Comparative Genomic and Transcriptomic Analysis Uncovers Metabolic Mechanisms Underlying Drought Tolerance Variation in Maize
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Drought Treatment
2.3. Determination of Plant Growth and Environmental Factors
2.4. Transcriptome Analysis
2.5. Genome Resequencing
2.6. Quantitative Real-Time PCR (qRT-PCR) Assay
2.7. Statistical Analysis
3. Results
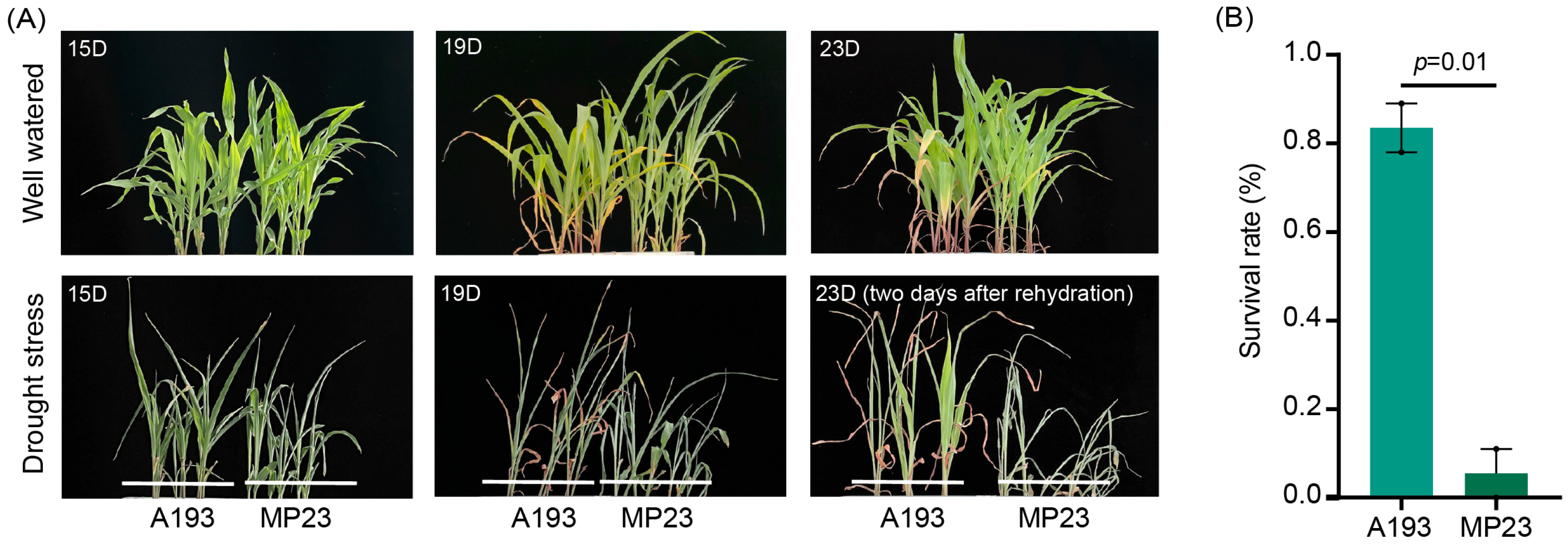
3.1. Comparison of Drought Tolerance Between the Two Inbred Maize Lines Under Different Water Supply Conditions
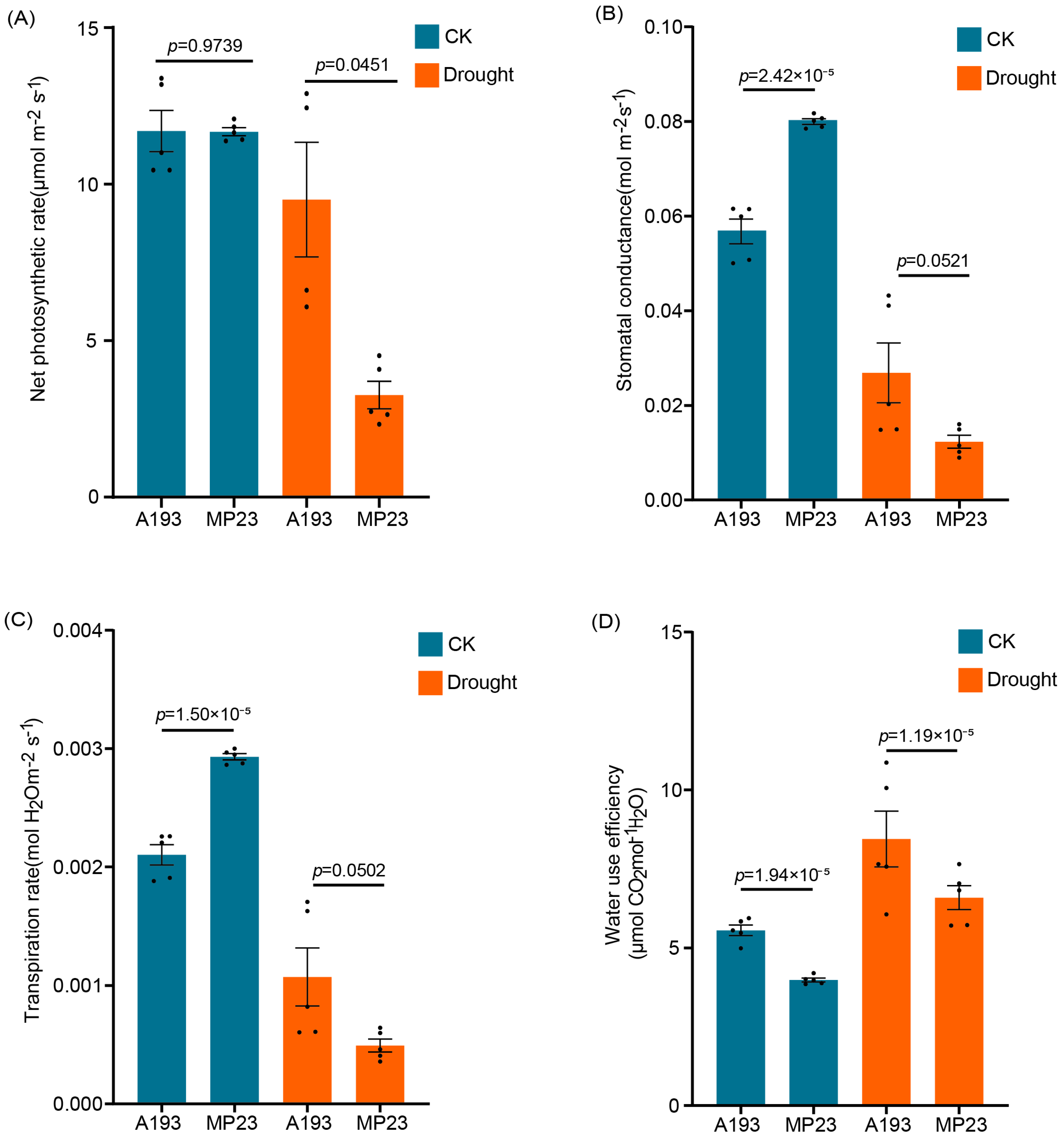
3.2. Photosynthetic Responses and Water Use Efficiency of Inbred Maize Lines Under Drought Stress
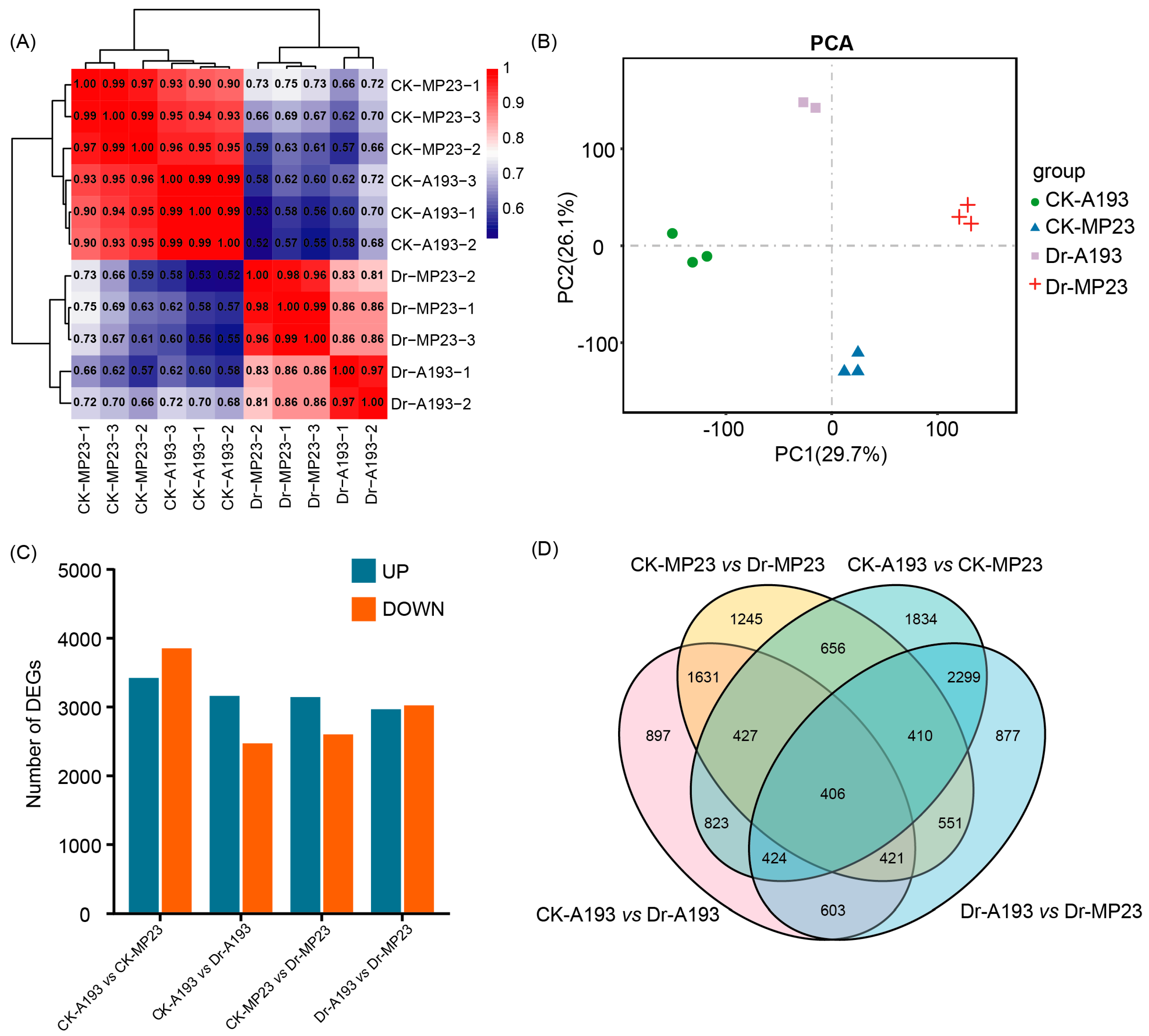
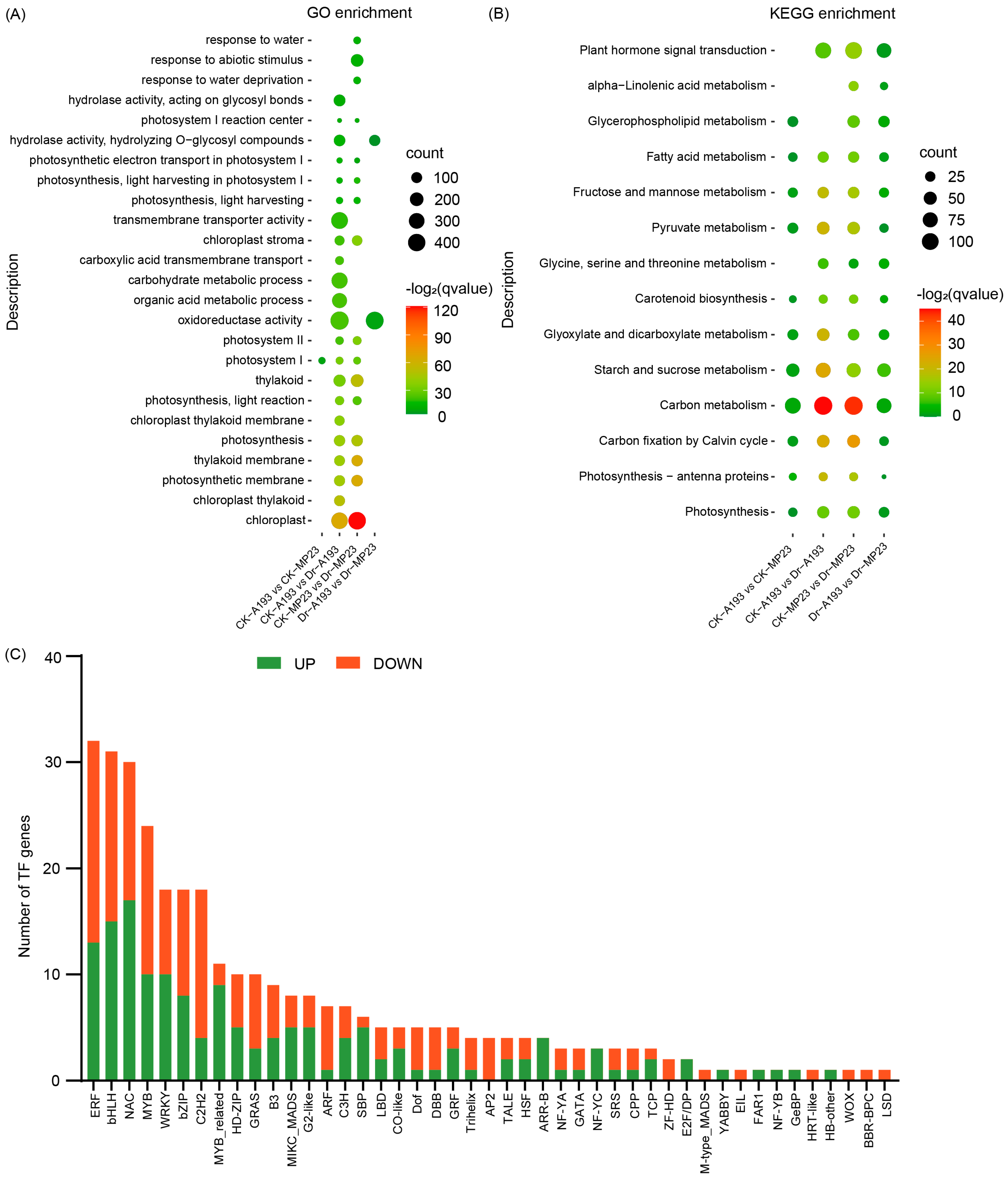
3.3. Transcriptome Analysis of the Two Inbred Maize Lines Under Different Water Supply Conditions
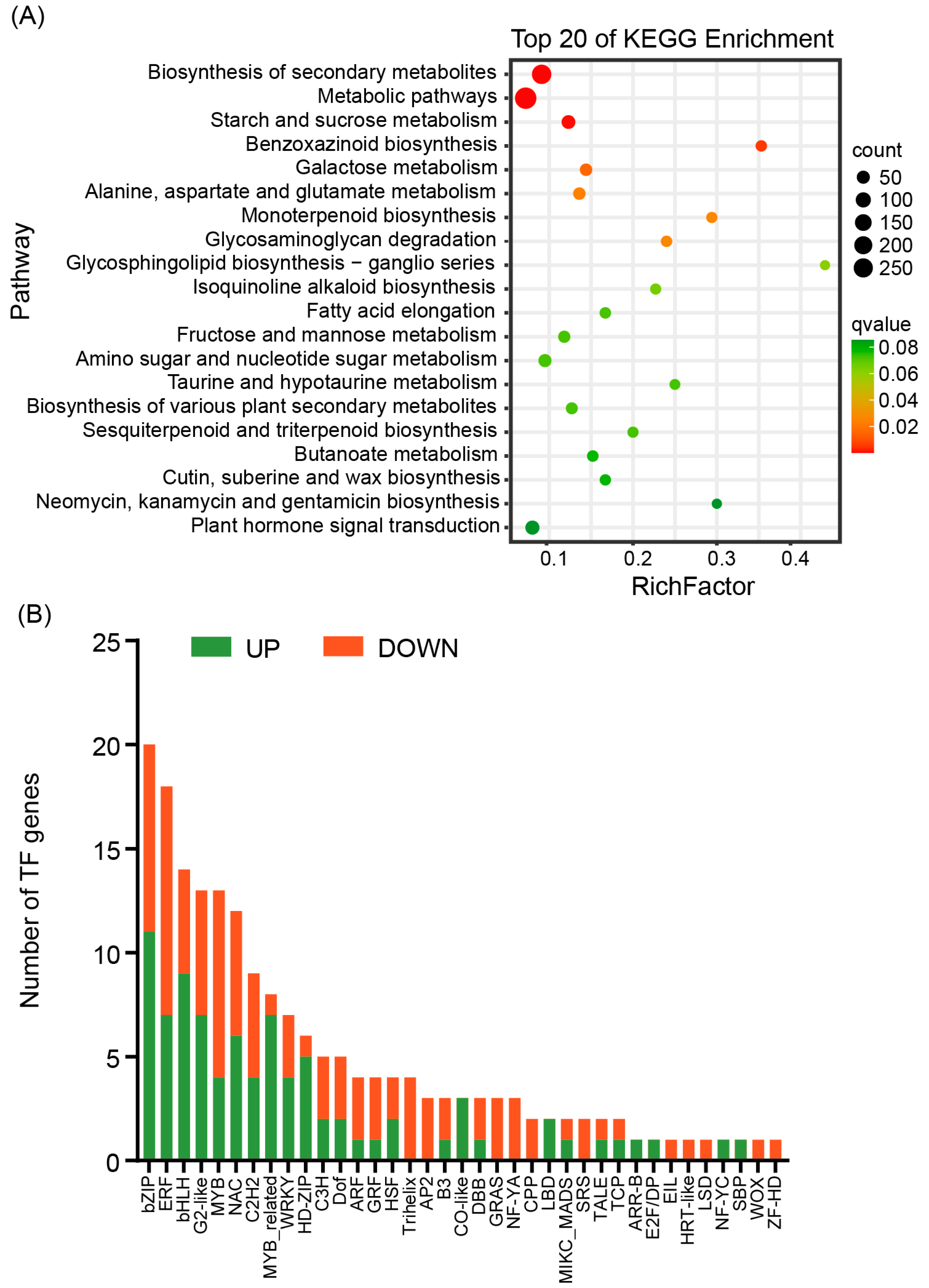
3.4. Differential Expression of Genes Between the Two Inbred Maize Lines Under Different Water Supply Conditions
3.5. Genomic Variations Between the Two Inbred Maize Lines Through Whole-Genome Resequencing
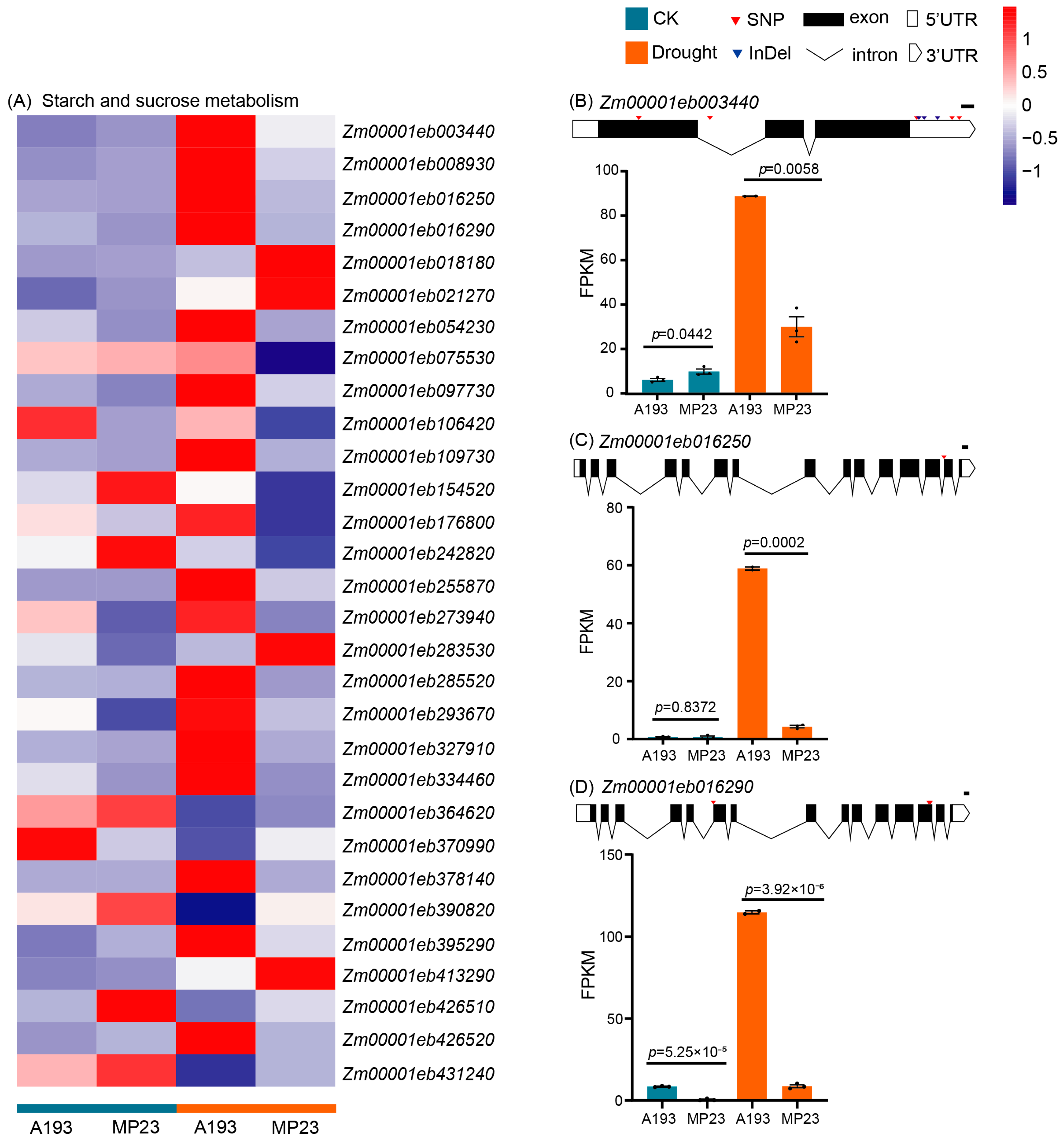
3.6. Analysis of DEVGs in Starch and Sucrose Metabolism
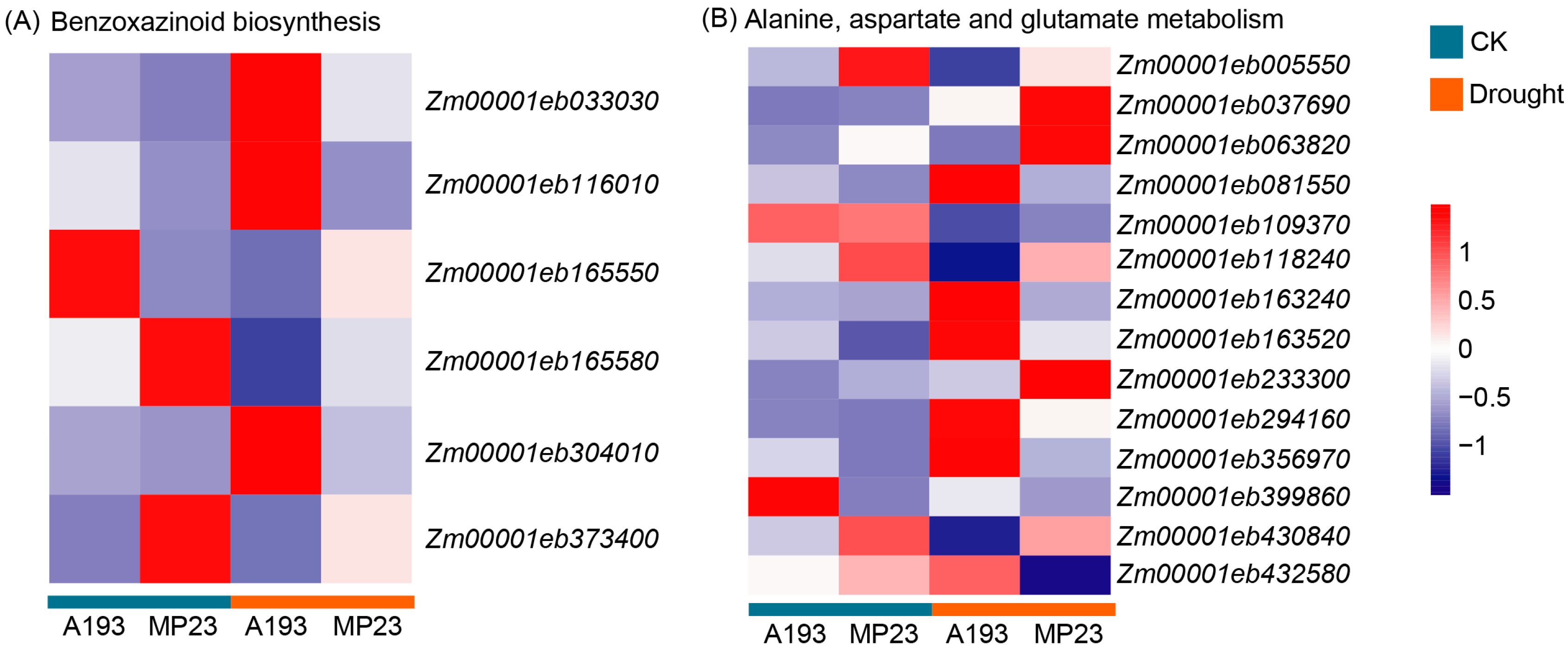
3.7. Analysis of DEVGs in Benzoxazinoid Biosynthesis and Alanine, Aspartate, and Glutamate Metabolism
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Bedoya, C.A.; Dreisigacker, S.; Hearne, S.; Franco, J.; Mir, C.; Prasanna, B.M.; Taba, S.; Charcosset, A.; Warburton, M.L. Genetic diversity and population structure of native maize populations in Latin America and the Caribbean. PLoS ONE 2017, 12, e0173488. [Google Scholar] [CrossRef]
- Luo, N.; Meng, Q.; Feng, P.; Qu, Z.; Yu, Y.; Liu, L.; Müller, C.; Wang, P. China can be self-sufficient in maize production by 2030 with optimal crop management. Nat. Commun. 2023, 14, 2637. [Google Scholar] [CrossRef]
- Kim, K.-H.; Lee, B.-M. Effects of Climate Change and Drought Tolerance on Maize Growth. Plants 2023, 12, 3548. [Google Scholar] [CrossRef]
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C.; et al. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 3890. [Google Scholar] [CrossRef]
- Liu, S.; Wu, W.; Yang, X.; Yang, P.; Sun, J. Exploring drought dynamics and its impacts on maize yield in the Huang-Huai-Hai farming region of China. Clim. Change 2020, 163, 415–430. [Google Scholar] [CrossRef]
- Sah, R.P.; Chakraborty, M.; Prasad, K.; Pandit, M.; Tudu, V.K.; Chakravarty, M.K.; Narayan, S.C.; Rana, M.; Moharana, D. Impact of water deficit stress in maize: Phenology and yield components. Sci. Rep. 2020, 10, 2944. [Google Scholar] [CrossRef]
