Analysis of Genetic Diversity in Polymers of Saccharum spontaneum L. and Their Hybrid Progenies
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Experimental Methods
2.2.1. DNA Extraction
2.2.2. PCR Amplification and Product Detection
2.2.3. Detection by Capillary Electrophoresis
| HIDI | 9.75 µL |
| Salmon 500 internal standard | 0.75 µL |
| Diluted PCR product | 1.00 µL |
2.3. Data Statistics and Analysis
3. Results
3.1. SSR Primer Polymorphism Analysis
3.2. Cluster Analysis
3.2.1. Genetic Distance Analysis
3.2.2. Cluster Analysis Based on SSR Molecular Markers
3.3. Genetic Diversity Analysis
3.4. Genetic Analysis of SSR Loci
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ren, S.; Hu, X.; Yao, L.; Zhao, L.; Qin, W.; Zan, F.; Liu, J. Research Progress on the Genetic Diversity of Saccharum spontaneum L. Sugar Crops China 2023, 45, 8–15. [Google Scholar]
- Yang, C.; Yang, L.; Li, Y. Origins and evolution of sugarcane. J. South. Agric. 2014, 45, 1744–1750. [Google Scholar]
- Da Silva, J.A. The importance of the wild cane Saccharum spontaneum for bioenergy genetic breeding. Sugar Tech 2017, 19, 229–240. [Google Scholar] [CrossRef]
- Deng, H. Pedigree analysis of the native spontaneum-derived varieties of sugarcane in mainland China. Guangdong Agric. Sci. 2012, 39, 167–170. [Google Scholar]
- Jeswiet, J. The Development of Selection and Breeding of the Sugar Cane in Java. Proc. Int. Soc. Sugar Cane Technol. 1930, 3, 44–57. [Google Scholar]
- Fang, J.; Que, Y.; Chen, R. A Review of Saccharum Origin and its Evolutionary Relationship with Related Genera. Chin. J. Trop. Crops 2014, 35, 816–822. [Google Scholar]
- Huang, Y.; Zhang, B.; Zhou, S.; Gao, Y.; Yang, C.; Zhang, G.; Lu, S.; Duan, W. Advances of sugarcane germplasm resources. Sci. Technol. Rev. 2023, 41, 43–49. [Google Scholar]
- Liang, H.; Cai, W.; Zhao, X.; Li, H.; Huang, W.; Shen, W. Study on the Physiological Characteristics of 14 Intergeneric Hybrids of BC3F1 Lines of Saccharum officinarum × Erianthus arundinaceus. Chin. J. Trop. Agric. 2023, 43, 18–27. [Google Scholar]
- Deng, Q.; Dou, Z.; Chen, J.; Wang, K.; Shen, W. Drought Tolerance Evaluation of Intergeneric Hybrids of BC3F1 Lines of Saccharum officinarum × Erianthus arundinaceus. Euphytica 2019, 215, 102. [Google Scholar] [CrossRef]
