Abstract
Arbuscular mycorrhizal (AM) fungi play an essential role for the nutrient uptake of the majority of land plants, including many important crop species. The extraradical mycelium of the fungus takes up nutrients from the soil, transfers these nutrients to the intraradical mycelium within the host root, and exchanges the nutrients against carbon from the host across a specialized plant-fungal interface. The contribution of the AM symbiosis to the phosphate nutrition has long been known, but whether AM fungi contribute similarly to the nitrogen nutrition of their host is still controversially discussed. However, there is a growing body of evidence that demonstrates that AM fungi can actively transfer nitrogen to their host, and that the host plant with its carbon supply stimulates this transport, and that the periarbuscular membrane of the host is able to facilitate the active uptake of nitrogen from the mycorrhizal interface. In this review, our current knowledge about nitrogen transport through the fungal hyphae and across the mycorrhizal interface is summarized, and we discuss the regulation of these pathways and major research gaps.
1. Introduction
The arbuscular mycorrhizal (AM) symbiosis plays a key role for the nutrient uptake of more than 60% of land plants, including many important crop species such as wheat, corn, and soybean [1]. AM fungi are ubiquitous and can account for up to 50% of the microbial biomass in soils [2]. The extraradical mycelium (ERM) of the fungus acts as an extension of the root system and increases the uptake of phosphate (P), nitrogen (N), sulfur, and magnesium but also of trace elements, such as copper and zinc. In addition, AM fungi also provide non-nutritional benefits to their host plant, as they improve the resistance of plants against several abiotic (drought, salinity, heavy metals) and biotic (pathogens, herbivores) stresses [3]. AM fungi therefore play a key role in the survival and fitness of plants and act as “ecosystem engineers” of plant communities [4]. However, the benefits for the plant are not free of charge, and the plant transfers between 4% to 22% of its assimilated carbon (C) to the AM fungus [5]. It has been suggested that the substantial C costs are responsible for the variability of mycorrhizal growth responses that have been described. These growth responses can range from highly beneficial to detrimental and follow a mutualism to parasitism continuum [6,7,8].
AM fungi belong to the fungal phylum Glomeromycota, and are obligate biotrophs that are unable to complete their life cycle without the carbon supply from their host. This dependency on the host and the observation that many host plants suppress their mycorrhizal colonization particularly under high nutrient supply conditions has led to the overall assumption that the host plant is in control of the symbiosis [9]. However, the long co-evolution of about 400 to 450 million years for both partners in the AM symbiosis also allowed the fungus to improve its strategies to control the nutrient transport to the host despite its obligate biotrophic life cycle [10,11]. It has been suggested that carbon to nutrient exchange in the AM symbiosis is controlled by biological market dynamics and that reciprocal reward mechanisms ensure a “fair trade” between both partners in the AM symbiosis [10].
AM fungi and their plant partners form a complex network of many-to-many interactions; each host plant is colonized by communities of AM fungi and fungal individuals colonize multiple host plants simultaneously and interconnect plants by common mycorrhizal networks (CMNs). These many-to-many interactions allow both partners in the symbiosis to choose among multiple trading partners but also force both partners to compete with other partners for nutrient or carbon resources [12,13]. CMNs play a key role for the long distance transport of nutrients, water, stress chemicals and allelochemicals and allow the interconnected host plants to “communicate” with other plants within their CMN [14,15,16,17,18]. CMNs have also been discussed as a pathway for the transport of N from donor to recipient plants [19]. It is clear that CMNs affect the survival and fitness, behavior and competitiveness of the plants and fungi that are linked via these networks, but our current understanding about how the nutrient or infochemical allocation among plants within a CMN is controlled, is very limited [11,20].
The contribution of AM fungi to the N nutrition of their host plant is still under debate [21], and it has been suggested that higher N contents in mycorrhizal plants are just a consequence of an improved supply with P [22]. However, it is clear that AM fungi transfer N to their host, and plants are able to take up N from the mycorrhizal interface. This review summarizes our current knowledge about N uptake, metabolism, and transport in the AM symbiosis and discusses major research gaps in our understanding.
2. Mycorrhizal Roots Have Two Uptake Pathways for Nutrients
Mycorrhizal roots have two uptake pathways for nutrients: the plant uptake pathway (PP) and the mycorrhizal uptake pathway (MP; Figure 1). The PP involves the uptake of nutrients via high- or low affinity uptake transporters in the epidermis or root hairs. Particularly for nutrients with a low mobility in the soil (e.g., P), the uptake via the PP is often limited by the development of depletion zones around the roots. By contrast, the MP involves the uptake by high affinity nutrient transporters in the ERM, followed by the translocation along the hyphae to the intraradical mycelium (IRM) in the root cortex, and the uptake from the mycorrhizal interface by mycorrhiza-inducible plant uptake transporters [23]. However, a plant is simultaneously colonized by communities of AM fungi that can differ in their efficiency with which their MP contributes to the total uptake of nutrients by the plant. The uptake and transport of nutrients via both pathways and their contribution to the nutrient supply of the plant has so far primarily been studied for P [21,23,24,25], but both pathways also play a role in the N uptake by plants.
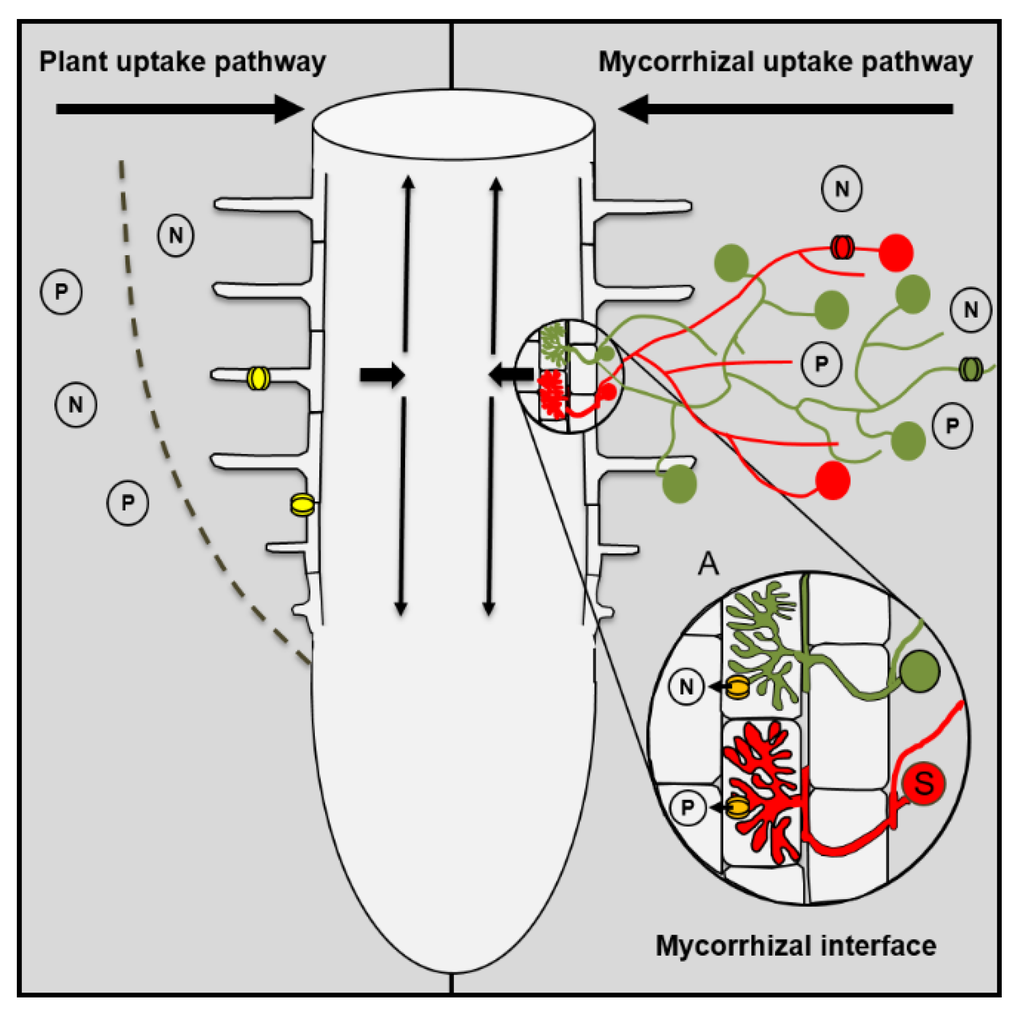
Figure 1.
Plant uptake and mycorrhizal uptake pathway. Plants can take up nutrients by transporters that are located in epidermis or root hairs (yellow symbols) or via the mycorrhizal uptake pathway that comprises the uptake of nutrients by fungal transporters in the extraradical mycelium (red or green symbols), the transport through the hyphae from the ERM to the IRM (see mycorrhizal interface), and the uptake from the mycorrhizal interface by mycorrhiza-inducible plant transporters in the periarbuscular membrane (orange symbols). Indicated by the red and green fungal structures is the colonization of one host root by multiple fungal species that can differ in their efficiency with which they are able to take up nutrients from the soil and transfer these nutrients to their host.
Figure 1.
Plant uptake and mycorrhizal uptake pathway. Plants can take up nutrients by transporters that are located in epidermis or root hairs (yellow symbols) or via the mycorrhizal uptake pathway that comprises the uptake of nutrients by fungal transporters in the extraradical mycelium (red or green symbols), the transport through the hyphae from the ERM to the IRM (see mycorrhizal interface), and the uptake from the mycorrhizal interface by mycorrhiza-inducible plant transporters in the periarbuscular membrane (orange symbols). Indicated by the red and green fungal structures is the colonization of one host root by multiple fungal species that can differ in their efficiency with which they are able to take up nutrients from the soil and transfer these nutrients to their host.
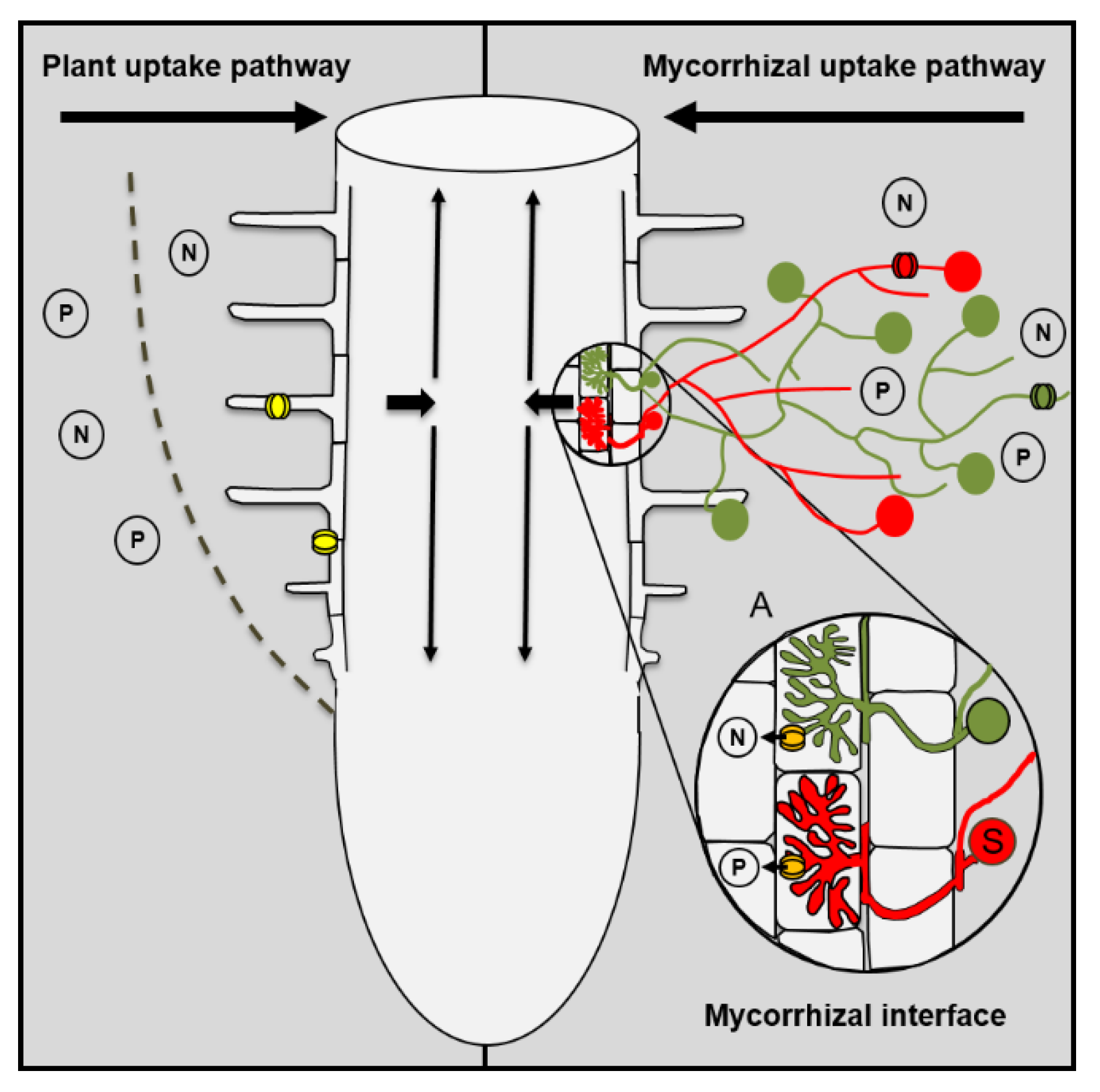
Ectomycorrhizal roots are enclosed by a more or less densely arranged fungal sheath that can represent a significant apoplastic barrier and restricts the nutrient uptake of ectomycorrhizal roots via the PP [26,27]. However, in contrast to ectomycorrhizal roots, AM roots are structurally unaltered and can theoretically use both pathways for nutrient uptake. Previously, it was generally believed that the PP is not affected by the symbiosis and that both uptake pathways act additively. This led to the overall assumption that the uptake via the MP was negligible in cases in which the mycorrhizal growth responses were neutral and the mycorrhizal plants did not differ in their P contents from non-mycorrhizal plants. However, there is increasing evidence that this view is oversimplified, and that mycorrhizal plants differ in their nutrient acquisition strategy from non-mycorrhizal plants [21,23,28].