- Gao, H.; Cui, J.; Liu, S.; Wang, S.; Lian, Y.; Bai, Y.; Zhu, T.; Wu, H.; Wang, Y.; Yang, S.; et al. Natural variations of ZmSRO1d modulate the trade-off between drought resistance and yield by affecting ZmRBOHC-mediated stomatal ROS production in maize. Mol. Plant 2022, 15, 1558–1574. [Google Scholar] [CrossRef]
- Tian, T.; Wang, S.; Yang, S.; Yang, Z.; Liu, S.; Wang, Y.; Gao, H.; Zhang, S.; Yang, X.; Jiang, C.; et al. Genome assembly and genetic dissection of a prominent drought-resistant maize germplasm. Nat. Genet. 2023, 55, 496–506. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Wang, H.; Liu, S.; Ferjani, A.; Li, J.; Yan, J.; Yang, X.; Qin, F. Genetic variation in ZmVPP1 contributes to drought tolerance in maize seedlings. Nat. Genet. 2016, 48, 1233–1241. [Google Scholar] [CrossRef]
- Chen, Y.; Leng, Y.N.; Zhu, F.Y.; Li, S.E.; Song, T.; Zhang, J. Water-saving techniques: Physiological responses and regulatory mechanisms of crops. Adv. Biotechnol. 2023, 1, 3. [Google Scholar] [CrossRef] [PubMed]
- Oguz, M.C.; Aycan, M.; Oguz, E.; Poyraz, I.; Yildiz, M. Drought Stress Tolerance in Plants: Interplay of Molecular, Biochemical and Physiological Responses in Important Development Stages. Physiologia 2022, 2, 180–197. [Google Scholar] [CrossRef]
- Haghpanah, M.; Hashemipetroudi, S.; Arzani, A.; Araniti, F. Drought Tolerance in Plants: Physiological and Molecular Responses. Plants 2024, 13, 2962. [Google Scholar] [CrossRef]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef]
- Zhou, W.; Yin, J.; Zhou, Y.; Li, Y.; He, H.; Yang, Y.; Wang, X.; Lian, X.; Dong, X.; Ma, Z.; et al. DSD1/ZmICEb regulates stomatal development and drought tolerance in maize. J. Integr. Plant Biol. 2025, 67, 1487–1500. [Google Scholar] [CrossRef]
- Hou, P.; Wang, F.; Luo, B.; Li, A.; Wang, C.; Shabala, L.; Ahmed, H.A.I.; Deng, S.; Zhang, H.; Song, P.; et al. Antioxidant Enzymatic Activity and Osmotic Adjustment as Components of the Drought Tolerance Mechanism in Carex duriuscula. Plants 2021, 10, 436. [Google Scholar] [CrossRef]
- Jia, Y.; Xiao, W.; Ye, Y.; Wang, X.; Liu, X.; Wang, G.; Li, G.; Wang, Y. Response of Photosynthetic Performance to Drought Duration and Re-Watering in Maize. Agronomy 2020, 10, 533. [Google Scholar] [CrossRef]