- Aitken, K.; Li, J.; Wang, L.; Qing, C.; Fan, Y.; Jackson, P. Characterization of Intergeneric Hybrids of Erianthus rockii and Saccharum Using Molecular Markers. Genet. Resour. Crop Evol. 2007, 54, 1395–1405. [Google Scholar] [CrossRef]
- Li, F. Studies on Hybridization Utilizing of Erianthus fulvus Wild Speciesand Its F, Hybrid Appraising for Germplasm Enhancement. Ph.D. Thesis, South China University of Tropical Agriculture, Danzhou, China, 2005. [Google Scholar]
- Jin, D.; Zhang, T.; Lou, H.; He, L.; Li, F.; Yang, Q.; Wang, X. Genetic Variation of BC1F1 Progenies of Sugarcane and Erianthus fulvus Based on Phenotypic and Cytologic Characteristics. Mol. Plant Breed. 2024, 1–25. [Google Scholar]
- Fan, Y.; Chen, H.; Shi, X.; Cai, Q.; Zhang, M.; Zhang, Y. RAPD Analysis of Saccharum spontaneum from Different Ecospecific Colonies in Yunnan. Acta Bot. Yunnanica 2001, 23, 298–308. [Google Scholar]
- Huang, Z.; Zhou, F.; Wang, Q.; Jin, Y.; Fu, C.; Hu, H.; Zhang, C.; Chang, H.; Ji, J.; Wu, Q.; et al. Genetic Diversity Assessment of Saccharum spontanuem L. Native of Domestic and Overseas with Phenotype Agronomic Traits. J. Plant Genet. Resour. 2012, 13, 825–829. [Google Scholar]
- Liu, J.; Bai, C.; Yan, L.; Yang, H.; Zhang, L.; Huang, C. Genetic Diversity in Saccharum spontaneum L. Based on ISSR Markers. Chin. J. Trop. Crops 2014, 35, 68–73. [Google Scholar]
- Qi, Y.; Fan, L.; He, H.; Chen, Y.; Ao, J.; Deng, H. Genetic Diversity Assessment of Saccharum spontaneum L. Native to Guangdong Area with Agronomic Traits. Chin. J. Trop. Crops 2009, 3, 7–10. [Google Scholar]
- Aitken, K.; Li, J.; Piperidis, G.; Qing, C.; Yuanhong, F.; Jackson, P. Worldwide Genetic Diversity of the Wild Species Saccharum spontaneum and Level of Diversity Captured within Sugarcane Breeding Programs. Crop Sci. 2018, 58, 218–229. [Google Scholar] [CrossRef]
- Cai, Q.; Wen, J.; Fan, Y.; Wang, L.; Ma, L. Chromosome analysis of Saccharum L. and related plants. Southwest China J. Agric. Sci. 2002, 15, 16–19. [Google Scholar]
- Li, S.; Wang, X.; Yang, Q. The Ploidy Identification of Saccharum spontaneum Collected from Myanmar Based on 5S rDNA-FISH Localization. Mol. Plant Breed. 2018, 16, 1229–1235. [Google Scholar]