Multiple studies have demonstrated that even in non-responsive plants the MP can contribute significantly to the P uptake of the plant [24,29]. Plant P transporters that are involved in the uptake via the PP are down-regulated in response to the AM symbiosis [30,31], while mycorrhiza-specific transporters that are involved in the P uptake from the mycorrhizal interface are induced [32,33,34]. The suppression of the PP by the AM symbiosis can also lead to negative mycorrhizal growth responses when the reduction in the P uptake via the PP is not fully compensated for by an increase in the P uptake via the MP. This can cause an overall reduction in total P uptake and lead to P deficiency of the plant [21]. AM fungi differ in their capability to suppress the PP [31]. A strong suppression of the PP will shift the ratio between the two uptake pathways towards the MP and will result in a higher mycorrhizal dependency of the host.
Whether the N uptake via the PP is affected by the AM symbiosis is currently unknown. However, similar to what has previously been described for P, there are indications that plant ammonium (NH4+) or nitrate (NO3−) transporters are down-regulated [35,36], while mycorrhiza-inducible N transporters are up-regulated by the AM symbiosis [36,37,38,39]. Arbuscules are prematurely degraded when the mycorrhiza-inducible P transporter PT4 is not expressed [40]. Recently, it was shown that the arbuscular life span in Medicago plants is not only influenced by PT4 but also by the mycorrhiza-inducible NH4+ transporter AMT2;3 [41]. There is increasing evidence that suggests that the N and P uptake and transport are tightly linked in the AM symbiosis and that the transport of both nutrients controls mycorrhizal functioning (see also below). More studies are necessary to understand the role that both uptake pathways play for the N uptake, and how the activity of both pathways is regulated.
Contribution of the Arbuscular Mycorrhizal Symbiosis to Plant Nutrition
While the positive effect of the AM symbiosis on P nutrition is long known [3,21], the contribution of AM fungi to the N nutrition of their host plant is still under debate [23]. The mobility of the inorganic N sources nitrate (NO3−) and ammonium (NH4+) in the soil is relatively high, and the rhizosphere is less likely to become N depleted. Unlike for P, where the access of the ERM to P sources beyond the root depletion zone is clearly advantageous, it is expected that the ERM does not increase the access to the N resources in the soil. Recent studies, however, suggest that the NH4+ uptake system of AM fungi has a five times higher affinity for NH4+ than typical uptake systems of plants, what would enable the fungus to take up NH4+ from the soil even under low N supply conditions [42].
In the literature, negative [43], neutral [44] or positive [45,46] effects of the AM symbiosis on N nutrition have been reported (for review [47]), and several authors postulated that an improved N status of AM plants is only the consequence of an improved P nutrition [22]. Hawkins and George [44], for example, reported that the hyphal N supply was not sufficient to sustain an adequate N nutrition of a host plant under N limitation. By contrast, other studies have shown that AM fungi can increase the N acquisition of plants compared to non-mycorrhizal controls that were supplied with additional P [48]. Recent results demonstrated that the ability of AM fungi to improve the N nutrition of their host is relatively widespread within the Glomeromycota, but that there is a high intraspecific diversity between different isolates of one AM fungal morphospecies [46]. The authors reported that out of the 31 fungal isolates that were tested, the colonization with six fungal isolates led on average to a mycorrhizal growth response of 191% and a 2.4-fold increase in the N tissue concentration of Medicago sativa. These positive effects were independent from the contribution of these fungal isolates to P supply. There is increasing evidence that a pathway for N through the fungal hyphae to the host plant exists, even if the percentage contribution to total N nutrition of the host plant can vary considerably and is context dependent [23,46]. In AM root organ cultures, 21% of the total N in the roots were taken up by the ERM [49]; and in similar experiments even higher proportions were observed [50]. In maize, 75% of the N in the leaves were taken up by the ERM of an AM fungus [51].
In several plant species mycorrhiza-inducible NO3− [52] or NH4+ [36,38,39,41,53], transporters have been identified that are able to facilitate the uptake of N sources from the mycorrhizal interface. Recent results demonstrated that both P and N are important determinants for the AM symbiosis and that the colonization of the plant host is controlled by feedback mechanisms between both nutrients [12]. For example, both, P and N starvation of the plant induce a nutrient stress transcriptome that is favorable for AM colonization. Under P and N stress, plant defense genes are down-regulated, while genes that are involved in the strigolactone biosynthesis are up-regulated [54]. Strigolactone serves as an important signal for AM fungi in the soil and stimulates hyphal branching during the presymbiotic growth stage [55]. High P availabilities often reduce the AM colonization of the plant, but N starvation triggers a signal that promotes AM colonization and reverses the inhibitory effects of high P availabilities on AM colonization [41,56]. However, the recovery of AM colonization did not lead to increased N levels in these plants [56]. When the fungal P transport to Medicago truncatula plants is inhibited by the suppression of the mycorrhiza-inducible plant P transporter MtPt4, arbuscules are prematurely degraded, and the symbiosis is not maintained by the host [40]. However, AM fungi can escape premature degeneration of their arbuscules in MtPt4 mutants under N starvation [41,57], and it has been demonstrated that the expression of mycorrhiza-induced plant P transporters are not critical for the AM symbiosis as long as the plant is grown under low N conditions [41]. All these results clearly demonstrate that N is transferred through the fungal hyphae to the host, and that N transport to the host may play a critical role for the maintenance and the nutrient efficiency of the AM symbiosis.
3. Nitrogen Uptake by Arbuscular Mycorrhizal Fungi
3.1. Uptake of Inorganic N Sources
Nitrate (NO3−) is the dominant form of N that is available to plants and fungi in most agricultural soils, whereas ammonium (NH4+) predominates in many undisturbed or very acidic soils, where NO3− can be almost entirely absent. The ERM of AM fungi can take up NH4+ [58,59,60,61,62] and NO3− [62,63], but NH4+ is generally preferred, because it is energetically more efficient than NO3− [49,50,62,64] (see also Table 1). Based on pH changes induced by the ERM, when hyphae were supplied with NO3− or NH4+, it has been hypothesized that the NO3− uptake by the hyphae is active and coupled to an H+-symport mechanism, while NH4+ is taken up by an antiport mechanism with a net H+ efflux [65,66]. Similar to plants and other fungi, AM fungi have high-affinity and low-affinity uptake systems for NH4+. The low Km value of 2.53 ± 0.25 μM for the high affinity uptake system of Rhizophagus irregularis demonstrates that the fungus is able to take up NH4+ from soils with very low concentrations [42]. Km values of high affinity NH4+ uptake systems of plants are typically higher than the estimated Km value for the fungal NH4+ uptake system [42,67,68].

Table 1.
Demonstrated metabolic pathways of nitrogen (N) in arbuscular mycorrhizal (AM) fungi. Shown are the fungal species or the tissues that were tested. Fungal species were renamed according to the reclassification of the Glomeromycota [69]. Species abbreviations and authorities: Acla—Acaulospora laevis (Gerd. & Trappe), Clet—Claroideoglomus etunicatum (W. N. Becker & Gerd.; previously Glomus etunicatum), Fumo—Funneliformis mosseae (T. H. Nicolson & Gerd., previously Glomus mosseae), Gima—Gigaspora margarita (W. N. Becker & I. R. Hall), Giro—Gigaspora rosea (T. H. Nicolson & N. C. Schenck), Glfa—Glomus fasciculatum (Gerd. & Trappe), Glma—Glomus macrocarpus (Tul. & C. Tul.), Glve—Glomus versiforme (P. Karst., S. M. Berch), Rhcl—Rhizophagus clarus (T. H. Nicolson & N. C. Schenck, previously Glomus clarum), Rhir—Rhizophagus irregularis (N.C. Schenck & G. S. Sm.), Rhma—Rhizophagus manihotis (R. H. Howeler, Sieverd. & N. C. Schenck).
| Pathway/Enzyme | Fungal Species | Tissue | References | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Spore | ERM | AM | IRM | ||||||
| Inorganic N uptake | |||||||||
| NH4+ uptake | Fumo, Rhir, Acla, Gima | + | + | nd | nd | [50,58,60,61,62,64,70,71] | |||
| NH4+ transporter | Rhir | + | + | + | + | [53,72,73] | |||
| NO3− uptake | Glfa, Rhir, Fumo | + | + | nd | nd | [62,63,64,65,70,71] | |||
| NO3− transporter | Rhir | + | + | + | + | [71,72,74,75,76] | |||
| Nitrate permease | Rhir | + | + | + | + | [76] | |||
| Organic N uptake | |||||||||
| Acla, Rhir, Gima | nd | +/− | nd | nd | [60] | ||||
| Fumo | nd | + | + | nd | [45,58] | ||||
| nd | − | nd | nd | [77] | |||||
| Amino acid transporter | + | + | + | + | [72] | ||||
| Amino acid permease | Fumo | − | + | + | nd | [78] | |||
| Peptide transporter | Rhir | nd | + | + | + | [79] | |||
| Alanine | Rhir | − | nd | nd | nd | [71] | |||
| Arginine | Rhir | − | + | nd | nd | [50,71] | |||
| Cysteine | Rhir | − | + | nd | nd | [80] | |||
| Glycine | Rhir, Fumo, un | + | + | nd | nd | [62,71] | |||
| Glutamate | Fumo, Rhir | + | + | nd | nd | [62,71] | |||
| Glutamine | Fumo, Rhir | + | + | nd | nd | [71,81] | |||
| Methionine | Rhir | − | + | nd | nd | [80] | |||
| Ornithine | Rhir | + | nd | nd | nd | [71] | |||
| Urea 4 | Gima | + | + | nd | nd | [71,82] | |||
| Nitrate reduction | |||||||||
| Nitrate reductase1 | Glfa, Rhir, Fumo, Glma, Glsp | + | nd | + | nd | [72,83,84,85,86] | |||
| Nitrate reductase (NADH) | Un, Fumo | + | + | + | nd | [52,87,88,89] | |||
| Nitrate reductase (NADPH) | Rhir | nd | + | + | + | [52,90] | |||
| Nitrite reductase | Rhir | nd | + | + | + | [72,84] | |||
| GDH pathway | |||||||||
| GDH1 | Clet | nd | nd | + | + | [91] | |||
| GDH (NADH) | Fumo, Rhir | + | + | + | + | [71,72,92] | |||
| GDH (NADPH) | Glfa | nd | nd | +/− | nd | [83,93] | |||
| GS/GOGAT pathway | |||||||||
| Glutamine synthetase | Glfa, Fumo, Rhir | + | + | + | + | [71,72,74,75,81,83,84,93,94] | |||
| Glutamate synthase (NADH) | Rhir | + | + | + | + | [64,72,74,75,84,94] | |||
| Glutamate synthase (NADPH) | Glfa, Rhir | nd | nd | + | + | [72,83] | |||
| Amino acid biosynthesis | |||||||||
| Transaminases | Glfa | nd | nd | + | nd | [83,95] | |||
| Asparagine synthase | Rhir | + | + | + | + | [72] | |||
| Arginine biosynthesis | |||||||||
| Carbamoyl-P synthase | Rhir | + | + | + | + | [72,74,75] | |||
| Argininosuccinate synthase | Rhir | + | + | + | + | [72,74,75] | |||
| Argininosuccinate lyase | Rhir | + | + | + | + | [72,74,75] | |||
| Arginine breakdown | |||||||||
| Arginase | Rhir | + | + | + | + | [72,74,75] | |||
| Urease | Rhcl, Rhir | + | + | + | + | [72,74,75] | |||
| Urease accessory protein | Rhir | + | + | + | + | [71,72,75,94] | |||
| Ornithine aminotransferase | Rhir | + | + | + | + | [71,72,74,75,94] | |||
| Polyamine biosynthesis | |||||||||
| Ornithine decarboxylase | Rhir | + | + | + | + | [72,74,75] | |||
| N uptake from interface | Plant species | ||||||||
| Plant NH4+ transporter | Medicago truncatula | na | na | + | na | [39,41] | |||
| Lotus japonicus | na | na | + | na | [38] | ||||
| Glycine max | na | na | + | na | [36] | ||||
| Sorghum bicolor | na | na | + | na | [37] | ||||
Abbreviations: AM—arbuscular mycorrhizal roots; ERM—extraradical mycelium; IRM—intraradical mycelium; na—not applicable; nd—not determined.