- Gulzar, F.; Fu, J.; Zhu, C.; Yan, J.; Li, X.; Meraj, T.A.; Shen, Q.; Hassan, B.; Wang, Q. Maize WRKY Transcription Factor ZmWRKY79 Positively Regulates Drought Tolerance through Elevating ABA Biosynthesis. Int. J. Mol. Sci. 2021, 22, 10080. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, X.; Ren, Z.; Abou-Elwafa, S.F.; Pu, X.; Zhu, Y.; Dou, D.; Su, H.; Cheng, H.; Liu, Z.; et al. ZmERF21 directly regulates hormone signaling and stress-responsive gene expression to influence drought tolerance in maize seedlings. Plant Cell Environ. 2022, 45, 312–328. [Google Scholar] [CrossRef]
- Yang, L.; Fountain, J.C.; Ji, P.; Ni, X.; Chen, S.; Lee, R.D.; Kemerait, R.C.; Guo, B. Deciphering drought-induced metabolic responses and regulation in developing maize kernels. Plant Biotechnol. J. 2018, 16, 1616–1628. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, X.; Wang, H.; Xin, H.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.; Shinozaki, K.; Yamaguchi-Shinozaki, K.; et al. Genome-wide analysis of ZmDREB genes and their association with natural variation in drought tolerance at seedling stage of Zea mays L. PLoS Genet. 2013, 9, e1003790. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Wang, H.; Liu, S.; Li, Z.; Yang, X.; Yan, J.; Li, J.; Tran, L.S.; Qin, F. A transposable element in a NAC gene is associated with drought tolerance in maize seedlings. Nat. Commun. 2015, 6, 8326. [Google Scholar] [CrossRef]
- Zhang, H.; Xiang, Y.; He, N.; Liu, X.; Liu, H.; Fang, L.; Zhang, F.; Sun, X.; Zhang, D.; Li, X.; et al. Enhanced Vitamin C Production Mediated by an ABA-Induced PTP-like Nucleotidase Improves Plant Drought Tolerance in Arabidopsis and Maize. Mol. Plant 2020, 13, 760–776. [Google Scholar] [CrossRef]
- Feng, X.; Jia, L.; Cai, Y.; Guan, H.; Zheng, D.; Zhang, W.; Xiong, H.; Zhou, H.; Wen, Y.; Hu, Y.; et al. ABA-inducible DEEPER ROOTING 1 improves adaptation of maize to water deficiency. Plant Biotechnol. J. 2022, 20, 2077–2088. [Google Scholar] [CrossRef]
- Zhang, X.; Mi, Y.; Mao, H.; Liu, S.; Chen, L.; Qin, F. Genetic variation in ZmTIP1 contributes to root hair elongation and drought tolerance in maize. Plant Biotechnol. J. 2020, 18, 1271–1283. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Du, Y.; He, C.; Dietrich, C.R.; Li, J.; Ma, X.; Wang, R.; Liu, Q.; Liu, S.; Wang, G.; et al. Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 2019, 70, 3089–3099. [Google Scholar] [CrossRef]