- Wen, J.; Cai, Q.; Fan, Y.; Zhang, M.; Chen, H. Studies on the Chromosome Numbers of Saccharum spontaneum and Related Plants-Sclerostachya, Narenga in China. Sugarcane Canesugar 2001, 3, 12–15. [Google Scholar]
- Wang, S.; Wang, Z.; Guo, C.; Pan, S.; Zeng, D. Studies on the Chromosome of Saccharum spontaneum from Fujian. Sugarcane Canesugar 1996, 5, 9–13. [Google Scholar]
- Dijoux, J.; Rio, S.; Hervouet, C.; Garsmeur, O.; Barau, L.; Dumont, T.; Rott, P.; D’Hont, A.; Hoarau, J.-Y. Unveiling the Predominance of Saccharum spontaneum Alleles for Resistance to Orange Rust in Sugarcane Using Genome-Wide Association. Theoret. Appl. Genet. 2024, 137, 81. [Google Scholar] [CrossRef]
- Zhao, J.; Li, S.; Xu, Y.; Ahmad, N.; Kuang, B.; Feng, M.; Wei, N.; Yang, X. The Subgenome Saccharum spontaneum Contributes to Sugar Accumulation in Sugarcane as Revealed by Full-Length Transcriptomic Analysis. J. Adv. Res. 2023, 54, 1–13. [Google Scholar] [CrossRef]
- Jiang, Q.; Hua, X.; Shi, H.; Liu, J.; Yuan, Y.; Li, Z.; Li, S.; Zhou, M.; Yin, C.; Dou, M.; et al. Transcriptome Dynamics Provides Insights into Divergences of the Photosynthesis Pathway between Saccharum officinarum and Saccharum spontaneum. Plant J. 2023, 113, 1278–1294. [Google Scholar] [CrossRef]
- Zhao, T.; Sun, T.; Wang, J.; Wang, W.; Feng, C.; Liu, X.; Gan, Y.; Zhao, P.; Xu, C.; Lin, J.; et al. Research Progress in Sugarcane Breeding. Sci. Sin. Vitae 2024, 54, 1814–1832. [Google Scholar]
- Wang, L.; Ma, L.; Xia, H.; Lu, X.; Cai, Q.; Fan, Y.; Chen, H.; Liu, X. Application of S. spontaneum in Sugarcane Cross-breeding. Sugar Crops China 2006, 1, 1–4. [Google Scholar]
- Liu, S.; Wang, Q.; Huang, Z.; Zhang, C.; Hu, H.; Fu, C.; Zhou, F. Utilization of Sugarcane Parents of Yacheng Series in Sugarcane Breeding of China. Sugarcane Canesugar 2011, 4, 5–10. [Google Scholar]
- Jing, Y.; Zhu, J.; Tao, L.; Dong, L.; An, R.; Yang, L.; Zhou, Q.; Duan, H. Main Advancement on Breeding Derived from China in Sugarcane Creation Parents in Using S. spontaneum. Mol. Plant Breed. 2011, 9, 1274–1283. [Google Scholar] [CrossRef]
- Liu, J.; Deng, Z.; Wu, C.; Tao, L.; Lu, X.; Zhao, P.; Zhang, Y. Progress and Discussion of Sugarcane Breeding using Saccharum spontaneum L. J. Plant Genet. Resour. 2021, 22, 1491–1497. [Google Scholar]