Despite the fungal preference for NH4+, Hawkins and George [70] reported that when NH4+ was the sole N source for mycorrhizal plants, root and shoot biomass, hyphal length densities and N transport via the hyphae to the plant were lower than after NO3− supply. Valentine et al. [96] also reported, that NH4+ fed AM roots had lower numbers of vesicles and arbuscules than NO3− supplied roots. An excess of NH4+ as sole N source is often considered to be toxic for plants, and inhibits root growth [97]. NH4+ assimilation occurs in the root, while the assimilation of NO3− is predominantly foliar [98,99]. When NH4+ is the sole N source, the assimilation of NH4+ could increase the consumption of C skeletons in the root, and reduce the carbon availability for the fungus. Hawkins and George [70] found that NH4+ reduced the hyphal length in the soil, but not the number of arbuscules, and assumed that high concentrations of NH4+ could also have a direct deleterious effect on the ERM.
Transcriptome studies revealed the expression of several fungal NH4+ and NO3− transporters in spores, ERM, and IRM [72]. The expression of GintAMT1, an NH4+ transporter of the AM fungus Rhizophagus irregularis (previously Glomus intraradices) is induced by low additions of NH4+ to the medium (in the presence of relatively high concentrations of NO3− in the medium) but suppressed under high NH4+ supply. This suggests that the expression of this transporter is substrate inducible, and is regulated by the NH4+ supply and fungal NH4+ status [73]. By contrast, the NH4+ transporter GintAMT2 is constitutively expressed in the ERM under N limiting conditions, and transitory induced when different N sources are added to the N deficient ERM [53]. The differential localization of high transcript levels of these transporters in colonized roots suggests that both transporters may differ in their role for N uptake and transport. The high expression levels of GintAMT1 in the ERM indicate that this transporter could be primarily involved in the NH4+ acquisition of fungal hyphae from the soil. By contrast, GintAMT2 is particularly expressed in the IRM, suggesting that this transporter could play a role in the re-uptake of NH4+ by the fungus from the symbiotic interface [53].
An exogenous supply of NO3− stimulates the expression of a fungal NO3− transporter in the ERM of Rhizophagus irregularis (also known as Rhizophagus intraradices or previously Glomus intraradices) [74]. However, the expression of this NO3− transporter is repressed by an increase in the internal levels of NH4+ or of a downstream metabolite, such as glutamine [75]. Nitrate transporters in many organisms are suppressed when more preferred N sources, such as NH4+ become available. This process is known as N catabolite repression, and this regulatory system also controls the expression of NH4+ transporters in other fungi [100]. In ectomycorrhizal fungi, the expression of high affinity NH4+ transporters is suppressed by high levels of intracellular glutamine [101], and NO3− transporter proteins in the yeast Hansenula polymorpha are degraded in response to glutamine [102]. GATA transcription factors have been shown to be involved in N catabolite repression and in the promoter sequence of the NH4+ transporter GintAMT2 two GATA core sequences have been identified [53]. The expression profiles of AM fungal NO3− and NH4+ transporters under different N availabilities suggest that N catabolite repression could also operate in AM fungi and could control the N uptake by AM fungi from the soil.
3.2. Uptake of Organic N by Hyphae
Organic N can represent in many soils a significant proportion of total soil N [103], but the obligate biotrophic life cycle of AM fungi has led to the overall assumption that AM fungi are unable to utilize organic N sources. Several studies, however, have demonstrated that the hyphae of AM fungi grow into organic patches of varying complexity and transfer 15N from these organic patches to their host plant [104,105], and that this can lead to higher plant N contents [106]. When the fungus had access to organic patches that were labelled with 15N and 13C, the fungal ERM got only enriched with 15N, but not with 13C. This confirms that AM fungi do not have saprophytic capabilities and that the fungus acquires 15N from these organic patches likely as a decomposition product [105]. However, even if AM fungi themselves do not act as decomposers, AM fungi accelerate the N mineralization from organic matter [107] and affect the carbon flow through soil microbial communities during decomposition [108].
Plant roots release fixed carbon into their rhizosphere, and this carbon flux into the root rhizosphere acts as an important trigger for decomposition processes and nutrient cycling in soils [109]. AM fungi form an extensive network of hyphae in soils (ERM), and the mycorrhizosphere, the large interface between the fungal hyphae and the soil, could play a similar role than the rhizosphere for decomposition and nutrient cycling in soils. The ERM of the AM fungus acts as a conduit between decomposing microbial communities and the host plant, and provides decomposers with plant-derived carbon inputs and transfers decomposition products to the plant. The mycorrhizosphere represents in soils an important ecological niche and provides nutritionally favorable conditions for diverse microbial communities, and it has been suggested that the presence of AM fungal hyphae plays an important role in the bacterial community assembly during decomposition [110].
Compared to ectomycorrhizal fungi, the capability of AM fungi to utilize organic N sources is considered to be relatively small but also AM fungi are able to take up organic N sources from the soil, such as amino acids (Table 1). Free amino acids can represent an important N source in soils, and AM fungi can take up several amino acids, e.g., aspartic acid, serine [111], glycine, glutamic acid [62,112], glutamine [81], cysteine or methionine [80]. Some amino acids are also taken up by germinating spores during the presymbiotic growth stage of the fungus [71]. Transcriptome studies have shown that fungal amino acid transporters are expressed in the ERM [72]. Cappellazzo and co-authors characterized an amino acid permease of the AM fungus Funnelliformis mosseae (previously Glomus mosseae) and found that this amino acid transporter binds preferably neutral, nonpolar and hydrophobic amino acids [78]. While some amino acids such as glycine, serine and glutamine competed strongly with proline for this transporter, positively charged amino acids such as arginine, histidine and lysine were only poor competitors. The mycorrhizal colonization of Sorghum bicolor led to an increase in the uptake of phenylalanine, methionine, asparagine, tryptophane, and cysteine, but also increased the uptake of the charged amino acids arginine, lysine, and histidine [113]. Other studies demonstrated that arginine can be taken up by the ERM and that arginine is not metabolized during its transport from the ERM to the IRM [50]. This suggests that multiple transporters are involved in the amino acid uptake by the ERM.
Recently, the putative dipeptide transporter RiPTR2 has been identified in the AM fungus Rhizophagus irregularis [79]. Short peptides can represent a greater proportion of N in soils than free amino acids, and this transporter facilitated in complementation assays the uptake of several dipeptides. This suggests that this transporter could play a role in the fungal uptake of small peptides from the soil. However, the transcript levels of this transporter were higher in the IRM than in the ERM, suggesting that RiPTR2 could also play a role in the reabsorption of peptides from the interfacial apoplast [79].
4. Nitrogen Assimilation and Transport by Arbuscular Mycorrhizal Fungi
4.1. Nitrate Reduction in Arbuscular Mycorrhizal Fungi
After NO3− is taken up by plants or fungi, NO3− is first reduced to nitrite by a nitrate reductase. Both NADH and NADPH can act as reductant for nitrite formation [52], but, in AM fungi, nitrate reductase activity is mainly driven by NADPH [90]. Nitrate reductase activity in both roots [44,83,84,87] and shoots [83,87,95] of AM plants is generally higher than in non-mycorrhizal control plants (for more information, see also Table 1). In nonmycorrhizal plants, NO3− reduction primarily takes place in the leaves, while in mycorrhizal plants NO3− is predominantly reduced in the roots [88,90]. A gene that encodes a fungal nitrate reductase is expressed in spores, ERM and IRM, but the transcript levels are particularly high in the IRM. This could indicate that NO3− that is not directly assimilated in the ERM can also be reduced in the fungal tissue within the host root [72,85,90].
The second step in N assimilation is the conversion of nitrite into NH4+ by nitrite reductase. A gene encoding a putative fungal nitrite reductase of the AM fungus Rhizophagus irregularis shows particularly high expression levels in spores and in the IRM [72]. This is consistent with the high transcript levels of a fungal nitrate reductase in the IRM of AM roots [72]. The expression of tbnir1, the nitrite reductase of the ectomycorrhizal (ECM) fungus Tuber borchii, is induced by NO3− but suppressed when more preferred N sources, such as NH4+ or glutamine become available [114]. It has been demonstrated in ECM roots that the activity of the fungal nitrite reductase controls the expression of the plant nitrite reductase gene. When the plant is colonized with a fungal wild type strain, the expression of the plant nitrite reductase is repressed, but the expression increased in roots that were colonized with nitrate reductase deficient fungal strains [115]. This is consistent with the finding that, in mycorrhizal roots, the transcript levels of plant nitrate reductase are lower than in non-mycorrhizal control plants [52,90] and suggests that the expression of the plant nitrite reductase is suppressed by the transfer of reduced N compounds from the fungus to the host [115].
4.2. Nitrogen Assimilation into Amino Acids in Arbuscular Mycorrhizal Fungi
Since excess N in its reduced form NH4+ is toxic for cells due to its ability to uncouple respiration, NH4+ is rapidly assimilated into amino acids [116]. Two pathways can be involved in the assimilation of NH4+ by fungi: the NAD(P)-glutamate dehydrogenase (GDH) or the glutamine synthetase—glutamate synthase (GS-GOGAT) pathway. GDH uses NH4+ and 2-oxoglutarate to produce glutamate (Glu), while in the GS-GOGAT pathway, first glutamine (Gln) is produced from Glu and NH4+ by glutamine synthetase, and then Gln and 2-oxoglutarate are converted by glutamate synthase (also known as glutamine oxoglutarate aminotransferase) into two molecules of Glu. In the cytoplasm of ascomycetes and basidiomycetes activities of NAD and NADP-dependent GDH can be detected [117]. However, Smith et al. [93] found no increased GDH activity in AM roots, and also in the ERM of Rhizophagus irregularis no GDH activity could be detected [49]. A putative NAD-dependent GDH gene of R. irregularis is down-regulated in the ERM in response to either NO3− or NH4+ supply, suggesting that GDH plays in AM fungi a more catabolic role rather than a role in N assimilation [49,71,83,94].
NH4+ is predominately assimilated via the GS-GOGAT pathway in AM fungi [64,81,83,94]. GS and GOGAT activities have been shown to be significantly higher in roots and shoots of AM plants [83,84,93,95] (see also Table 1), and the decrease in the levels of free amino acids in the ERM of the AM fungus R. irregularis after incubation with the GOGAT inhibitor albizzine confirms the activity of a functional GS-GOGAT pathway in AM fungi [64]. Tian and co-authors [74] identified two different functional GS isoforms of R. irregularis, GiGS1 and GiGS2. GiGS1 has a lower Km than GiGS2 and is constitutively expressed at high levels in the ERM, while GiGS2 is strongly induced by an addition of NO3− to the ERM. This suggests that GiGS1 is the main functional enzyme for N assimilation at low N availabilities, and that GiGS2 may play a more significant role for N assimilation under high N supply conditions [74]. Breuninger and co-authors [81] also found that the fungal GS genes of Funneliformis mosseae (GmGln1) and R. irregularis (GiGln1) are constitutively expressed but that the GS activities in the ERM are modulated in response to different N availabilities. Based on this observation, the authors concluded that the fungal GS activity is not controlled on a transcriptional level, but is subjected to post-transcriptional regulation [81]. However, the GS transcript that was examined by these authors has a closer sequence similarity to GiGS1 (see above), and only GiGS2 has been shown to be strongly up-regulated after NO3− supply [74].
Consistent with the GS-GOGAT pathway, Gln becomes highly labelled when 15NH4+ is supplied to AM fungi, and represents one of the major N sinks [50,71,83,95,118]. Gln plays a central role in N metabolism (1) as key N donor; (2) as precursor of many essential metabolites such as nucleic acids, amino sugars, and other amino acids, such as histidine, tyrosine and asparagine; and (3) as key effector for N metabolite repression and regulator of genes involved in N metabolism [67,101,102,119,120]. Due to these important functions, the free levels of Gln in AM fungi are tightly controlled [71]. In addition to Gln, Glu, asparagine (Asn), aspartate (Asp) and alanine (Ala) are abundant free amino acids in germinating spores [71], in the ERM [64] or in AM roots [50,95,118]. Ornithine, serine and glycine are also detectable but in much lower concentrations [64].
Arginine levels (Arg) were not determined in several of the earlier investigations [64,84,95], but more recent studies demonstrate that Arg is the most abundant free amino acid and can represent more than 90% of the total free amino acids in the ERM [118]. Arg levels of up to 200 nM·mg−1 dry weight have been reported in the ERM [50]. Due to its low carbon-to-nitrogen ratio of 6:4, Arg plays an important role for N storage and N transfer from the ERM to the IRM [50,121]. For example, Arg serves as a N storage molecule in quiescent spores and its catabolic breakdown during spore germination provides the N and C skeletons for the biosynthesis of other amino acids or proteins for the presymbiotic growth of the AM fungus [71].