- Yang, Y.; Shi, J.; Chen, L.; Xiao, W.; Yu, J. ZmEREB46, a maize ortholog of Arabidopsis WAX INDUCER1/SHINE1, is involved in the biosynthesis of leaf epicuticular very-long-chain waxes and drought tolerance. Plant Sci. Int. J. Exp. Plant Biol. 2022, 321, 111256. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Bourgault, R.; Galli, M.; Strable, J.; Chen, Z.; Feng, F.; Dong, J.; Molina, I.; Gallavotti, A. The FUSED LEAVES1-ADHERENT1 regulatory module is required for maize cuticle development and organ separation. New Phytol. 2021, 229, 388–402. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Liu, M.; Zhao, H.; Tan, Z.; Liang, K.; Sun, Q.; Gong, D.; He, H.; Zhou, W.; Qiu, F. ZmSRL5 is involved in drought tolerance by maintaining cuticular wax structure in maize. J. Integr. Plant Biol. 2020, 62, 1895–1909. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, B.; Yang, Z.; Liu, Y.; Yang, S.; Shi, Y.; Jiang, C.; Qin, F. Manipulating ZmEXPA4 expression ameliorates the drought-induced prolonged anthesis and silking interval in maize. Plant Cell 2021, 33, 2058–2071. [Google Scholar] [CrossRef]
- Sun, X.; Xiang, Y.; Dou, N.; Zhang, H.; Pei, S.; Franco, A.V.; Menon, M.; Monier, B.; Ferebee, T.; Liu, T.; et al. The role of transposon inverted repeats in balancing drought tolerance and yield-related traits in maize. Nat. Biotechnol. 2023, 41, 120–127. [Google Scholar] [CrossRef]
- Wei, S.; Xia, R.; Chen, C.; Shang, X.; Ge, F.; Wei, H.; Chen, H.; Wu, Y.; Xie, Q. ZmbHLH124 identified in maize recombinant inbred lines contributes to drought tolerance in crops. Plant Biotechnol. J. 2021, 19, 2069–2081. [Google Scholar] [CrossRef]
- Kaderbek, T.; Huang, L.; Yue, Y.; Wang, Z.; Lian, J.; Ma, Y.; Li, J.; Zhuang, J.; Chen, J.; Lai, J.; et al. Identification of the maize drought-resistant gene Zinc-finger Inflorescence Meristem 23 through high-resolution temporal transcriptome analysis. Int. J. Biol. Macromol. 2025, 308, 142347. [Google Scholar] [CrossRef]
- Li, Y.; Su, Z.; Lin, Y.; Xu, Z.; Bao, H.; Wang, F.; Liu, J.; Hu, S.; Wang, Z.; Yu, X.; et al. Utilizing transcriptomics and metabolomics to unravel key genes and metabolites of maize seedlings in response to drought stress. BMC Plant Biol. 2024, 24, 34. [Google Scholar] [CrossRef]
- Privitera, G.F.; Treccarichi, S.; Nicotra, R.; Branca, F.; Pulvirenti, A.; Lo Piero, A.R.; Sicilia, A. Comparative transcriptome analysis of B. oleracea L. var. italica and B. macrocarpa Guss. genotypes under drought stress: De novo vs reference genome assembly. Plant Stress 2024, 14, 100657. [Google Scholar] [CrossRef]
- Xu, H.; Chen, L.; Song, B.; Fan, X.; Yuan, X.; Chen, J. De novo transcriptome sequencing of pakchoi (Brassica rapa L. chinensis) reveals the key genes related to the response of heat stress. Acta Physiol. Plant. 2016, 38, 252. [Google Scholar] [CrossRef]