- Liu, X.; Li, X.; Xu, C.; Lin, X.; Deng, Z. Genetic Diversity of Populations of Saccharum spontaneum with Different Ploidy Levels Using SSR Molecular Markers. Sugar Tech 2015, 18, 365–372. [Google Scholar] [CrossRef]
- Holland, M.; Parson, W. GeneMarker® HID: A Reliable Software Tool for the Analysis of Forensic STR Data. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Zhang, S.; Gao, Q.; Liu, F.; Wang, J.; Wang, X. Genetic Diversity and Population Structure Analysis in a Large Collection of Vicia Amoena in China with Newly Developed SSR Markers. BMC Plant Biol. 2021, 21, 544. [Google Scholar] [CrossRef] [PubMed]
- Kimura, M.; Crow, J. The Number of Alleles That Can Be Maintained in a Finite Population. Genetics 1964, 49, 725–738. [Google Scholar] [CrossRef]
- Lewontin, R. The Apportionment of Human Diversity. Evol. Biol. 1972, 6, 381–398. [Google Scholar]
- Cao, Z.; Zhang, X.; Pang, J.; Qin, X.; Zhang, F.; Yang, Z.; Hao, L. Extraction of DNA and Establishment Application of SSR System in Pugionium cornutum (L.) Gaertn. Mol. Plant Breed. 2023, 1–12. [Google Scholar]
- Harisha, R.; Bhadru, D.; Vanisri, S.; Gourishanakar, V.; Satish, L. SSR and Morphological Traits Based Fingerprints and DNA Barcodes for Varietal Identification in Rice. Biotechnol. Biotechnol. Equip. 2021, 35, 1461–1473. [Google Scholar] [CrossRef]
- Parthiban, S.; Govindaraj, P.; Senthilkumar, S. Comparison of relative efficiency of genomic SSR and EST-SSR markers in estimating genetic diversity in sugarcane. 3 Biotech 2018, 8, 144. [Google Scholar] [CrossRef]
- Luzaran, R.T.; Pan, Y.B. Cultivar-specific SSR markers as revealed through fluorescence-labeling and capillary electrophoresis in sugarcane (Saccharum hybrids spp.). Philipp. J. Crop Sci. 2020, 45, 47–57. [Google Scholar]
- Wu, J.F. SNP and SSR Analysis and Genome-Wide Association Study of Drought Tolerance Traits in Brassica napus. Master’s Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2014. [Google Scholar]
- Ellegren, H. Microsatellites: Simple Sequences with Complex Evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Cui, G.; Wu, L.; Zhang, Y.; Ming, J.; Wang, J.; Wang, J. Identification of ISSR in Lily Hybrids. Acta Hortic. Sin. 2009, 36, 749–754. [Google Scholar]
