_g04830 | Ma10_g18710 | |
| Ma02_g14160 | Ma01_g21640 | Ma03_g18040 | ||
| Ma04_g18750 | Ma04_g24480 | Ma08_g09460 | ||
| Ma04_g22630 | Ma07_g22610 | Ma03_g24180 | ||
| Ma02_g09730 | Ma05_g28740 | |||
| Root | ||||
| Calcium-Dependent Protein Kinase | Calcium Ion Transport | Calcium Exchanger | Calcium-Binding Protein | |
| Ma02_g14160 | Ma05_g19800 | Ma04_g05840 | Ma04_g24480 | |
| Ma05_g29940 | Ma06_g24990 | Ma09_g21740 | Ma03_g18040 | |
| Ma03_g12240 | Ma06_g17570 | Ma05_g07830 | Ma06_g25970 | |
| Ma02_g09730 | Ma03_g19230 | |||
| Ma01_g04260 | Ma05_g23960 | |||
| SPX Domain-Containing Protein | Pi Transporters | |
|---|---|---|
| Ma04_g37210 | Ma03_g26260 | Ma05_g15460 |
| Ma10_g26920 | Ma02_g06130 | Ma01_g13800 |
| Ma01_g15270 | Ma01_g01890 | Ma04_g36790 |
| Ma04_g30530 | Ma06_g38500 | Ma04_g33330 |
| Ma07_g09970 | Ma01_g14000 | Ma03_g03540 |
| Ma05_g15470 | Ma04_g09790 | |
| Root | ||||
| WRKY Transcription Factor 26 | WRKY Transcription Factor 71 | WRKY Transcription Factor 31 | ||
| Ma06_g01150 | Ma06_g28830 | Ma02_g16960 | ||
| Ma07_g26900 | ||||
| Ma04_g15010 | ||||
| Aboveground | ||||
| WRKY Transcription Factor 50 | WRKY Transcription Factor 70 | WRKY Transcription Factor 26 | WRKY Transcription Factor 40 | WRKY Transcription Factor 31 |
| Ma03_g21270 | Ma06_g17380 | Ma08_g01650 | Ma07_g20830 | Ma10_g26000 |
| Ma06_g15160 | ||||
| Ma06_g07520 | ||||
| Aboveground | |||
| Gene ID | Function | Forward Primer (5′→3′) | Reverse Primer (5′→3′) |
| Ma03_g26260 | Inorganic phosphate transporter 1–11 | CGTGCTGTCGTTTACCAA | GCCGCTGACTCTTTCATTCTC |
| Ma01_g01890 | Inorganic phosphate transporter 1–6 | CGACCTGCCACGGTATCTC | CGGAACCAGGAAGGTGAAG |
| Ma04_g36790 | Probable inorganic phosphate transporter 1–4 | GCCCAACAGCACCACCTT | TCCAGCGATTTGCCCTTC |
| Ma08_g01650 | Probable WRKY transcription factor 26 | AAGTACGGTGAGAAACAGGTC | CGGCTTTGAGTGGCTATG |
| Root | |||
| Ma10_g25380 | Phosphoenolpyruvate carboxylase | TGTCGGAAGAGGAGGTGG | CGGTGAAATCGGAGGGTG |
| Ma07_g26900 | Putative WRKY11 | CACCAAATCCCGCAACTT | TGCAGGAGAGCGTAGTCG |
| Ma06_g38500 | Phosphate transporter PHO1–2 | GGTGCTTAGGCTTGCTTG | TGTTCAGGTGCTCATTCTCC |
| Ma01_g14000 | Phosphate transporter PHO1 homolog 1 | CTTGTGCTTCGCCTTGCT | AGTTCCAATGCCCTCGTC |
| Ma02_g061′ | Inorganic phosphate transporter 1–6 | TGCCCGGTTACTGGTTCA | AAGCCGATGTGGTTTCCCT |
| Ma01_g01890 | Inorganic phosphate transporter 1–6 | GTGCCCGGCTACTGGTTT | AGGCGTACATGACGACGAA |
| Ma04_g18750 | Putative phosphoenolpyruvate carboxylase kinase 2 | TCGGATCGAGAGCTATCC | GAGTGTCTCGGCTTGCTT |
| Ma03_g26260 | Inorganic phosphate transporter 1–11 | CGTGCTGGCGTTTACCAA | GCCGCTGACTCTTTCATTCTC |
| Ma07_g01650 | Purple acid phosphatase 2 | TCTTGAAGGGCTTGCTAA | TCCATCCTGGTTTCTGTG |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xiong, R.; Tang, H.; Xu, M.; Zeng, C.-B.; Peng, Y.; He, R.; Yan, Z.; Qi, Z.; Cheng, Y. Transcriptomic Analysis of Banana in Response to Phosphorus Starvation Stress. Agronomy 2018, 8, 141. https://doi.org/10.3390/agronomy8080141
Xiong R, Tang H, Xu M, Zeng C-B, Peng Y, He R, Yan Z, Qi Z, Cheng Y. Transcriptomic Analysis of Banana in Response to Phosphorus Starvation Stress. Agronomy. 2018; 8(8):141. https://doi.org/10.3390/agronomy8080141
Chicago/Turabian StyleXiong, Rui, Hua Tang, Min Xu, Can-Bin Zeng, Yun Peng, Rui He, Zhen Yan, Zhao Qi, and Yu Cheng. 2018. "Transcriptomic Analysis of Banana in Response to Phosphorus Starvation Stress" Agronomy 8, no. 8: 141. https://doi.org/10.3390/agronomy8080141
APA StyleXiong, R., Tang, H., Xu, M., Zeng, C.-B., Peng, Y., He, R., Yan, Z., Qi, Z., & Cheng, Y. (2018). Transcriptomic Analysis of Banana in Response to Phosphorus Starvation Stress. Agronomy, 8(8), 141. https://doi.org/10.3390/agronomy8080141