- Lee, S.-G.; Na, D.; Park, C. Comparability of reference-based and reference-free transcriptome analysis approaches at the gene expression level. BMC Bioinform. 2021, 22, 310. [Google Scholar] [CrossRef]
- Todaka, D.; Zhao, Y.; Yoshida, T.; Kudo, M.; Kidokoro, S.; Mizoi, J.; Kodaira, K.S.; Takebayashi, Y.; Kojima, M.; Sakakibara, H.; et al. Temporal and spatial changes in gene expression, metabolite accumulation and phytohormone content in rice seedlings grown under drought stress conditions. Plant J. Cell Mol. Biol. 2017, 90, 61–78. [Google Scholar] [CrossRef]
- Dwivedi, A.K.; Singh, V.; Anwar, K.; Pareek, A.; Jain, M. Integrated transcriptome, proteome and metabolome analyses revealed secondary metabolites and auxiliary carbohydrate metabolism augmenting drought tolerance in rice. Plant Physiol. Biochem. PPB 2023, 201, 107849. [Google Scholar] [CrossRef]
- Zhang, F.; Wu, J.; Sade, N.; Wu, S.; Egbaria, A.; Fernie, A.R.; Yan, J.; Qin, F.; Chen, W.; Brotman, Y.; et al. Genomic basis underlying the metabolome-mediated drought adaptation of maize. Genome Biol. 2021, 22, 260. [Google Scholar] [CrossRef]
- Pelleschi, S.; Leonardi, A.; Rocher, J.P.; Cornic, G.; de Vienne, D.; Thévenot, C.; Prioul, J.L. Analysis of the Relationships between Growth, Photosynthesis and Carbohydrate Metabolism Using Quantitative Trait Loci (QTLs) in Young Maize Plants Subjected to Water Deprivation. Mol. Breed. 2006, 17, 21–39. [Google Scholar] [CrossRef]
- Liu, S.; Zenda, T.; Li, J.; Wang, Y.; Liu, X.; Duan, H. Comparative transcriptomic analysis of contrasting hybrid cultivars reveal key drought-responsive genes and metabolic pathways regulating drought stress tolerance in maize at various stages. PLoS ONE 2020, 15, e0240468. [Google Scholar] [CrossRef]
- Wang, Y.; Guo, H.; Wu, X.; Wang, J.; Li, H.; Zhang, R. Transcriptomic and physiological responses of contrasting maize genotypes to drought stress. Front. Plant Sci. 2022, 13, 928897. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Xu, C.; Wang, X.; Wang, P.; Huang, S. Abscisic acid collaborates with lignin and flavonoid to improve pre-silking drought tolerance by tuning stem elongation and ear development in maize (Zea mays L.). Plant J. Cell Mol. Biol. 2023, 114, 437–454. [Google Scholar] [CrossRef]
- Sutour, S.; Doan, V.C.; Mateo, P.; Züst, T.; Hartmann, E.R.; Glauser, G.; Robert, C.A.M. Isolation and Structure Determination of Drought-Induced Multihexose Benzoxazinoids from Maize (Zea mays). J. Agric. Food Chem. 2024, 72, 3427–3435. [Google Scholar] [CrossRef]