4.3. Nitrogen Transport from the Extraradical Mycelium to the Intraradical Mycelium
The ERM takes up N from the soil in a significant distance from the root, and transfers N from the ERM to the IRM. The transport of N through the hyphae of the AM symbiosis can be very fast and flux rates similar to those of P have been observed [121,122]. Fungal vacuoles often contain polyphosphates (polyP) and basic amino acids in equimolar concentrations [123,124,125], and it has been suggested that N could move in the form of Arg with fungal polyP from the ERM to the IRM [50,94,121]. PolyP are negatively charged polyanions and the basic amino acid Arg could serve together with other cations such as K+ and Mg2+ as counter charge and contribute to the required charge balance [126]. Studies on the N transport in the AM symbiosis of Agropyron repens, however, also suggest that in addition to Arg, other amino acids such as Gln or Glu could be involved in the translocation of N from the ERM to the IRM [127].
Gene expression studies are consistent with the biosynthesis of Arg in the ERM. Shortly after NO3− supply, the transcript levels of a fungal carbamoyl-phosphate synthetase (CPS), argininosuccinate synthase (ASS), and argininosuccinate lyase (AL) are induced in the ERM (Figure 2). All of these enzymes are involved in Arg biosynthesis; CPS catalyzes the formation of carbamoylphosphate from CO2, ATP, and NH3, which is converted together with ornithine to citrulline and Pi by an ornithine transcarbamoylase (OTC). Citrulline and aspartate are converted to argininosuccinate (AS) by ASS, and AL converts argininosuccinate to fumarate and Arg. In contrast, in the IRM a fungal arginase (CAR1) and urease (URE) are up-regulated that are involved in the catabolic breakdown of Arg [74]. The biosynthesis of Arg in the ERM followed by the subsequent breakdown of Arg in the IRM are spatially separated but synchronized processes, and confirm the function of the anabolic (ERM) and catabolic part (IRM) of the urea cycle in the AM symbiosis (Figure 2). The synchronization of these processes suggests that Arg plays an important role in the N translocation from the ERM to the IRM [50,74,121].
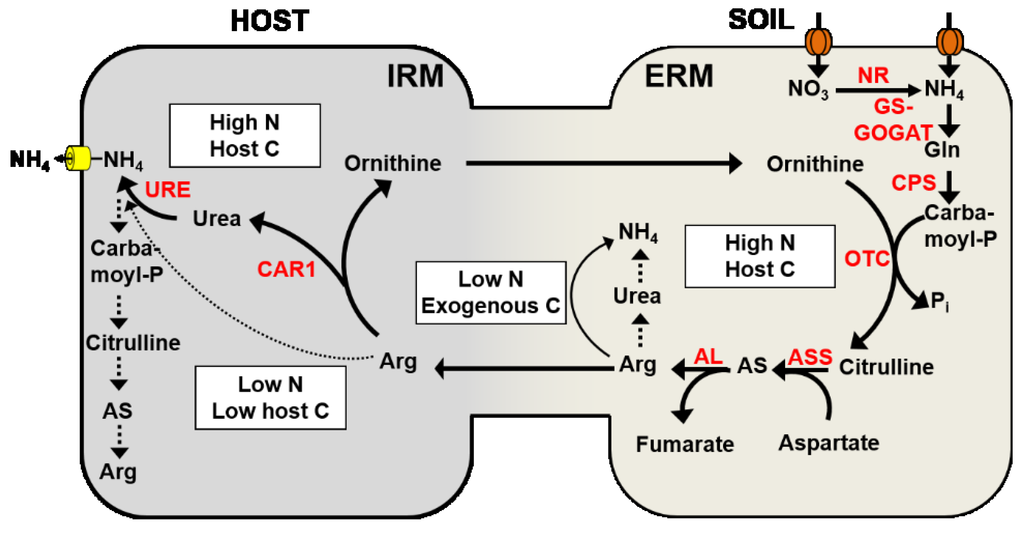
Figure 2.
Function and regulation of the anabolic and catabolic arm of the urea cycle in the ERM or the IRM, respectively. The activities of the following enzymes are indicated in red: nitrate reductase/nitrite reductase (NR), glutamine synthetase—glutamate synthase (GS-GOGAT), carbamoyl-phosphate synthetase (CPS), ornithine transcarbamoylase (OTC), argininosuccinate synthase (ASS), argininosuccinate lyase (AL), arginase (CAR1), and urease (URE). Favored pathways are indicated by bold lines, suppressed pathways by dotted lines. White boxes provide additional information about the putative regulation of these pathways.
Figure 2.
Function and regulation of the anabolic and catabolic arm of the urea cycle in the ERM or the IRM, respectively. The activities of the following enzymes are indicated in red: nitrate reductase/nitrite reductase (NR), glutamine synthetase—glutamate synthase (GS-GOGAT), carbamoyl-phosphate synthetase (CPS), ornithine transcarbamoylase (OTC), argininosuccinate synthase (ASS), argininosuccinate lyase (AL), arginase (CAR1), and urease (URE). Favored pathways are indicated by bold lines, suppressed pathways by dotted lines. White boxes provide additional information about the putative regulation of these pathways.
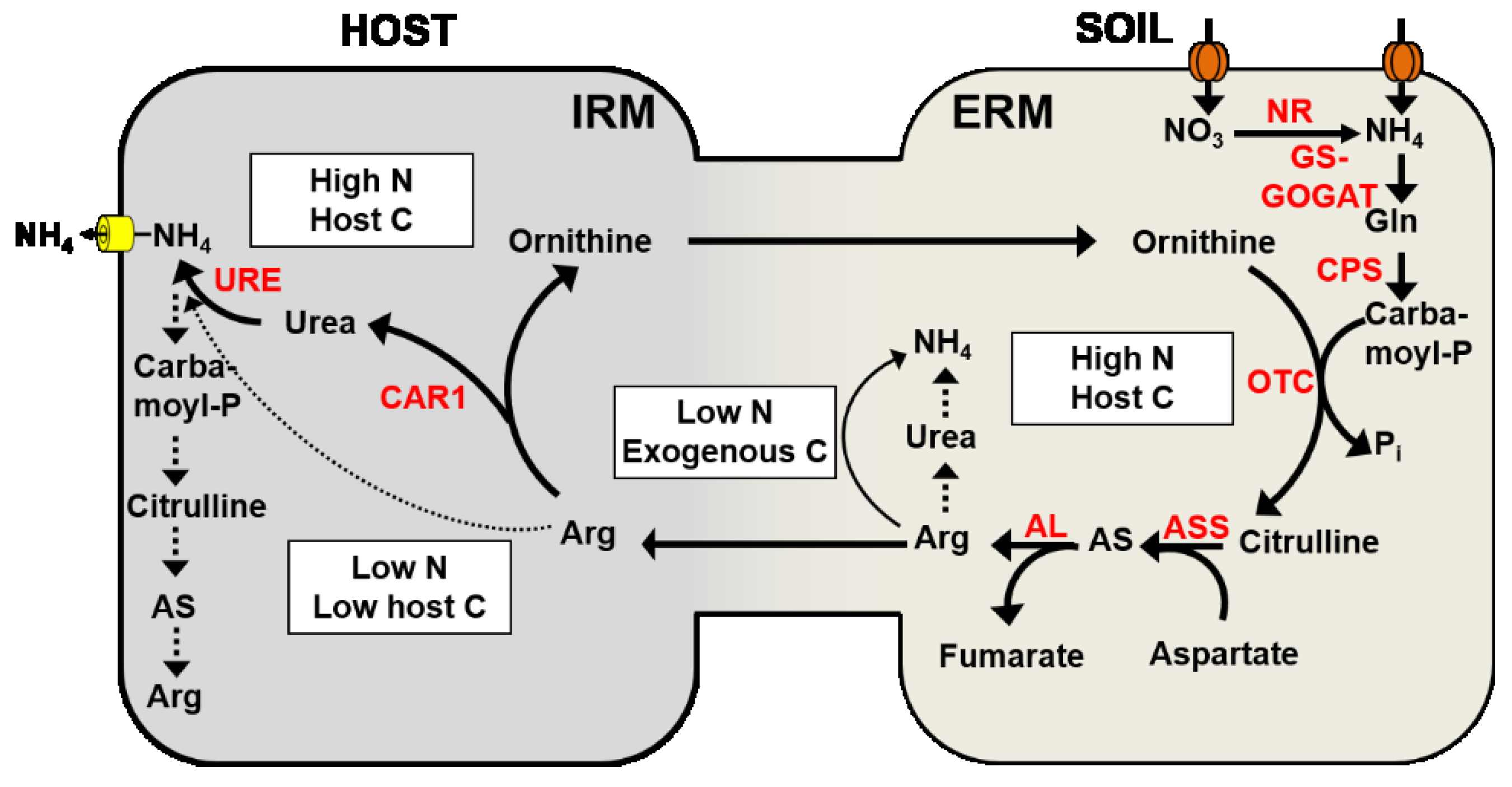
The C supply of the host plant plays a significant role in the regulation of these processes [12,75]. An increase in the C supply from the host plant led to an up-regulation of genes that are involved in Arg biosynthesis (AL, ASS, CPS), but to a down-regulation of a fungal urease (URE) in the ERM [75]. This will lead to increasing levels of Arg in the ERM (stimulation in biosynthesis and reduced catabolic breakdown) and stimulate the transfer of Arg from the ERM to the IRM. In contrast, in the IRM, an increase in the fungal arginase and urease activity and a down-regulation of genes involved in Arg biosynthesis were observed. This will favor the catabolic breakdown of Arg but prevent the re-assimilation of the released NH4+ in the IRM [75]. This demonstrates that the host plant with its C supply is able to regulate fungal gene expression and to trigger fungal N transport. However, when the fungus had access to an exogenous C source (acetate supply to the ERM), the transcript levels of genes involved in N assimilation and Arg biosynthesis were reduced, but the expression of a fungal arginase was induced in the ERM. This could indicate that the Arg transport from the ERM to the IRM is suppressed when the fungus has access to an exogenous carbon source and is not solely dependent on its host for its carbon supply [75].
The active hydrolysis of polyP will release Pi and Arg (for subsequent breakdown into NH3 and NH4+ by the catabolic arm of the urea cycle) into the fungal cytoplasm of the IRM and will facilitate the efflux of P and of N into the mycorrhizal interface. AM and ECM fungi regulate the nutrient transport to the host by the accumulation or remobilization of polyP, and it has been shown that the carbon supply of the host plant triggers polyP hydrolysis [10,75,128,129]. The regulation of polyP and Arg biosynthesis in the ERM and of polyP and Arg breakdown in the IRM would allow the fungus to control its P and N transport across the mycorrhizal interface.
4.4. Nitrogen Transport across the Mycorrhizal Interface
In Figure 3, a model of the current N transport pathway and its associations to the P and C flux in the AM symbiosis is shown. According to this model, the N transport pathway in the AM symbiosis includes the following steps: (1) uptake of P and of N by the fungal ERM through Pi, NO3− or NH4+ transporters; (2) N assimilation into Arg via the anabolic arm of the urea cycle and conversion of Pi into polyp; (3) transport of Arg into the fungal vacuole and binding to polyp; (4) transport of polyP from the ERM to the IRM; (5) polyP hydrolysis and release of Arg and Pi in the IRM; (6) Arg breakdown to NH4+ via the catabolic arm of the urea cycle; (7) facilitated Pi, NH4+, and potential amino acid efflux into the interfacial apoplast; (8) plant uptake from the interface through mycorrhiza-inducible Pi or NH4+ transporters; (9) stimulation in photosynthesis by improved nutrient supply and facilitated efflux of sucrose into the interfacial apoplast; (10) sucrose hydrolysis via an apoplastic plant invertase, and uptake of hexoses by the AM fungus through fungal monosaccharide transporters.
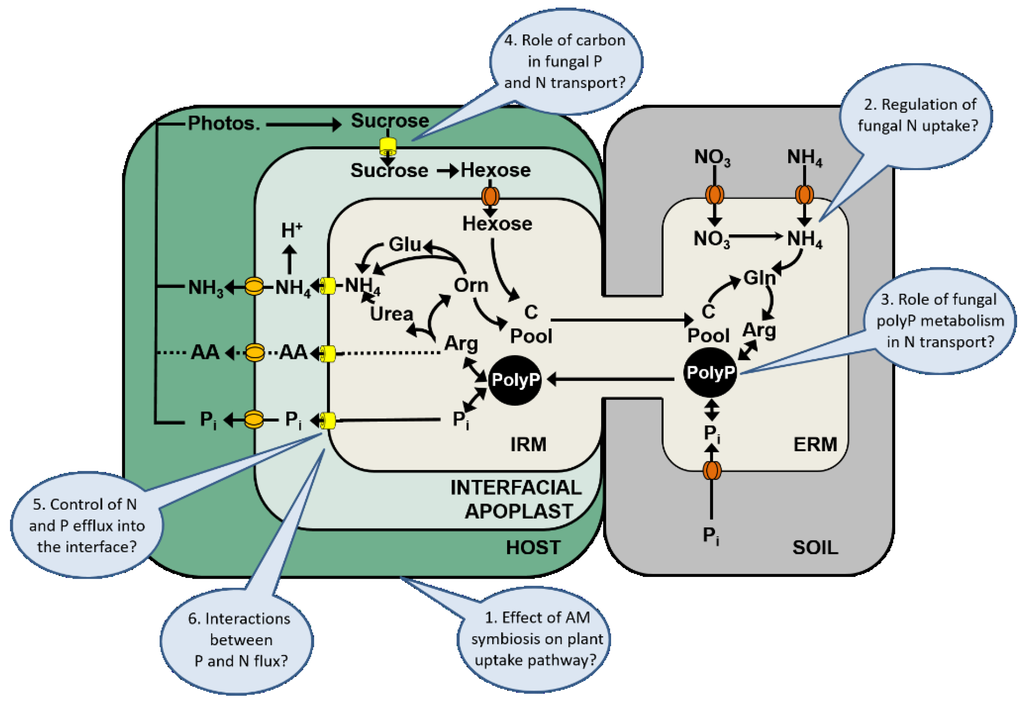
Figure 3.