- Ayliffe, M.; Lawrence, G.; Ellis, J.; Pryor, A. Heteroduplex Molecules Formed between Allelic Sequences Cause Nonparental RAPD Bands. Nucleic Acids Res. 1994, 22, 1632–1636. [Google Scholar] [CrossRef]
- Gao, X.; Tan, G.; Zhu, C.; He, D. Genetic Diversity of F1 Hybrids Population of Different Broccoli Varieties. North. Hortic. 2022, 1–8. [Google Scholar]
- Tian, C.; Bian, X.; Dong, L.; Wu, C.; Lang, R.; Yu, H.; Zhang, Y.; Tao, L.; Jing, Y. Identification and Genetic Analysis of F1 Hybrids from Wild Saccharum spontaneum L. of Sugarcane Using SSR Markers. Chin. J. Trop. Crops 2022, 43, 2021–2029. [Google Scholar]
- Mu, P.; Chai, J.; Su, W.; Zhang, H.; Zhao, G. Phenotype and genetic variation analysis of forward and reverse hybrid progeny from different oat crosses. Acta Prataculturae Sin. 2024, 33, 73–86. [Google Scholar]
- Zheng, Y. SSR Analysis of Sugarcane Parents and Construction of DNA Fingerprinting. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2012. [Google Scholar]
- You, Q. Genetic Diversity Analysis and Database Construction of DNA Fingerprintings in Sugarcane Based on SSR Fluorescence Markers. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou, China, 2014. [Google Scholar]
- Huang, Y.X.; Zhang, B.Q.; Gao, Y.J.; Duan, W.X.; Zhou, S.; Yang, C.F.; Zhang, G.M.; Wang, Z.P. Genetic Diversity Analysis of Phenotypic Traits in Saccharum arundinaceum from Guangxi. Chin. J. Trop. Crops 2019, 40, 1706–1712. [Google Scholar]
- Ali, A.; Pan, Y.-B.; Wang, Q.-N.; Wang, J.-D.; Chen, J.-L.; Gao, S.-J. Genetic Diversity and Population Structure Analysis of Saccharum and Erianthus Genera Using Microsatellite (SSR) Markers. Sci. Rep. 2019, 9, 395. [Google Scholar] [CrossRef] [PubMed]
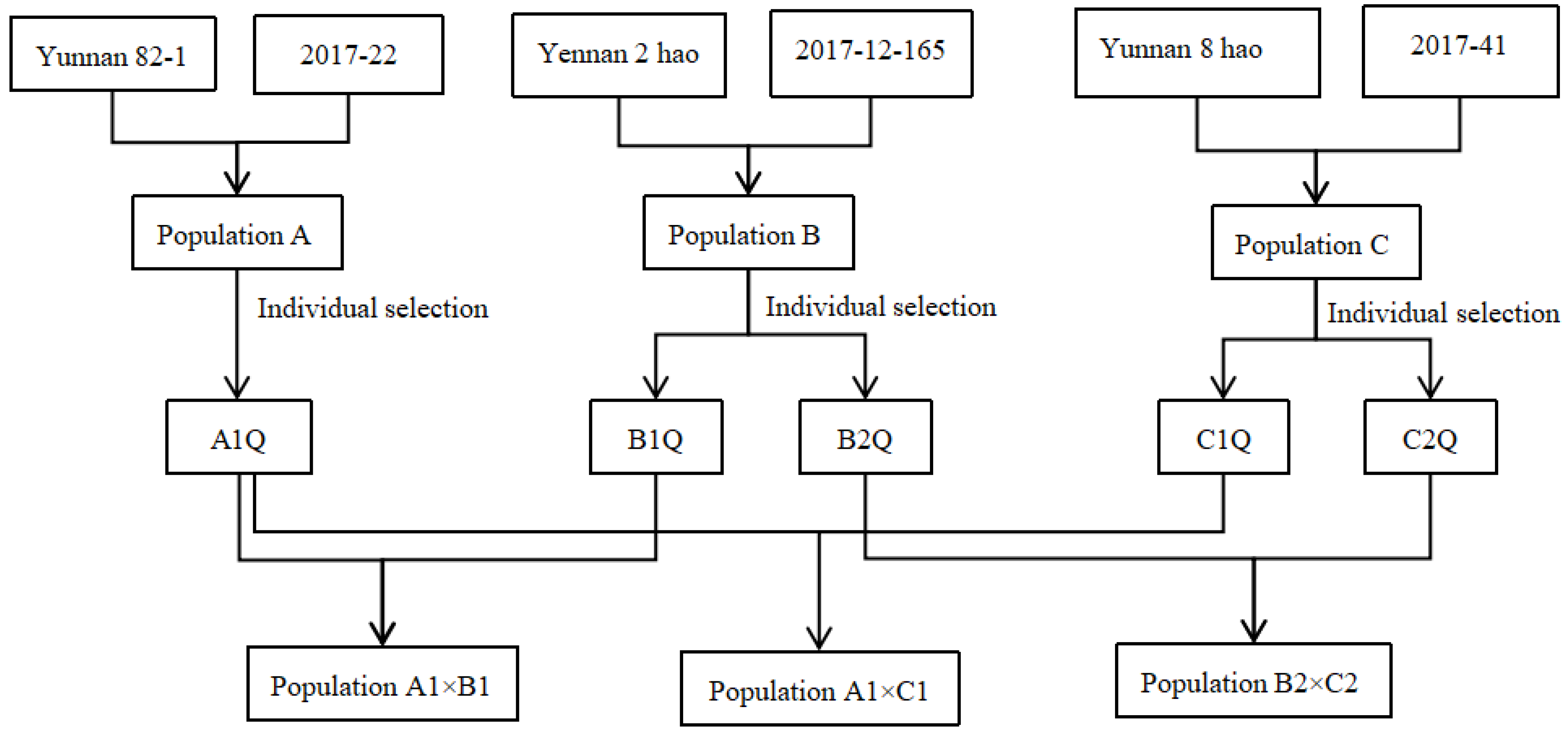

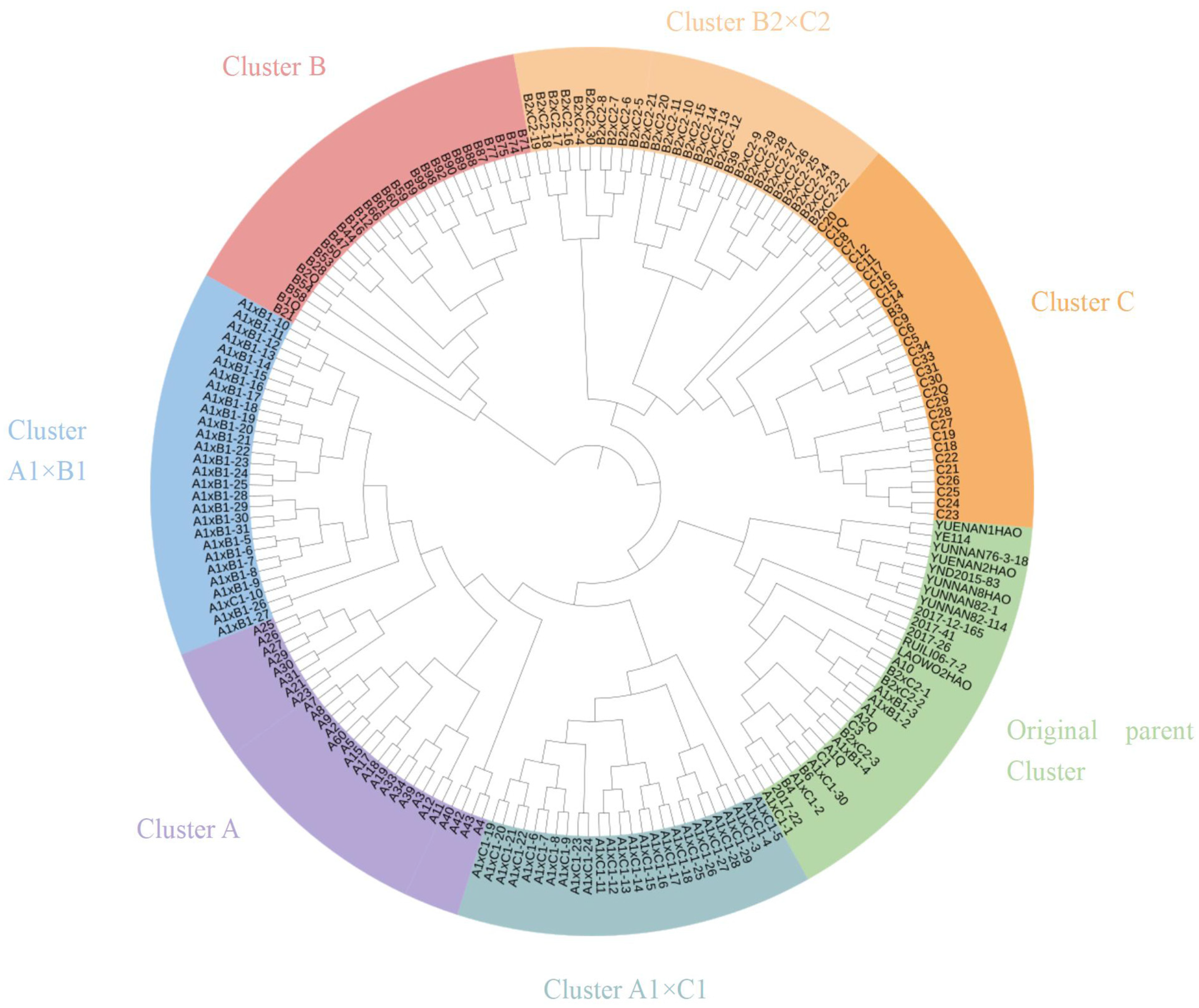
| Parent | F1 | F2 | ||||
|---|---|---|---|---|---|---|
| A | B | C | A1 × B1 | A1 × C1 | B2 × C2 | |
| A1Q | A1 | B6 | C1 | A1 × B1-2 | A1 × C1-1 | B2 × C2-1 |
| A2Q | A2 | B3 | C3 | A1 × B1-3 | A1 × C1-2 | B2 × C2-2 |
| B1Q | A3 | B4 | C5 | A1 × B1-4 | A1 × C1-3 | B2 × C2-3 |
| B2Q | A4 | B8 | C6 | A1 × B1-5 | A1 × C1-4 | B2 × C2-4 |
| C1Q | A5 | B9 | C7 | A1 × B1-6 | A1 × C1-5 | B2 × C2-5 |
| C2Q | A7 | B12 | C8 | A1 × B1-7 | A1 × C1-6 | B2 × C2-6 |
| Yunnan82-1 | A8 | B16 | C9 | A1 × B1-8 | A1 × C1-7 | B2 × C2-7 |