- Xu, C.; Li, F.; Zhuang, Y.; Li, Q.; Zhang, Z.; Zhang, L.; Zhao, H.; Bian, S.; Wang, H.; Zhao, R.; et al. The Effect of Drip Irrigation Quota on Biochemical Activities and Yield-Related Traits in Different Drought-Tolerant Maize Varieties. Agriculture 2023, 13, 1682. [Google Scholar] [CrossRef]
- Li, P.; Zhu, T.; Wang, Y.; Zhang, X.; Yang, X.; Fang, S.; Li, W.; Rui, W.; Yang, A.; Duan, Y.; et al. Natural variation in a cortex/epidermis-specific transcription factor bZIP89 determines lateral root development and drought resilience in maize. Sci. Adv. 2025, 11, eadt1113. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Wang, L.; Feng, Z.; Wang, X.; Wang, X.; Zhang, X. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinform 2010, 26, 136–138. [Google Scholar] [CrossRef]
- Niu, L.; Wu, X.; Liu, H.; Hu, X.; Wang, W. Leaf starch degradation by β-amylase ZmBAM8 influences drought tolerance in maize. Carbohydr. Polym. 2024, 345, 122555. [Google Scholar] [CrossRef]
- Xiao, N.; Ma, H.; Wang, W.; Sun, Z.; Li, P.; Xia, T. Overexpression of ZmSUS1 increased drought resistance of maize (Zea mays L.) by regulating sucrose metabolism and soluble sugar content. Planta 2024, 259, 43. [Google Scholar] [CrossRef]
- Vaughan, M.M.; Block, A.; Christensen, S.A.; Allen, L.H.; Schmelz, E.A. The effects of climate change associated abiotic stresses on maize phytochemical defenses. Phytochem. Rev. 2018, 17, 37–49. [Google Scholar] [CrossRef]
- Daryanto, S.; Wang, L.; Jacinthe, P.A. Global Synthesis of Drought Effects on Maize and Wheat Production. PLoS ONE 2016, 11, e0156362. [Google Scholar] [CrossRef]
- Jiang, Y.; Su, S.; Chen, H.; Li, S.; Shan, X.; Li, H.; Liu, H.; Dong, H.; Yuan, Y. Transcriptome analysis of drought-responsive and drought-tolerant mechanisms in maize leaves under drought stress. Physiol. Plant. 2023, 175, e13875. [Google Scholar] [CrossRef]
- Dong, A.; Wang, N.; Zenda, T.; Zhai, X.; Zhong, Y.; Yang, Q.; Xing, Y.; Duan, H.; Yan, X. ZmDnaJ-ZmNCED6 module positively regulates drought tolerance via modulating stomatal closure in maize. Plant Physiol. Biochem. PPB 2025, 218, 109286. [Google Scholar] [CrossRef]
- Zhang, Z.; Xie, X.; Naseer, M.A.; Zhou, H.; Cheng, W.; Xie, H.; Qin, L.; Yang, X.; Jiang, Y.; Zhou, X. Screening and Physiological Responses of Maize Inbred Lines to Drought Stress in South China. Sustainability 2024, 16, 7366. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Wang, X.; Jin, H.; Liu, G.; Duan, H. Comparative Proteomic and Physiological Analyses of Two Divergent Maize Inbred Lines Provide More Insights into Drought-Stress Tolerance Mechanisms. Int. J. Mol. Sci. 2018, 19, 3225. [Google Scholar] [CrossRef]