Model of the N and P transport in the AM symbiosis. The model is based on previous models presented in the literature [50,74,75,94]. The model shows the nutrient uptake by the fungal ERM through Pi, NO3− or NH4+ transporters (red), N assimilation into Arg and the conversion of Pi into polyP, the transport of polyP from the ERM to the IRM, polyP hydrolysis and the release of Arg and Pi in the IRM, Arg breakdown to NH4+ via the catabolic arm of the urea cycle, and the Pi, NH4+, and potential amino acid efflux (yellow) into the interfacial apoplast, and the plant uptake from the interface through mycorrhiza-inducible Pi or NH4+ transporters. The blue ovals indicate gaps in our current understanding of these processes.
Figure 3.
Model of the N and P transport in the AM symbiosis. The model is based on previous models presented in the literature [50,74,75,94]. The model shows the nutrient uptake by the fungal ERM through Pi, NO3− or NH4+ transporters (red), N assimilation into Arg and the conversion of Pi into polyP, the transport of polyP from the ERM to the IRM, polyP hydrolysis and the release of Arg and Pi in the IRM, Arg breakdown to NH4+ via the catabolic arm of the urea cycle, and the Pi, NH4+, and potential amino acid efflux (yellow) into the interfacial apoplast, and the plant uptake from the interface through mycorrhiza-inducible Pi or NH4+ transporters. The blue ovals indicate gaps in our current understanding of these processes.
Previous models suggested that N is transferred from the fungus to the host in form of amino acids, but this would lead to a substantial reflux of C from the obligately biotrophic fungus to the host. The uptake of organic N from the mycorrhizal interface would also require the presence of e.g., amino acid transporters in the periarbuscular membrane that could facilitate an uptake of organic N from the mycorrhizal interface. In Lotus japonicus a mycorrhiza-inducible plant amino acid transporter (LjLHT1.2) has been identified that shows a high expression in arbusculated cells but also in non-colonized cells of the root cortex. The expression of this amino acid transporter in the periarbuscular membrane could indicate that this transporter is involved in the uptake of amino acids from the mycorrhizal interface [130]. However, the expression of this transporter in non-colonized cells suggests that the transporter could also play a role for the re-absorption of amino acids by the cortical cells.
Based on the elucidation of the N transport pathway [74,75,94] (see above and Figure 3), and the identification of several mycorrhiza-inducible NH4+ transporters in the periarbuscular membrane [36,37,38], newer models of N transport in the AM symbiosis now conclude that NH4+ is the main form in which N is transferred into the mycorrhizal interface and is taken up by the host. Studies after labeling with 13C also indicate that the resulting CO2 or HCO3− from the breakdown of Arg is not transferred to the plant and remains within the fungal compartment [50,94].
The release of nutrients from free living fungi is normally regarded as slow, and membrane transport processes are generally favoring fungal re-absorption [131]. Therefore, it is likely that conditions at the mycorrhizal interface exist that facilitate the efflux of nutrients from the fungus or reduce the level of competing fungal uptake systems. It is currently unknown how the N transport from the fungal hyphae into the mycorrhizal interface is facilitated, but several different mechanisms have been discussed. For example, in arbusculated cells and in the ERM of maize roots, two functional fungal aquaporins have been identified that are particularly expressed under drought stress [132]. Aquaporins are integral membrane channels that facilitate the concentration gradient-driven water transport through the plasma membrane. However, in addition to their function as water channels, aquaporins have also been shown to facilitate the diffusion across membranes of low molecular weight neutral solutes such as glycerol, ammonia, and carbon dioxide [133]. This has led to the hypothesis that aquaporins could play a role in the leakage of NH4+ into the interface [134]. The continuous breakdown of Arg in the IRM and the re-assimilation of NH4+ in the root cells will lead to a concentration gradient that could facilitate the movement of NH4+ into the mycorrhizal interface, and reduce the re-absorption of NH4+ by the AM fungus.
Plant NH4+ transporters in the periarbuscular membrane compete with fungal transporters for the NH4+ that has been released into the interfacial apoplast. The expression of fungal NH4+ transporters in arbusculated cells indicates that the fungus is also able to re-absorb released NH4+, and the fungus could modulate the amount of N that is delivered to the host by the expression of these transporters in the IRM [53]. Several plant NH4+ transporters have been identified that are induced in arbusculated cells during the AM symbiosis and that have been implicated in the NH4+ uptake from the mycorrhizal interface [37,38,39,41]. One of these mycorrhiza-inducible NH4+ transporters has been identified in Lotus japonicus (LjAMT2;2) [38]. Interestingly, when LjAMT2;2 is expressed in oocytes of Xenopus laevis the uptake of NH4+ does not result in a flow of current. Based on this observation, the authors concluded that the transporter binds NH4+ externally but transfers uncharged NH3 across the periarbuscular membrane. It is unlikely that this high-affinity transporter could recruit NH3 from the interfacial apoplast, because NH4+ should be the dominant form of NH3/NH4+ in the interfacial apoplast due to the assumed low pH conditions in this compartment (pKa of protonation 9.25) [38].
Recently, two plant NH4+ transporters of Medicago truncatula (AMT2;3 and AMT2;4) have been identified that play a role for the arbuscular life span [41]. Arbuscules are prematurely degraded in mutants in which pt4, the mycorrhiza-inducible plant P transporter that is critical for the uptake of P from the mycorrhizal interface, is not expressed [40,57]. This premature degeneration of arbuscules is suppressed by an expression of AMT2;3 when the plant is grown under N stress, but not by AMT2;4. However, only AMT2;4 has been shown to be a functional NH4+ transporter and was able to support in complementation assays the growth of a yeast NH4+ transporter mutant [41]. This suggests that AMT2;3 and AMT2;4 differ in their function, and that AMT2;3 could play more a sensing or signaling function that is not present in other AMT transporters [41]. It has been hypothesized that some mycorrhiza-inducible nutrient transporters that are localized in the periarbuscular membrane could act as transceptors, transporter-like proteins with a receptor function [135].
5. Conclusions
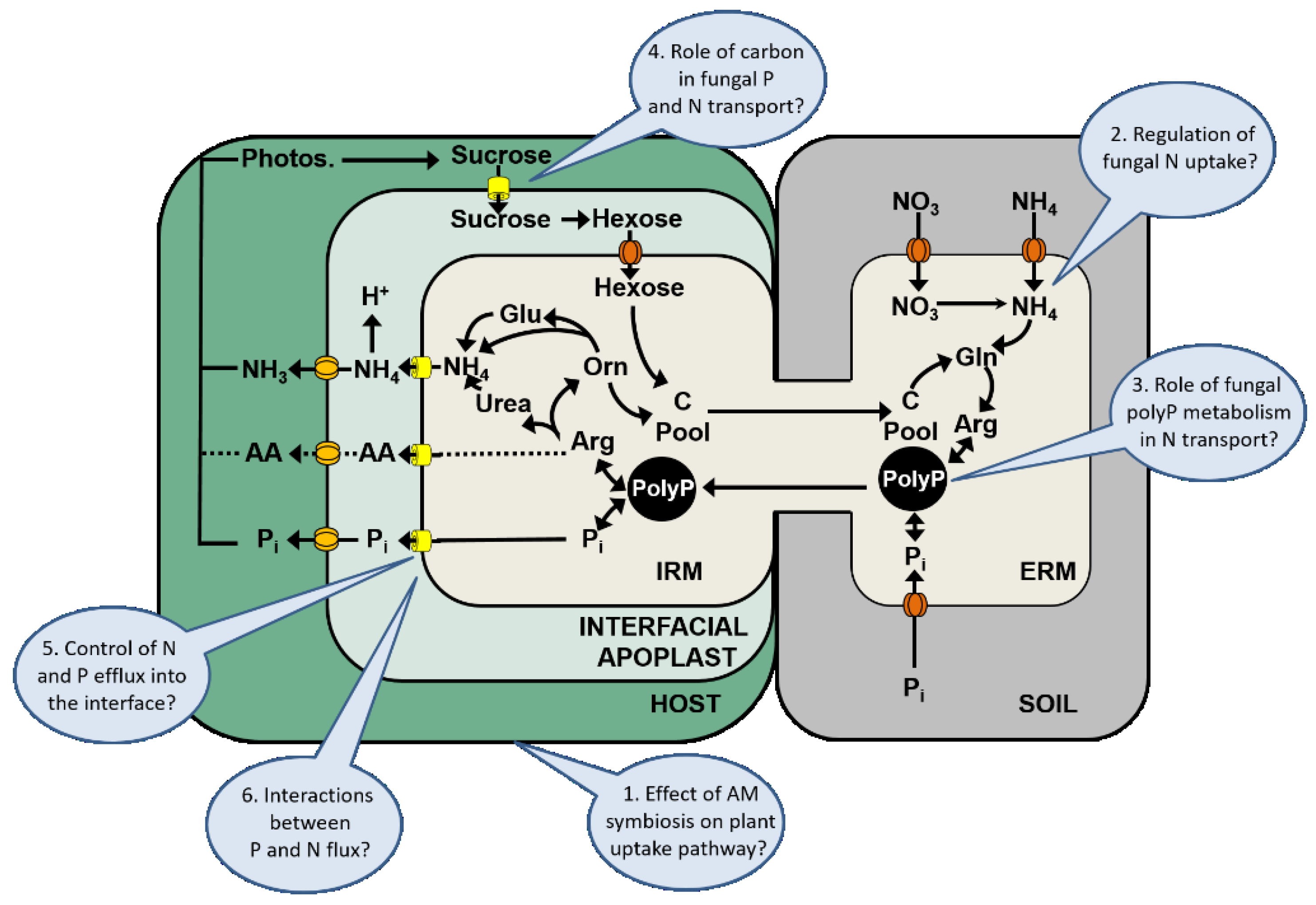
In recent years, significant progress has been made in the elucidation of the N transport pathway and the identification of fungal or plant genes that are involved in the N transport from the ERM via the IRM to the host. In this review, we summarized our current understanding of N transport and its regulation in the AM symbiosis that is primarily based on 13C and 15N labeling studies and more recent data on fungal and plant gene expression. However, despite the progress that has been made, many critical questions are still unanswered and should be addressed in future studies. Some of these major research gaps are also indicated in Figure 3, and are listed below.
5.1. Effect of the Arbuscular Mycorrhizal Symbiosis on the Plant Uptake Pathway
There is increasing evidence that the AM symbiosis changes the nutrient uptake strategy of the plant, and that P transporters of the plant uptake pathway are downregulated in response to the AM symbiosis [31]. It has been shown that even in non-responsive plants, the uptake via the mycorrhizal pathway can dominate the overall P uptake by the plant [29]. Whether and how the plant uptake pathway for N is affected by the colonization with AM fungi is currently unknown, but there is evidence that some plant N transporters are downregulated in colonized roots [35,36].
5.2. Regulation of Fungal N Uptake
Fungal N uptake systems have a high affinity for the uptake of N from the soil [42], but how the expression of these transporters is regulated is largely unknown. The transcript levels of some of these transporters are substrate inducible, and regulated by the NH4+ supply and/or fungal NH4+ status [73]. However, our current understanding of the N transport pathways in the AM symbiosis, is still limited by the fact that these transport pathways were only studied in a small number of fungal species (see also Table 1), and by the lack of molecular tools in AM research. There is evidence that there is a high intraspecific diversity in the efficiency with which fungi are able to improve the N nutrition of their host [46]. So far, the transcriptome and genome of only one AM fungal species is known [72,76], and attempts to develop fungal mutants for research studies were unsuccessful.
5.3. Role of Fungal PolyP Metabolism in N Transport
PolyP play a significant role for the P storage in fungal hyphae, for the long distance transport of P and other nutrients from the ERM to the IRM, and in the regulation of the nutrient transport to the host [10,126,129]. There are indications that the polyP metabolism may play a role in the efficiency with which AM fungi are able to provide nutritional benefits to their host [46]. The processes that are involved in fungal Pi homeostasis and polyP biosynthesis or remobilization have so far mainly been studied in yeasts (e.g., [136]), but the regulatory mechanisms that control the rates of polyP turnover in mycorrhizal fungal hyphae are largely unknown.
5.4. Role of Carbon in Fungal P and N Transport
Some studies have demonstrated that the carbon supply of the host acts as an important trigger for P and N transport in the AM symbiosis [10,12,75]. However, there are also other reports in which the carbon to nutrient resource exchange was not directly correlated [13]. Currently, we have only limited information how the carbon to nutrient exchange across the mycorrhizal interface is regulated [47]. It has been shown that the compatibility between host plants and AM fungi plays a significant role for the nutritional benefits in the AM symbiosis [25], but it is currently unknown why some AM fungi are only beneficial to certain host plant species, despite their ability to colonize a variety of host plants.