| Yuenan 2 hao | A9 | B21 | C11 | A1 × B1-9 | A1 × C1-8 | B2 × C2-8 |
| 2017-12-165 | A10 | B22 | C12 | A1 × B1-10 | A1 × C1-9 | B2 × C2-9 |
| 2017-26 | A11 | B28 | C14 | A1 × B1-11 | A1 × C1-10 | B2 × C2-10 |
| Yunnan 8 hao | A12 | B39 | C15 | A1 × B1-12 | A1 × C1-11 | B2 × C2-11 |
| YND 2015-83 | A15 | B44 | C16 | A1 × B1-13 | A1 × C1-12 | B2 × C2-12 |
| Yuenan 1 hao | A17 | B47 | C17 | A1 × B1-14 | A1 × C1-13 | B2 × C2-13 |
| Ye114 | A18 | B50 | C18 | A1 × B1-15 | A1 × C1-14 | B2 × C2-14 |
| Yunnan 82-114 | A19 | B53 | C19 | A1 × B1-16 | A1 × C1-15 | B2 × C2-15 |
| Laowo 2 hao | A21 | B54 | C20 | A1 × B1-17 | A1 × C1-16 | B2 × C2-16 |
| Ruili 06-7-2 | A23 | B58 | C21 | A1 × B1-18 | A1 × C1-17 | B2 × C2-17 |
| 2017-41 | A25 | B59 | C22 | A1 × B1-19 | A1 × C1-18 | B2 × C2-18 |
| Yunnan 76-3-18 | A26 | B60 | C23 | A1 × B1-20 | A1 × C1-19 | B2 × C2-19 |
| 2017-22 | A27 | B61 | C24 | A1 × B1–21 | A1 × C1-20 | B2 × C2-20 |
| A29 | B66 | C25 | A1 × B1-22 | A1 × C1-21 | B2 × C2-21 | |
| A30 | B71 | C26 | A1 × B1-23 | A1 × C1-22 | B2 × C2-22 | |
| A31 | B74 | C27 | A1 × B1-24 | A1 × C1-23 | B2 × C2-23 | |
| A33 | B75 | C28 | A1 × B1-25 | A1 × C1-24 | B2 × C2-24 | |
| A34 | B77 | C29 | A1 × B1-26 | A1 × C1-25 | B2 × C2-25 | |
| A39 | B87 | C30 | A1 × B1-27 | A1 × C1-26 | B2 × C2-26 | |
| A40 | B89 | C31 | A1 × B1-28 | A1 × C1-27 | B2 × C2-27 | |
| A42 | B90 | C33 | A1 × B1-29 | A1 × C1-28 | B2 × C2-28 | |
| A43 | B92 | C34 | A1 × B1-30 | A1 × C1-29 | B2 × C2-29 | |
| A60 | B99 | A1 × B1-31 | A1 × C1-30 | B2 × C2-30 | ||
| Names of Primers | Forward Primer Sequence (5′-3′) | Reverse Primer Sequence (5′-3′) | Tm (°C) |
|---|---|---|---|
| mSSCIR3 | ATAGCTCCCACACCAAATGC | GGACTACTCCACAATGATGC | 54 |
| mSSCIR9 | TCTCTATGCACCCTATCGT | TAACTTGACCCCCTCTTGA | 50 |
| mSSCIR16 | TGGGGAGGGCTGACTAGA | GGCGGTATATATGCTGTG | 54 |
| mSSCIR19 | GGTTCCAAAATACACAAA | CAATCTTATCTACGCACTT | 52 |
| mSSCIR21 | CGCCAGCCACATAAAAGG | CGACCAGGAGTTCATCAA | 54 |
| mSSCIR26 | AAAATCAGACAAACAGCAT | AGAAGAAGCAGATACAGGT | 48 |
| mSSCIR34 | ATCGCCTCCACTAAATAAT | TTGTCTTTGCTTCCTCCTC | 54 |
| mSSCIR36 | CAACAATAACTTAACTGGTA | CTGTCCTTTTTATTCTCTTT | 54 |
| mSSCIR47 | GCAATGGAGGTAGGAATG | TAGAATCACCCAAAAATAAA | 48 |
| mSSCIR53 | TGGTCTACTGAAGTTCGTG | TGCTTCTAAGTCAACCAAA | 50 |
| mSSCIR56 | ATTTGACGCTACGATGGTG | ATCCGTTTTTCAGCAGAGC | 52 |