- Hao, B.; Xue, Q.; Marek, T.H.; Jessup, K.E.; Becker, J.D.; Hou, X.; Xu, W.; Bynum, E.D.; Bean, B.W.; Colaizzi, P.D.; et al. Grain yield, evapotranspiration, and water-use efficiency of maize hybrids differing in drought tolerance. Irrig. Sci. 2019, 37, 25–34. [Google Scholar] [CrossRef]
- Zhang, K.; Xue, M.; Qin, F.; He, Y.; Zhou, Y. Natural polymorphisms in ZmIRX15A affect water-use efficiency by modulating stomatal density in maize. Plant Biotechnol. J. 2023, 21, 2560–2573. [Google Scholar] [CrossRef]
- Xu, J.; Yuan, Y.; Xu, Y.; Zhang, G.; Guo, X.; Wu, F.; Wang, Q.; Rong, T.; Pan, G.; Cao, M.; et al. Identification of candidate genes for drought tolerance by whole-genome resequencing in maize. BMC Plant Biol. 2014, 14, 83. [Google Scholar] [CrossRef]
- Shikha, M.; Kanika, A.; Rao, A.R.; Mallikarjuna, M.G.; Gupta, H.S.; Nepolean, T. Genomic Selection for Drought Tolerance Using Genome-Wide SNPs in Maize. Front. Plant Sci. 2017, 8, 550. [Google Scholar] [CrossRef]
- Ahmad, S.; Belwal, V.; Punia, S.S.; Ram, M.; Dalip; Rajput, S.S.; Kunwar, R.; Meena, M.K.; Gupta, D.; Kumawat, G.L.; et al. Role of Plant Secondary Metabolites and Phytohormones in Drought Tolerance: A Review. Gesunde Pflanz. 2023, 75, 729–746. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, F.; Hong, Y.; Yao, J.; Ren, Z.; Shi, H.; Zhu, J.-K. The Flowering Repressor SVP Confers Drought Resistance in Arabidopsis by Regulating Abscisic Acid Catabolism. Mol. Plant 2018, 11, 1184–1197. [Google Scholar] [CrossRef]
- Sheng, M.; Xia, H.; Ding, H.; Pan, D.; He, J.; Li, Z.; Liu, J. Long-Term Soil Drought Limits Starch Accumulation by Altering Sucrose Transport and Starch Synthesis in Sweet Potato Tuberous Root. Int. J. Mol. Sci. 2023, 24, 3053. [Google Scholar] [CrossRef]
- Zanella, M.; Borghi, G.L.; Pirone, C.; Thalmann, M.; Pazmino, D.; Costa, A.; Santelia, D.; Trost, P.; Sparla, F. β-amylase 1 (BAM1) degrades transitory starch to sustain proline biosynthesis during drought stress. J. Exp. Bot. 2016, 67, 1819–1826. [Google Scholar] [CrossRef]
- Yang, Z.; Xu, S.; Wang, Y.; Zhang, E.; Xu, C. Molecluar Evolution and Expression Patterns Under Abiotic Stresses of Beta-amylase Gene Family in Grasses. Sci. Technol. Rev. 2014, 32, 29–36. [Google Scholar] [CrossRef]
- Yue, W.; Pengjun, D.; Junru, C.; Ruilei, W.; Kaiwen, Z.; Mengzhen, L.; Zhaoxue, H. Evolutionary analysis of plant SUS gene family and expressionpatterns of maize SUS genes under drought stress. Agric. Res. Arid Areas 2023, 41, 71–79. [Google Scholar]