5.5. Control of Fungal N and P Efflux into the Interface
The processes by which nutrients are released into the mycorrhizal interface, or the mechanisms that control this efflux of nutrients through the fungal plasma membrane, are currently unknown. However, a better knowledge of these processes is key to understand the variability or context-dependency of nutritional benefits or mycorrhizal growth responses that have been described for the AM symbiosis.
5.6. Interactions between P and N Flux
There is a growing body of evidence that suggests that there are interactions between the P and N transport in the AM symbiosis [121] and that the maintenance of the AM symbiosis is more determined by the sum of nutritional benefits that are provided via the mycorrhizal interface [54,56,57,137]. It has been speculated that the transport of P or NH4+ through the respective mycorrhiza-inducible transporters in the periarbuscular membrane is not only important for the uptake of these nutrients from the mycorrhizal interface but also initiates signaling that controls the arbuscular life span [41]. Currently, the processes by which plants control the symbiosis in order to maximize their nutritional gains or control their C costs are largely unknown. These considerations are also complicated by the fact that, in addition to P and N, other nutrients are also transferred from the fungus to the host and that the symbiosis also provides a variety of non-nutritional benefits to host plants [3]. However, the transport of other nutrients in the AM symbiosis is under-researched and it is currently unknown how other nutrient fluxes or non-nutritional benefits contribute to the C costs of the symbiosis for the host plant.
Acknowledgments
We thank the National Science Foundation (IOS award 1051397) and the South Dakota Soybean Research and Promotion Council for financial support.
Author Contributions
The manuscript was primarily written by Heike Bücking with contributions by Arjun Kafle.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Wang, B.; Qiu, Y.L. Phylogenetic distribution and evolution of mycorrhizae in land plants. Mycorrhiza 2006, 16, 299–363. [Google Scholar] [CrossRef] [PubMed]
- Ryan, M.H.; Graham, J.H. Is there a role for arbuscular mycorrhizal fungi in production agriculture? Plant Soil 2002, 244, 263–271. [Google Scholar] [CrossRef]
- Smith, S.E.; Read, D.J. Mycorrhizal Symbiosis, 3rd ed.; Academic Press: New York, NY, USA, 2008. [Google Scholar]
- Cameron, D.D. Arbuscular mycorrhizal fungi as (agro)ecosystem engineers. Plant Soil 2010, 333, 1–5. [Google Scholar] [CrossRef]
- Wright, D.P.; Read, D.J.; Scholes, J.D. Mycorrhizal sink strength influences whole plant carbon balance of Trifolium repens L. Plant Cell Environ. 1998, 21, 881–891. [Google Scholar] [CrossRef]
- Smith, F.A.; Smith, S.E. How useful is the mutualism-parasitism continuum of arbuscular mycorrhizal functioning? Plant Soil 2013, 363, 7–18. [Google Scholar] [CrossRef]
- Johnson, N.C.; Graham, J.H. The continuum concept remains a useful framework for studying mycorrhizal functioning. Plant Soil 2013, 363, 411–419. [Google Scholar] [CrossRef]
- Johnson, N.C.; Graham, J.H.; Smith, F.A. Functioning of mycorrhizal associations along the mutualism-parasitism continuum. New Phytol. 1997, 135, 575–585. [Google Scholar] [CrossRef]
- Smith, S.E.; Smith, F.A. Fresh perspectives on the roles of arbuscular mycorrhizal fungi in plant nutrition and growth. Mycologia 2012, 104, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Kiers, E.T.; Duhamel, M.; Beesetty, Y.; Mensah, J.A.; Franken, O.; Verbruggen, E.; Fellbaum, C.R.; Kowalchuk, G.A.; Hart, M.M.; Bago, A.; et al. Reciprocal rewards stabilize cooperation in the mycorrhizal symbiosis. Science 2011, 333, 880–882. [Google Scholar] [CrossRef] [PubMed]
- Bücking, H.; Mensah, J.A.; Fellbaum, C.R. Common mycorrhizal networks and their effect on the bargaining power of the fungal partner in the arbuscular mycorrhizal symbiosis. Comp. Integr. Biol. 2015, in press. [Google Scholar]
- Fellbaum, C.R.; Mensah, J.A.; Cloos, A.J.; Strahan, G.D.; Pfeffer, P.E.; Kiers, E.T.; Bücking, H. Fungal nutrient allocation in common mycelia networks is regulated by the carbon source strength of individual host plants. New Phytol. 2014, 203, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Walder, F.; Niemann, H.; Natarajan, M.; Lehmann, M.F.; Boller, T.; Wiemken, A. Mycorrhizal networks: Common goods of plants shared under unequal terms of trade. Plant Physiol. 2012, 159, 789–797. [Google Scholar] [CrossRef] [PubMed]
- Babikova, Z.; Gilbert, L.; Bruce, T.J.A.; Birkett, M.; Caulfield, J.C.; Woodcock, C.; Pickett, J.A.; Johnson, D. Underground signals carried through common mycelial networks warn neighbouring plants of aphid attack. Ecol. Lett. 2013, 16, 835–843. [Google Scholar] [CrossRef] [PubMed]
- Teste, F.P.; Veneklaas, E.J.; Dixon, K.W.; Lambers, H. Is nitrogen transfer among plants enhanced by contrasting nutrient-acquisition strategies? Plant Cell Environ. 2015, 38, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Gorzelak, M.A.; Asay, A.K.; Pickles, B.J.; Simard, S.W. Inter-plant communication through mycorrhizal networks mediates complex adaptive behaviour in plant communities. AOB Plants 2015. [Google Scholar] [CrossRef] [PubMed]
- Barto, E.K.; Hilker, M.; Muller, F.; Mohney, B.K.; Weidenhamer, J.D.; Rillig, M.C. The fungal fast lane: Common mycorrhizal networks extend bioactive zones of allelochemicals in soils. PLoS ONE 2011, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Weremijewicz, J.; Janos, D.P. Common mycorrhizal networks amplify size inequality in Andropogon gerardii monocultures. New Phytol. 2013, 198, 203–213. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Xu, M.; Qiu, G.Y.; Zhou, J. Use of 15N stable isotope to quantify nitrogen transfer between mycorrhizal plants. J. Plant Ecol. 2009, 2, 107–118. [Google Scholar] [CrossRef]
- Babikova, Z.; Johnson, D.; Bruce, T.J.A.; Pickett, J.; Gilbert, L. Underground allies: How and why do mycelial networks help plants defend themselves? Bioassays 2013, 36, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Jakobsen, I.; Grønlund, M.; Smith, F.A. Roles of arbuscular mycorrhizas in plant phosphorus nutrition: Interactions between pathways of phosphorus uptake in arbuscular mycorrhizal roots have important implications for understanding and manipulating plant phosphorus acquisition. Plant Physiol. 2011, 156, 1050–1057. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, H.L.; Hartley, A.E.; Vogelsang, K.M.; Bever, J.D.; Schultz, P.A. Arbuscular mycorrhizal fungi do not enhance nitrogen acquisition and growth of old-field perennials under low nitrogen supply in glasshouse culture. New Phytol. 2005, 167, 869–880. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A. Roles of arbuscular mycorrhizas in plant nutrition and growth: New paradigms from cellular to ecosystem scales. Annu. Rev. Plant Biol. 2011, 62, 227–250. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A.; Jakobsen, I. Mycorrhizal fungi can dominate phosphate supply to plants irrespective of growth responses. Plant Physiol. 2003, 133, 16–20. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.E.; Smith, F.A.; Jakobsen, I. Functional diversity in arbuscular mycorrhizal (AM) symbioses: The contribution of the mycorrhizal P uptake pathway is not correlated with mycorrhizal responses in growth or total P uptake. New Phytol. 2004, 162, 511–524. [Google Scholar] [CrossRef]
- Bücking, H.; Kuhn, A.J.; Schröder, W.H.; Heyser, W. The fungal sheath of ectomycorrhizal pine roots: An apoplastic barrier for the entry of calcium, magnesium, and potassium into the root cortex? J. Exp. Bot. 2002, 53, 1659–1669. [Google Scholar] [CrossRef] [PubMed]
- Behrmann, P.; Heyser, W. Apoplastic transport through the fungal sheath of Pinus sylvestris/Suillus bovinus ectomycorrhizae. Bot. Acta 1992, 105, 427–434. [Google Scholar] [CrossRef]
- Smith, S.E.; Facelli, E.; Pope, S.; Smith, F.A. Plant performance in stressful environments: Interpreting new and established knowledge of the roles of arbuscular mycorrhizas. Plant Soil 2010, 326, 3–20. [Google Scholar] [CrossRef]
- Li, H.Y.; Smith, S.E.; Holloway, R.E.; Zhu, Y.G.; Smith, F.A. Arbuscular mycorrhizal fungi contribute to phosphorus uptake by wheat grown in a phosphorus-fixing soil even in the absence of positive growth responses. New Phytol. 2006, 172, 536–543. [Google Scholar] [CrossRef] [PubMed]
- Chiou, T.J.; Liu, H.; Harrison, M.J. The spatial expression patterns of a phosphate transporter (MtPt1) from Medicago truncatula indicate a role in phosphate transport at the root/soil interface. Plant J. 2001, 25, 281–293. [Google Scholar] [CrossRef] [PubMed]
- Grunwald, U.; Guo, W.B.; Fischer, K.; Isayenkov, S.; Ludwig-Müller, J.; Hause, B.; Yan, X.L.; Küster, H.; Franken, P. Overlapping expression patterns and differential transcript levels of phosphate transporter genes in arbuscular mycorrhizal, Pi-fertilised and phytohormone-treated Medicago truncatula roots. Planta 2009, 229, 1023–1034. [Google Scholar] [CrossRef] [PubMed]
- Harrison, M.J.; Dewbre, G.R.; Liu, J. A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 2002, 14, 2413–2429. [Google Scholar] [CrossRef] [PubMed]
- Paszkowski, U.; Kroken, U.; Roux, C.; Briggs, S.P. Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2002, 99, 13324–13329. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.H.; Chague, V.; Melamed-Bessudo, C.; Kapulnik, Y.; Jain, A.; Raghothama, K.G.; Levy, A.A.; Silber, A. Functional characterization of LePt4: A phosphate transporter in tomato with mycorrhiza-enhanced expression. J. Exp. Bot. 2007, 58, 2491–2501. [Google Scholar] [CrossRef] [PubMed]
- Burleigh, S.H. Relative quantitative rt-pcr to study the expression of plant nutrient transporters in arbuscular mycorrhizas. Plant Sci. 2001, 160, 899–904. [Google Scholar] [CrossRef]
- Kobae, Y.; Tamura, Y.; Takai, S.; Banba, M.; Hata, S. Localized expression of arbuscular mycorrhiza-inducible ammonium transporters in soybean. Plant Cell Physiol. 2010, 51, 1411–1415. [Google Scholar] [CrossRef] [PubMed]
- Koegel, S.; Lahmidi, N.A.; Arnould, C.; Chatagnier, O.; Walder, F.; Ineichen, K.; Boller, T.; Wipf, D.; Wiemken, A.; Courty, P.E. The family of ammonium transporters (AMT) in Sorghum bicolor: Two AMT members are induced locally, but not systemically in roots colonized by arbuscular mycorrhizal fungi. New Phytol. 2013, 198, 853–865. [Google Scholar] [CrossRef] [PubMed]
- Guether, M.; Neuhauser, B.; Balestrini, R.; Dynowski, M.; Ludewig, U.; Bonfante, P. A mycorrhizal-specific ammonium transporter from Lotus japonicus acquires nitrogen released by arbuscular mycorrhizal fungi. Plant Physiol. 2009, 150, 73–83. [Google Scholar] [CrossRef] [PubMed]
- Gomez, S.K.; Javot, H.; Deewatthanawong, P.; Torres-Jerez, I.; Tang, Y.; Blancaflor, E.B.; Udvardi, M.K.; Harrison, M.J. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009, 9, 10. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Penmetsa, R.V.; Terzaghi, N.; Cook, D.R.; Harrison, M.J. A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2007, 104, 1720–1725. [Google Scholar] [CrossRef] [PubMed]