| SMC1047 | TGAGCCTAAGCCAGAAAGAAG | GGAACTAATTTCCTACGAGAACAC | 50 |
| SMC119CG | TTCATCTCTAGCCTACCCCAA | AGCAGCCATTTACCCAGGA | 54 |
| SMC1237 | TTCACGAACACCCCACCTA | GCGCCAGGTAACCTACTGAA | 58 |
| SMC1752 | GGCTGATTTACATGAAACTGTTCT | AAAGCTGGTATCCCAGCATACT | 64 |
| SMC1814 | GGTTGACGATGAGAAGGACGTG | CACCCACATAGTGCCCAACG | 64 |
| SMC22 | CCATTCGACGAAAGCGTCCT | CAAGCGTTGTGCTGCCGAGT | 62 |
| SMC278 | TTCTAGTGCCAATCCATCTCAGA | CATGCCAACTTCCAAACAGACT | 50 |
| SMC640 | TTAAGAGACCCGCCTTTGGAA | TGCCAGAAGTGGTTGTGCTCA | 62 |
| SMC720BS | CGCACCGACGCACGTCT | GCCAATGGAACGGGTCTA | 58 |
| Primer Name | Total Loci | Number of Polymorphic Locis | PPB/% | PIC |
|---|---|---|---|---|
| mSSCIR1 | 10 | 10 | 100.00 | 0.8507 |
| mSSCIR3 | 9 | 9 | 100.00 | 0.8409 |
| mSSCIR9 | 9 | 9 | 100.00 | 0.7909 |
| mSSCIR16 | 8 | 8 | 100.00 | 0.8261 |
| mSSCIR19 | 17 | 17 | 100.00 | 0.9605 |
| mSSCIR21 | 16 | 16 | 100.00 | 0.9749 |
| mSSCIR26 | 12 | 12 | 100.00 | 0.9149 |
| mSSCIR34 | 7 | 7 | 100.00 | 0.8162 |
| mSSCIR36 | 13 | 13 | 100.00 | 0.9096 |
| mSSCIR52 | 15 | 15 | 100.00 | 0.8971 |
| mSSCIR53 | 9 | 9 | 100.00 | 0.8752 |
| mSSCIR56 | 19 | 19 | 100.00 | 0.8989 |
| SMC1047 | 13 | 13 | 100.00 | 0.8977 |
| SMC1752 | 10 | 9 | 90.00 | 0.8656 |
| SMC22 | 12 | 12 | 100.00 | 0.8898 |
| SMC119CG | 21 | 21 | 100.00 | 0.9451 |
| SMC278 | 12 | 11 | 91.67 | 0.8704 |
| SMC286 | 13 | 13 | 100.00 | 0.8615 |
| SMC851 | 10 | 10 | 100.00 | 0.8783 |
| SMC2055 | 12 | 12 | 100.00 | 0.8884 |
| total | 247 | 245 | ||
| average | 12.35 | 12.25 | 99.08 | 0.8826 |
| Population | Number of Population | Number of Cluster | Same Number | Similarity Rate |
|---|---|---|---|---|
| original parent | 14 | 31 | 14 | 100.00 |
| Progeny population parent | 6 | 2 | 30.00 | |
| A | 30 | 28 | 28 | 93.33 |
| B | 30 | 28 | 26 | 86.67 |
| C | 29 | 30 | 28 | 96.55 |
| A1 × B1 | 30 | 28 | 27 | 90.00 |
| A1 × C1 | 30 | 26 | 26 | 86.67 |
| B2 × C2 | 30 | 28 | 27 | 90.00 |
| Population | Number of Accessions | NPB | PPB (%) | Na | Ne | H | I |
|---|---|---|---|---|---|---|---|
| parent | 14 | 225 | 88.93% | 1.8893 | 1.5163 | 0.3041 a | 0.4578 a |
| A | 32 | 223 | 88.14% | 1.8814 | 1.4226 | 0.2568 b | 0.3969 b |
| B | 32 | 211 | 83.40% | 1.8340 | 1.3821 | 0.2329 bc | 0.3624 bc |