- Robert, C.A.M.; Mateo, P. The Chemical Ecology of Benzoxazinoids. Chimia 2022, 76, 928–938. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, S.; Liu, K.; Abbasi, I.H.R.; Cai, C.; Yao, J. Molecular mechanisms relating to amino acid regulation of protein synthesis. Nutr. Res. Rev. 2019, 32, 183–191. [Google Scholar] [CrossRef]
- Dellero, Y. Manipulating Amino Acid Metabolism to Improve Crop Nitrogen Use Efficiency for a Sustainable Agriculture. Front. Plant Sci. 2020, 11, 602548. [Google Scholar] [CrossRef]
- Batista-Silva, W.; Heinemann, B.; Rugen, N.; Nunes-Nesi, A.; Araújo, W.L.; Braun, H.P.; Hildebrandt, T.M. The role of amino acid metabolism during abiotic stress release. Plant Cell Environ. 2019, 42, 1630–1644. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, T.; Zhang, C.; Zhang, W.; Deng, N.; Dirk, L.M.A.; Downie, A.B.; Zhao, T. Raffinose positively regulates maize drought tolerance by reducing leaf transpiration. Plant J. Cell Mol. Biol. 2023, 114, 55–67. [Google Scholar] [CrossRef] [PubMed]
- Radwan, A.; Kleinwächter, M.; Selmar, D. Impact of drought stress on specialised metabolism: Biosynthesis and the expression of monoterpene synthases in sage (Salvia officinalis). Phytochemistry 2017, 141, 20–26. [Google Scholar] [CrossRef]
- Konno, H.; Yamasaki, Y.; Sugimoto, M.; Takeda, K. Differential changes in cell wall matrix polysaccharides and glycoside-hydrolyzing enzymes in developing wheat seedlings differing in drought tolerance. J. Plant Physiol. 2008, 165, 745–754. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zhu, T.; Wang, Y.; Sun, Y.; Li, P.; Wang, H. Comparative Genomic and Transcriptomic Analysis Uncovers Metabolic Mechanisms Underlying Drought Tolerance Variation in Maize. Agronomy 2025, 15, 2189. https://doi.org/10.3390/agronomy15092189
Li Y, Zhu T, Wang Y, Sun Y, Li P, Wang H. Comparative Genomic and Transcriptomic Analysis Uncovers Metabolic Mechanisms Underlying Drought Tolerance Variation in Maize. Agronomy. 2025; 15(9):2189. https://doi.org/10.3390/agronomy15092189
Chicago/Turabian StyleLi, Yuxuan, Tianze Zhu, Yunyun Wang, Ye Sun, Pengcheng Li, and Houmiao Wang. 2025. "Comparative Genomic and Transcriptomic Analysis Uncovers Metabolic Mechanisms Underlying Drought Tolerance Variation in Maize" Agronomy 15, no. 9: 2189. https://doi.org/10.3390/agronomy15092189
APA StyleLi, Y., Zhu, T., Wang, Y., Sun, Y., Li, P., & Wang, H. (2025). Comparative Genomic and Transcriptomic Analysis Uncovers Metabolic Mechanisms Underlying Drought Tolerance Variation in Maize. Agronomy, 15(9), 2189. https://doi.org/10.3390/agronomy15092189