- Breuillin-Sessoms, F.; Floss, D.S.; Gomez, S.K.; Pumplin, N.; Ding, Y.; Levesque-Tremblay, V.; Noar, R.D.; Daniels, D.A.; Bravo, A.; Eaglesham, J.B.; et al. Suppression of arbuscule degeneration in Medicago truncatula phosphate transporter 4 mutants is dependent on the ammonium transporter 2 family protein AMT2;3. Plant Cell 2015. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tienda, J.; Valderas, A.; Camañes, G.; García-Agustín, P.; Ferrol, N. Kinetics of NH4+ uptake by the arbuscular mycorrhizal fungus Rhizophagus irregularis. Mycorrhiza 2012, 22, 485–491. [Google Scholar] [CrossRef] [PubMed]
- George, E.; Marschner, H.; Jakobsen, I. Role of arbuscular mycorrhizal fungi in uptake of phosphorus and nitrogen from soil. Crit. Rev. Biotechnol. 1995, 15, 257–270. [Google Scholar] [CrossRef]
- Hawkins, H.J.; George, E. Effect of plant nitrogen status on the contribution of arbuscular mycorrhizal hyphae to plant nitrogen uptake. Physiol. Plant. 1999, 105, 694–700. [Google Scholar] [CrossRef]
- Saia, S.; Benitéz, E.; Garcia-Garrido, J.M.; Settanni, L.; Amato, G.; Giambalvo, D. The effect of arbuscular mycorrhizal fungi on total plant nitrogen uptake and nitrogen recovery from soil organic material. J. Agric. Sci. 2014, 152, 370–378. [Google Scholar] [CrossRef]
- Mensah, J.A.; Koch, A.M.; Antunes, P.M.; Hart, M.M.; Kiers, E.T.; Bücking, H. High functional diversity within arbuscular mycorrhizal fungal species is associated with differences in phosphate and nitrogen uptake and fungal phosphate metabolism. Mycorrhiza 2015, 25, 533–546. [Google Scholar] [CrossRef] [PubMed]
- Corrêa, A.; Cruz, C.; Ferrol, N. Nitrogen and carbon/nitrogen dynamics in arbuscular mycorrhiza: The great unknown. Mycorrhiza 2015, 25, 499–515. [Google Scholar] [CrossRef] [PubMed]
- Azcon-Aguilar, C.; Alba, C.; Montilla, M.; Barea, J.M. Isotopic (15N) evidence of the use of less available N forms by VA mycorrhizas. Symbiosis 1993, 15, 39–48. [Google Scholar]
- Toussaint, J.P.; St-Arnaud, M.; Charest, C. Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and RI t-DNA roots of Daucus carota l. In an vitro compartmented system. Can. J. Microbiol. 2004, 50, 251–260. [Google Scholar] [PubMed]
- Jin, H.; Pfeffer, P.E.; Douds, D.D.; Piotrowski, E.; Lammers, P.J.; Shachar-Hill, Y. The uptake, metabolism, transport and transfer of nitrogen in an arbuscular mycorrhizal symbiosis. New Phytol. 2005, 168, 687–696. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, Y.; Yano, K. Nitrogen delivery to maize via mycorrhizal hyphae depends on the form of N supplied. Plant Cell Environ. 2005, 28, 1247–1254. [Google Scholar] [CrossRef]
- Hildebrandt, U.; Schmelzer, E.; Bothe, H. Expression of nitrate transporter genes in tomato colonized by an arbuscular mycorrhizal fungus. Physiol. Plant. 2002, 115, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tienda, J.; Testillano, P.S.; Balestrini, R.; Fiorilli, V.; Azcón-Aguilar, C.; Ferrol, N. GintAmt2, a new member of the ammonium transporter family in the arbuscular mycorrhizal fungus Glomus intraradices. Fungal Genet. Biol. 2011, 48, 1044–1055. [Google Scholar] [CrossRef] [PubMed]
- Bonneau, L.; Huguet, S.; Wipf, D.; Pauly, N.; Truong, H.N. Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol. 2013, 199, 188–202. [Google Scholar] [CrossRef] [PubMed]
- Besserer, A.; Puech-Pagès, V.; Kiefer, P.; Gomez-Roldan, V.; Jauneau, A.; Roy, S.; Portais, J.C.; Roux, C.; Bécard, G.; Séjalon-Delmas, N. Strigolactones stimulate arbuscular mycorrhizal fungi by activating mitochondria. PLoS Biol. 2006, 4, 1239–1247. [Google Scholar] [CrossRef] [PubMed]
- Nouri, E.; Breuillin-Sessoms, F.; Feller, U.; Reinhardt, D. Phosphorus and nitrogen regulate arbuscular mycorrhizal symbiosis in Petunia hybrida. PLoS ONE 2014, 9, e90841. [Google Scholar] [CrossRef] [PubMed]
- Javot, H.; Penmetsa, R.V.; Breuillin, F.; Bhattarai, K.K.; Noar, R.D.; Gomez, S.K.; Zhang, Q.; Cook, D.R.; Harrison, M.J. Medicago truncatula MtPt4 mutants reveal a role for nitrogen in the regulation of arbuscule degeneration in arbuscular mycorrhizal symbiosis. Plant J. 2011, 68, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Ames, R.N.; Reid, C.P.P.; Porter, L.K.; Cambardella, C. Hyphal uptake and transport of nitrogen from two 15N-labelled sources by Glomus mosseae, a vesicular-arbuscular mycorrhizal fungus. New Phytol. 1983, 95, 381–396. [Google Scholar] [CrossRef]
- Barea, J.M.; Azcón-Aguilar, C.; Azcón, R. Vesicular-arbuscular mycorrhiza improve both symbiotic N2 fixation and N uptake from soil as assessed with a 15N technique under field conditions. New Phytol. 1987, 106, 717–725. [Google Scholar] [CrossRef]
- Frey, B.; Schüepp, H. Acquisition of nitrogen by external hyphae of arbuscular mycorrhizal fungi associated with Zea mays L. New Phytol. 1993, 124, 221–230. [Google Scholar] [CrossRef]
- Johansen, A.; Jakobsen, I.; Jensen, E.S. Hyphal transport by a vesicular-arbuscular mycorrhizal fungus of N applied to the soil as ammonium or nitrate. Biol. Fertil. Soils 1993, 16, 66–70. [Google Scholar] [CrossRef]
- Hawkins, H.J.; Johansen, A.; George, E. Uptake and transport of organic and inorganic nitrogen by arbuscular mycorrhizal fungi. Plant Soil 2000, 226, 275–285. [Google Scholar] [CrossRef]
- Tobar, R.; Azcón, R.; Barea, J.M. Improved nitrogen uptake and transport from 15N-labelled nitrate by external hyphae of arbuscular mycorrhiza under water-stressed conditions. New Phytol. 1994, 126, 119–122. [Google Scholar] [CrossRef]
- Johansen, A.; Finlay, R.D.; Olsson, P.A. Nitrogen metabolism of external hyphae of the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 1996, 133, 705–712. [Google Scholar] [CrossRef]
- Bago, B.; Vierheilig, H.; Piché‚, Y.; Azcón-Aguilar, C. Nitrate depletion and pH changes induced by the extraradical mycelium of the arbuscular mycorrhizal fungus Glomus intraradices grown in monoxenic cultures. New Phytol. 1996, 133, 273–280. [Google Scholar] [CrossRef]
- Bago, B.; Azcón-Aguilar, C. Changes in the rhizospheric pH induced by arbuscular mycorrhiza formation in onion (Allium cepa L.). Z. Pflanzenernähr. Bodenkd. 1997, 160, 333–339. [Google Scholar] [CrossRef]
- Howitt, S.M.; Udvardi, M.K. Structure, function and regulation of ammonium transporters in plants. Biochim. Biophys. Acta 2000, 1465, 152–170. [Google Scholar] [CrossRef]
- D’Apuzzo, E.; Rogato, A.; Simon-Rosin, U.; El Alaoui, H.; Barbulova, A.; Betti, M.; Dimou, M.; Katinakis, P.; Marquez, A.; Marini, A.M.; et al. Characterization of three functional high-affinity ammonium transporters in Lotus japonicus with differential transcriptional regulation and spatial expression. Plant Physiol. 2004, 134, 1763–1774. [Google Scholar] [CrossRef] [PubMed]
- Schüßler, A.; Walker, C. The Glomeromycota. A species list with new families and new genera; Libraries at the Royal Botanic Garden Edinburgh, The Royal Botanic Garden Kew, Botanische Staatssammlung Munich, and Oregon State University: Gloucester, UK, 2010. [Google Scholar]
- Hawkins, H.J.; George, E. Reduced 15N-nitrogen transport through arbuscular mycorrhizal hyphae to Triticum aestivum L. supplied with ammonium vs. nitrate nutrition. Ann. Bot. 2001, 87, 303–311. [Google Scholar] [CrossRef]
- Gachomo, E.; Allen, J.W.; Pfeffer, P.E.; Govindarajulu, M.; Douds, D.D.; Jin, H.R.; Nagahashi, G.; Lammers, P.J.; Shachar-Hill, Y.; Bücking, H. Germinating spores of Glomus intraradices can use internal and exogenous nitrogen sources for de novo biosynthesis of amino acids. New Phytol. 2009, 184, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, E.; Kohler, A.; Dozolme-Seddas, P.; Balestrini, R.; Benabdellah, K.; Colard, A.; Croll, D.; da Silva, C.; Gomez, S.K.; Koul, R.; et al. The transcriptome of the arbuscular mycorrhizal fungus Glomus intraradices (DAOM 197198) reveals functional tradeoffs in an obligate symbiont. New Phytol. 2012, 193, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Pedrosa, A.; González-Guerrero, M.; Valderas, A.; Azcón-Aguilar, C.; Ferrol, N. GintAmt1 encodes a functional high-affinity ammonium transporter that is expressed in the extraradical mycelium of Glomus intraradices. Fungal Genet. Biol. 2006, 43, 102–110. [Google Scholar] [CrossRef] [PubMed]
- Tian, C.; Kasiborski, B.; Koul, R.; Lammers, P.J.; Bücking, H.; Shachar-Hill, Y. Regulation of the nitrogen transfer pathway in the arbuscular mycorrhizal symbiosis: Gene characterization and the coordination of expression with nitrogen flux. Plant Physiol. 2010, 153, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Fellbaum, C.R.; Gachomo, E.W.; Beesetty, Y.; Choudhari, S.; Strahan, G.D.; Pfeffer, P.E.; Kiers, E.T.; Bücking, H. Carbon availability triggers fungal nitrogen uptake and transport in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. USA 2012, 109, 2666–2671. [Google Scholar] [CrossRef] [PubMed]
- Tisserant, E.; Malbreil, M.; Kuo, A.; Kohler, A.; Symeonidi, A.; Balestrini, R.; Charron, P.; Duensing, N.; Frey, N.F.D.; Gianinazzi-Pearson, V.; et al. Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proc. Natl. Acad. Sci. USA 2014, 111, 563–563. [Google Scholar] [CrossRef] [PubMed]
- Hodge, A.; Robinson, D.; Fitter, A.H. An arbuscular mycorrhizal inoculum enhances root proliferation in, but not nitrogen capture from, nutrient-rich patches in soil. New Phytol. 2000, 145, 575–584. [Google Scholar] [CrossRef]
- Cappellazzo, G.; Lanfranco, L.; Fitz, M.; Wipf, D.; Bonfante, P. Characterization of an amino acid permease from the endomycorrhizal fungus Glomus mosseae. Plant Physiol. 2008, 147, 429–437. [Google Scholar] [CrossRef] [PubMed]
- Belmondo, S.; Fiorilli, V.; Pérez-Tienda, J.; Ferrol, N.; Marmeisse, R.; Lanfranco, L. A dipeptide transporter from the arbuscular mycorrhizal fungus Rhizophagus irregularis is upregulated in the intraradical phase. Front. Plant Sci. 2014, 5, 436. [Google Scholar] [CrossRef] [PubMed]
- Allen, J.W.; Shachar-Hill, Y. Sulfur transfer through an arbuscular mycorrhiza. Plant Physiol. 2009, 149, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Breuninger, M.; Trujillo, C.G.; Serrano, E.; Fischer, R.; Requena, N. Different nitrogen sources modulate activity but not expression of glutamine synthetase in arbuscular mycorrhizal fungi. Fungal Genet. Biol. 2004, 41, 542–552. [Google Scholar] [CrossRef] [PubMed]
- Nakano, A.; Takahashi, K.; Koide, R.T.; Kimura, M. Determination of the nitrogen source for arbuscular mycorrhizal fungi by 15N application to soil and plants. Mycorrhiza 2001, 10, 267–273. [Google Scholar] [CrossRef]
- Cliquet, J.B.; Stewart, G.R. Ammonia assimilation in Zea mays L. infected with a vesicular-arbuscular mycorrhizal fungus Glomus fasciculatum. Plant Physiol. 1993, 101, 865–871. [Google Scholar] [PubMed]
- Subramanian, K.S.; Charest, C. Arbuscular mycorrhizae and nitrogen assimilation in maize after drought and recovery. Physiol. Plant. 1998, 102, 285–296. [Google Scholar] [CrossRef]
- Kaldorf, M.; Zimmer, W.; Bothe, H. Genetic evidence for the occurence of assimilatory nitrate reductase in arbuscular mycorrhizal and other fungi. Mycorrhiza 1994, 5, 23–28. [Google Scholar] [CrossRef]
- Ho, I.; Trappe, J.M. Nitrate reducing capacity of two vesicular-arbuscular mycorrhizal fungi. Mycologia 1975, 67, 886–888. [Google Scholar] [CrossRef] [PubMed]