| C | 31 | 221 | 83.40% | 1.8340 | 1.4030 | 0.2423 bc | 0.374 bc |
| A1 × B1 | 30 | 186 | 73.52% | 1.7352 | 1.3043 | 0.1931 d | 0.3061 d |
| A1 × C1 | 30 | 223 | 88.14% | 1.8814 | 1.3554 | 0.223 cd | 0.3536 c |
| B2 × C2 | 30 | 226 | 89.33% | 1.8933 | 1.4977 | 0.2910 a | 0.4390 a |
| total | 199 | 253 | 100.00% | 2.0000 | 1.5787 | 0.3390 | 0.5082 |
| Population | ML | No. SPL | No. MIL | No. PIL | APL | TPL | MR (%) | MH (%) | PH (%) |
|---|---|---|---|---|---|---|---|---|---|
| A | 17.22 | 18.66 | 17.84 | 21.94 | 75.66 | 150.00 | 22.61 ± 1.71 | 48.67 | 36.24 |
| B | 16.13 | 24.31 | 6.75 | 12.66 | 59.84 | 121.00 | 26.81 ± 2.19 | 37.42 | 38.91 |
| C | 23.03 | 25.00 | 31.94 | 11.13 | 91.10 | 144.00 | 23.62 ± 1.42 | 49.94 | 45.16 |
| average | 18.79 | 22.66 | 18.84 | 15.24 | 75.53 | 138.33 | 24.35 | 45.34 | 40.11 |
| A1 × B1 | 11.97 | 27.27 | 30.53 | 5.53 | 75.30 | 150.00 | 16.52 ± 1.82 | 46.61 | 42.60 |
| A1 × C1 | 9.80 | 29.37 | 35.60 | 5.20 | 79.97 | 150.00 | 12.02 ± 1.58 | 52.39 | 49.38 |
| B2 × C2 | 24.43 | 23.60 | 18.53 | 18.17 | 84.73 | 122.00 | 28.51 ± 1.51 | 52.67 | 52.21 |
| average | 15.40 | 26.75 | 28.22 | 9.630 | 80.00 | 140.67 | 19.02 | 50.56 | 48.06 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, S.; Zhao, L.; Tao, L.; Zhang, Y.; Zan, F.; Lu, X.; Zhao, Y.; Zhang, J.; Liu, J. Analysis of Genetic Diversity in Polymers of Saccharum spontaneum L. and Their Hybrid Progenies. Agronomy 2025, 15, 2221. https://doi.org/10.3390/agronomy15092221
Ren S, Zhao L, Tao L, Zhang Y, Zan F, Lu X, Zhao Y, Zhang J, Liu J. Analysis of Genetic Diversity in Polymers of Saccharum spontaneum L. and Their Hybrid Progenies. Agronomy. 2025; 15(9):2221. https://doi.org/10.3390/agronomy15092221
Chicago/Turabian StyleRen, Shenlin, Liping Zhao, Lian’an Tao, Yuebin Zhang, Fenggang Zan, Xin Lu, Yong Zhao, Jing Zhang, and Jiayong Liu. 2025. "Analysis of Genetic Diversity in Polymers of Saccharum spontaneum L. and Their Hybrid Progenies" Agronomy 15, no. 9: 2221. https://doi.org/10.3390/agronomy15092221
APA StyleRen, S., Zhao, L., Tao, L., Zhang, Y., Zan, F., Lu, X., Zhao, Y., Zhang, J., & Liu, J. (2025). Analysis of Genetic Diversity in Polymers of Saccharum spontaneum L. and Their Hybrid Progenies. Agronomy, 15(9), 2221. https://doi.org/10.3390/agronomy15092221