- Oliver, A.J.; Smith, S.E.; Nicholas, D.J.D.; Wallace, W. Activity of nitrate reductase in Trifolium subterraneum: Effects of mycorrhizal infection and phosphate nutrition. New Phytol. 1983, 94, 63–79. [Google Scholar] [CrossRef]
- Vázquez, M.; Barea, J.; Azcón, R. Impact of soil nitrogen concentration on Glomus spp.-Sinorhizobium interactions as affecting growth, nitrate reductase activity and protein content of Medicago sativa. Biol. Fertil. Soils 2001, 34, 57–63. [Google Scholar]
- Hawkins, H.J.; Cramer, M.D.; George, E. Root respiratory quotient and nitrate uptake in hydroponically grown non-mycorrhizal and mycorrhizal wheat. Mycorrhiza 1999, 9, 57–60. [Google Scholar] [CrossRef]
- Kaldorf, M.; Schmelzer, E.; Bothe, H. Expression of maize and fungal nitrate reductase genes in arbuscular mycorrhiza. MPMI 1998, 11, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Nemec, S. Histochemical characterization of Glomus etunicatus infection of Citrus limon fibrous roots. Can. J. Bot. 1981, 59, 609–617. [Google Scholar]
- MacDonald, R.M.; Lewis, M. The occurence of some acid phosphatases and dehydrogenases in the vesicular-arbuscular mycorrhizal fungus Glomus mosseae. New Phytol. 1978, 80, 135–141. [Google Scholar] [CrossRef]
- Smith, S.E.; St John, B.J.; Smith, F.A.; Nicholas, D.J.D. Activity of glutamine synthetase and glutamate dehydrogenase in Trifolium subterraneum L. and Allium cepa L.: Effects of mycorrhizal infection and phosphate nutrition. New Phytol. 1985, 99, 211–227. [Google Scholar] [CrossRef]
- Govindarajulu, M.; Pfeffer, P.E.; Jin, H.R.; Abubaker, J.; Douds, D.D.; Allen, J.W.; Bücking, H.; Lammers, P.J.; Shachar-Hill, Y. Nitrogen transfer in the arbuscular mycorrhizal symbiosis. Nature 2005, 435, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Faure, S.; Cliquet, J.B.; Thephany, G.; Boucaud, J. Nitrogen assimilation in Lolium perenne colonized by the arbuscular mycorrhizal fungus Glomus fasciculatum. New Phytol. 1998, 138, 411–417. [Google Scholar] [CrossRef]
- Valentine, A.J.; Osborne, B.A.; Mitchell, D.T. Form of inorganic nitrogen influences mycorrhizal colonisation and photosynthesis of cucumber. Sci. Hortic. 2002, 92, 229–239. [Google Scholar] [CrossRef]
- Liu, Y.; Lai, N.; Gao, K.; Chen, F.; Yuan, L.; Mi, G. Ammonium inhibits primary root growth by reducing the length of meristem and elongation zone and decreasing elemental expansion rate in the root apex in Arabidopsis thaliana. PLoS ONE 2013, 8, e61031. [Google Scholar] [CrossRef] [PubMed]
- Cramer, M.D.; Lewis, O.A.M. The influence of NO3− and NH4+ nutrition on the carbon and nitrogen partitioning characteristics of wheat (Triticum aestivum) and maize (Zea mays) plants. Plant Soil 1993, 154, 289–300. [Google Scholar] [CrossRef]
- Cramer, M.D.; Lewis, O.A.M.; Lips, S.H. Inorganic carbon fixation and metabolism in maize roots as affected by nitrate and ammonium nutrition. Physiol. Plant. 1993, 89, 632–639. [Google Scholar] [CrossRef]
- Cooper, T.G.; Sumrada, R.A. What is the function of nitrogen catabolite repression in Saccharomyces cerevisiae? J. Bacteriol. 1983, 155, 623–627. [Google Scholar] [PubMed]
- Javelle, A.; Morel, M.; Rodriguez-Pastrana, B.R.; Botton, B.; Andr‚, B.; Marini, A.M.; Brun, A.; Chalot, M. Molecular characterization, function and regulation of ammonium transporters (Amt) and ammonium-metabolizing enzymes (GS, NADP-GDH) in the ectomycorrhizal fungus Hebeloma cylindrosporum. Mol. Microbiol. 2003, 47, 411–430. [Google Scholar] [CrossRef] [PubMed]
- Navarro, F.J.; Machín, F.; Martín, Y.; Siverio, J.M. Down-regulation of eukaryotic nitrate transporter by nitrogen-dependent ubiquitinylation. J. Biol. Chem. 2006, 281, 13268–13274. [Google Scholar] [CrossRef] [PubMed]
- Schulten, H.-R.; Schnitzer, M. The chemistry of soil organic nitrogen: A review. Biol. Fertil. Soils 1998, 26, 1–15. [Google Scholar] [CrossRef]
- Leigh, J.; Hodge, A.; Fitter, A.H. Arbuscular mycorrhizal fungi can transfer substantial amounts of nitrogen to their host plant from organic material. New Phytol. 2009, 181, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Hodge, A.; Fitter, A.H. Substantial nitrogen acquisition by arbuscular mycorrhizal fungi from organic material has implications for N cycling. Proc. Natl. Acad. Sci. USA 2010, 107, 13754–13759. [Google Scholar] [CrossRef] [PubMed]
- Thirkell, J.D.; Cameron, D.D.; Hodge, A. Resolving the “nitrogen paradox” of arbuscular mycorrhizas: Fertilization with organic matter brings considerable benefits for plant nutrition and growth. Plant Cell Environ. 2015. [Google Scholar] [CrossRef] [PubMed]
- Atul-Nayyar, A.; Hamel, C.; Hanson, K.; Germida, J. The arbuscular mycorrhizal symbiosis links N mineralization to plant demand. Mycorrhiza 2009, 19, 239–246. [Google Scholar] [CrossRef] [PubMed]
- Herman, D.J.; Firestone, M.K.; Nuccio, E.; Hodge, A. Interactions between an arbuscular mycorrhizal fungus and a soil microbial community mediating litter decomposition. FEMS Microbiol. Ecol. 2012, 80, 236–247. [Google Scholar] [CrossRef] [PubMed]
- Finzi, A.C.; Abramoff, R.Z.; Spiller, K.S.; Brzostek, E.R.; Darby, B.A.; Kramer, M.A.; Phillips, R.P. Rhizosphere processes are quantitatively important components of terrestrial carbon and nutrient cycles. Glob. Chang. Biol. 2015, 21, 2082–2094. [Google Scholar] [CrossRef] [PubMed]
- Nuccio, E.E.; Hodge, A.; Pett-Ridge, J.; Herman, D.J.; Weber, P.K.; Firestone, M.K. An arbuscular mycorrhizal fungus significantly modifies the soil bacterial community and nitrogen cycling during litter decomposition. Environ. Microbiol. 2013, 15, 1870–1881. [Google Scholar] [CrossRef] [PubMed]
- Cliquet, J.B.; Murray, P.J.; Boucaud, J. Effect of the arbuscular mycorrhizal fungus Glomus fasciculatum on the uptake of amino nitrogen by Lolium perenne. New Phytol. 1997, 137, 345–349. [Google Scholar] [CrossRef]
- Whiteside, M.D.; Digman, M.A.; Gratton, E.; Treseder, K.K. Organic nitrogen uptake by arbuscular mycorrhizal fungi in a boreal forest. Soil Biol. Biochem. 2012, 55. [Google Scholar] [CrossRef] [PubMed]
- Whiteside, M.D.; Garcia, M.O.; Treseder, K.K. Amino acid uptake in arbuscular mycorrhizal plants. PLoS ONE 2012, 7, e47643. [Google Scholar] [CrossRef] [PubMed]
- Guescini, M.; Zeppa, S.; Pierleoni, R.; Sisti, D.; Stocchi, L.; Stocchi, V. The expression profile of the Tuber borchii nitrite reductase suggests its positive contribution to host plant nitrogen nutrition. Curr. Genet. 2007, 51, 31–41. [Google Scholar] [CrossRef] [PubMed]
- Bailly, J.; Debaud, J.C.; Verner, M.C.; Plassard, C.; Chalot, M.; Marmeisse, R.; Fraissinet-Tachet, L. How does a symbiotic fungus modulate expression of its host-plant nitrite reductase? New Phytol. 2007, 175, 155–165. [Google Scholar] [CrossRef] [PubMed]
- Temple, S.J.; Vance, C.P.; Gantt, S.J. Glutamate synthase and nitrogen assimilation. Trends Plant Sci. 1998, 3, 51–56. [Google Scholar] [CrossRef]
- Botton, B.; Chalot, M. Nitrogen assimilation: Enzymology in ectomycorrhizas. In Mycorrhiza; Varma, A., Hock, B., Eds.; Springer-Verlag: Berlin, Germany, 1995; pp. 325–363. [Google Scholar]
- Rolin, D.; Pfeffer, P.E.; Douds, D.D.; Farrell, H.M.; Shachar-Hill, Y. Arbuscular mycorrhizal symbiosis and phosphorus nutrition: Effects on amino acid production and turnover in leek. Symbiosis 2001, 30, 1–14. [Google Scholar]
- Forde, B.G. Nitrate transporters in plants: Structure, function and regulation. Biochim. Biophys. Acta 2000, 1465, 219–235. [Google Scholar] [CrossRef]
- Marzluf, G.A. Genetic regulation of nitrogen metabolism in the fungi. Microbiol. Mol. Biol. Rev. 1997, 61, 17–32. [Google Scholar] [PubMed]
- Cruz, C.; Egsgaard, H.; Trujillo, C.; Ambus, P.; Requena, N.; Martins-Loucao, M.A.; Jakobsen, I. Enzymatic evidence for the key role of arginine in nitrogen translocation by arbuscular mycorrhizal fungi. Plant Physiol. 2007, 144, 782–792. [Google Scholar] [CrossRef] [PubMed]
- Cox, G.; Moran, K.J.; Sanders, F.; Nockolds, C.; Tinker, P.B. Translocation and transfer of nutrients in vesicular-arbuscular mycorrhizas. III. Polyphosphate granules and phosphorus translocation. New Phytol. 1980, 84, 649–659. [Google Scholar] [CrossRef]
- Cramer, C.L.; Davis, H.H. Polyphosphate-cation interaction in the amino acid-containing vacuole of Neurospora crassa. J. Biol. Chem. 1984, 259, 5152–5157. [Google Scholar] [PubMed]
- Cramer, C.L.; Vaughn, L.E.; Davis, R.H. Basic amino acids and inorganic polyphosphates in Neurospora crassa: Independent regulation of vacuolar pools. J. Bacteriol. 1980, 142, 945–952. [Google Scholar] [PubMed]
- Westenberg, B.; Boller, T.; Wiemken, A. Lack of arginine- and polyphosphate-storage pools in a vacuole-deficient mutant (end1) of Saccharomyces cerevisiae. FEBS Lett. 1989, 254, 133–136. [Google Scholar] [CrossRef]
- Bücking, H.; Heyser, W. Elemental composition and function of polyphosphates in ectomycorrhizal fungi - an X-ray microanalytical study. Mycol. Res. 1999, 103, 31–39. [Google Scholar] [CrossRef]
- George, E.; Häussler, K.U.; Vetterlein, D.; Gorgus, E.; Marschner, H. Water and nutrient translocation by hyphae of Glomus mosseae. Can. J. Bot. 1992, 70, 2130–2137. [Google Scholar] [CrossRef]
- Bücking, H.; Heyser, W. Uptake and transfer of nutrients in ectomycorrhizal associations: Interactions between photosynthesis and phosphate nutrition. Mycorrhiza 2003, 13, 59–68. [Google Scholar] [CrossRef] [PubMed]
- Bücking, H.; Shachar-Hill, Y. Phosphate uptake, transport and transfer by the arbuscular mycorrhizal fungus Glomus intraradices is stimulated by increased carbohydrate availability. New Phytol. 2005, 165, 899–912. [Google Scholar] [CrossRef] [PubMed]
- Guether, M.; Volpe, V.; Balestrini, R.; Requena, N.; Wipf, D.; Bonfante, P. LjLht1.2—A mycorrhiza-inducible plant amino acid transporter from Lotus japonicus. Biol. Fertil. Soils 2011, 47, 925–936. [Google Scholar] [CrossRef]
- Beever, R.E.; Burns, D.J.W. Phosphorus uptake, storage and utilization by fungi. Adv. Bot. Res. 1980, 8, 127–219. [Google Scholar]
- Li, T.; Hu, Y.J.; Hao, Z.P.; Li, H.; Wang, Y.S.; Chen, B.D. First cloning and characterization of two functional aquaporin genes from an arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2013, 197, 617–630. [Google Scholar] [CrossRef] [PubMed]
- Maurel, C.; Plassard, C. Aquaporins: For more than water at the plant–fungus interface? New Phytol. 2011, 190, 815–817. [Google Scholar] [CrossRef] [PubMed]
- Aroca, R.; Bago, A.; Sutka, M.; Paz, J.A.; Cano, C.; Amodeo, G.; Ruiz-Lozano, J.M. Expression analysis of the first arbuscular mycorrhizal fungi aquaporin described reveals concerted gene expression between salt-stressed and nonstressed mycelium. Mol. Plant Microb. Interact. 2009, 22, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.A.; Huang, W.; Liu, F.C.; Tang, N.W.; Liu, Y.; Lin, H.; Zhao, B. Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPt1 and Pht1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 2013, 198, 836–852. [Google Scholar] [CrossRef] [PubMed]
- Secco, D.; Wang, C.; Shou, H.; Whelan, J. Phosphate homeostasis in the yeast Saccharomyces cerevisiae, the key role of the SPX domain-containing proteins. FEBS Lett. 2012, 586, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Carbonnel, S.; Gutjahr, C. Control of arbuscular mycorrhiza development by nutrient signals. Front. Plant Sci. 2014, 5, 462. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
