rolB Promotes Adventitious Root Development in Pyrus betulaefolia by Modulating Endogenous Hormones and Gene Expression
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Adventitious Root Morphology and Growth Index Measurement
2.3. Determination of the Endogenous Hormone Content and Production of Paraffin Sections
2.4. Transcriptome Analysis
2.5. qRT-PCR Validation
2.6. Data Analysis
3. Results and Analysis
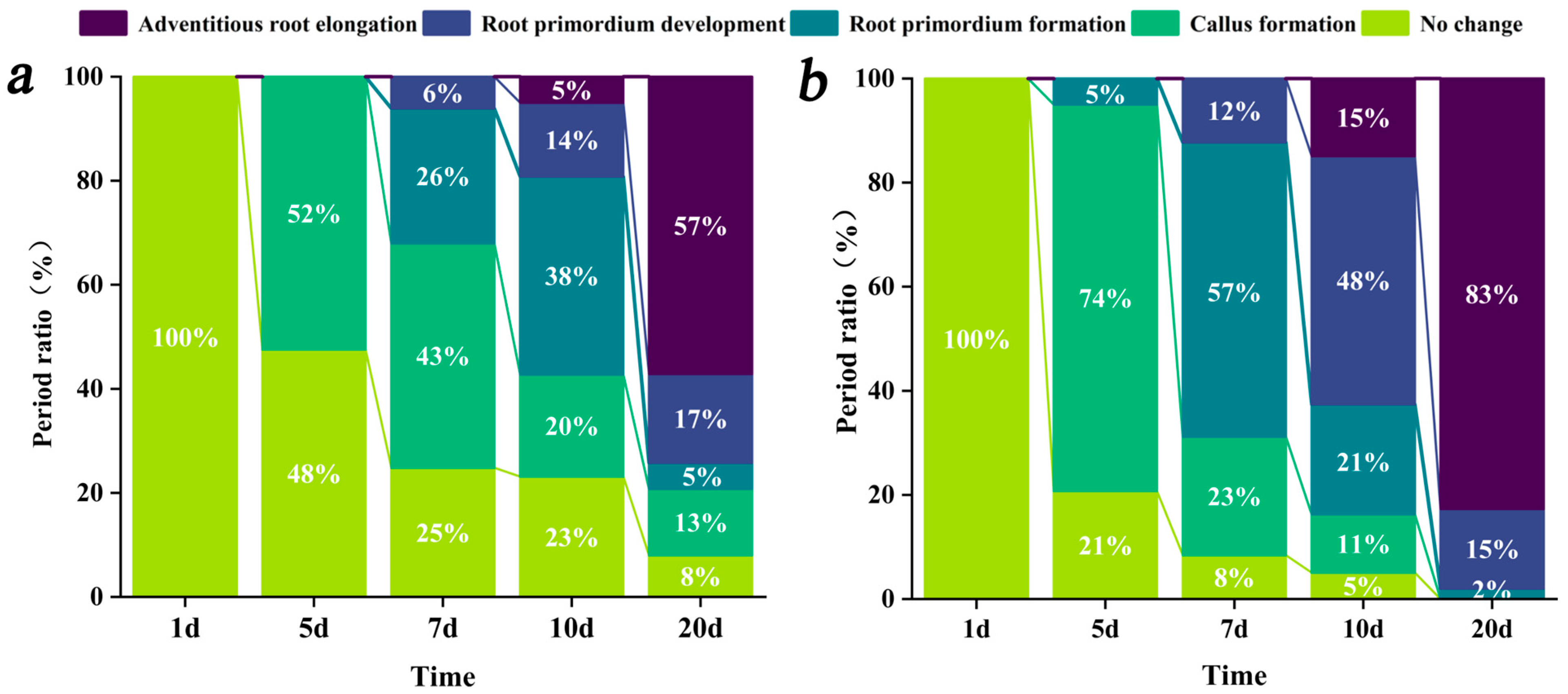
3.1. Cytological Changes and Developmental Progression of Adventitious Root Formation in rolB-Transformed ‘Duli’
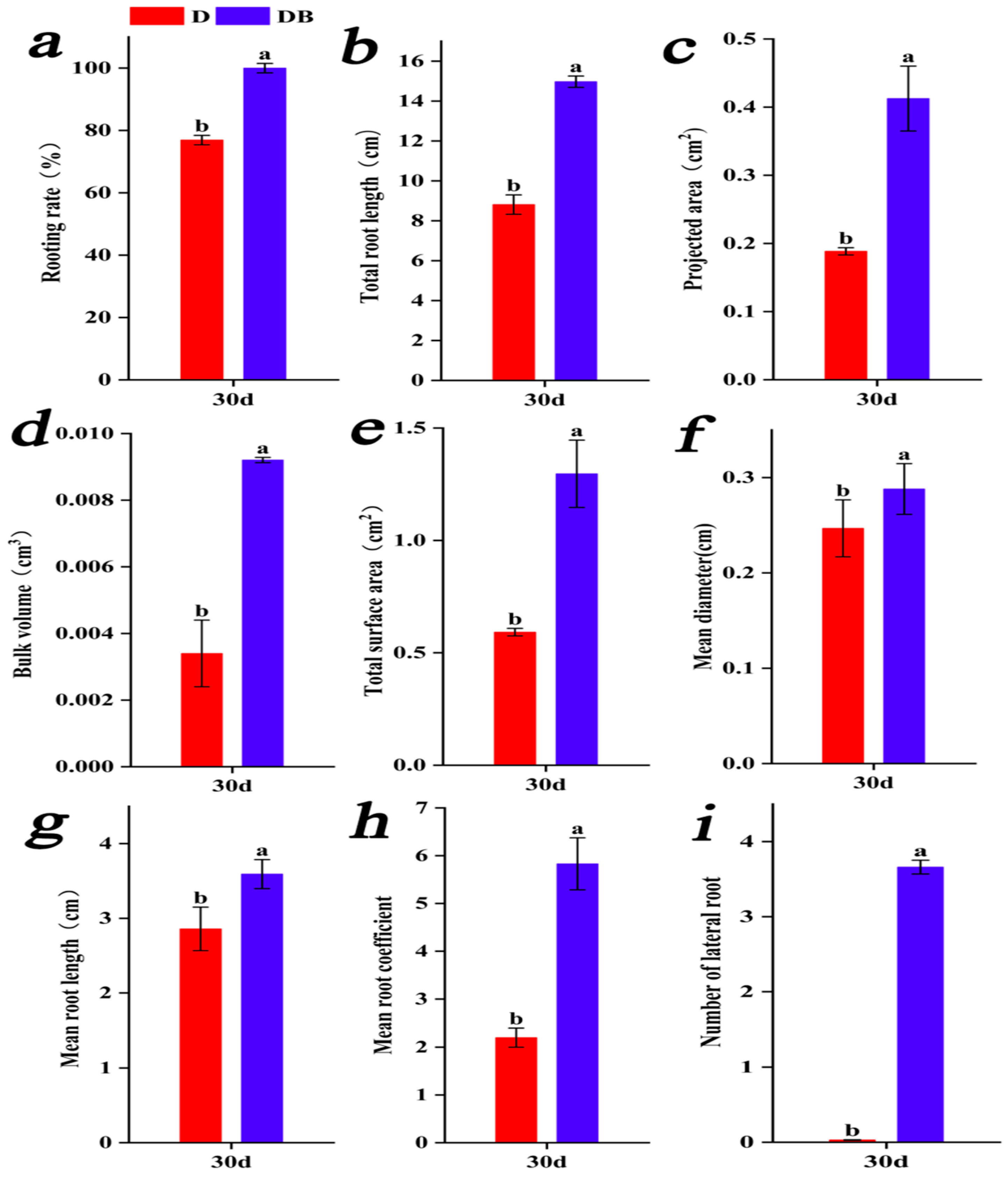
3.2. Morphological Analysis of Root System Development During Adventitious Root Formation in rolB-Transformed ‘Duli’
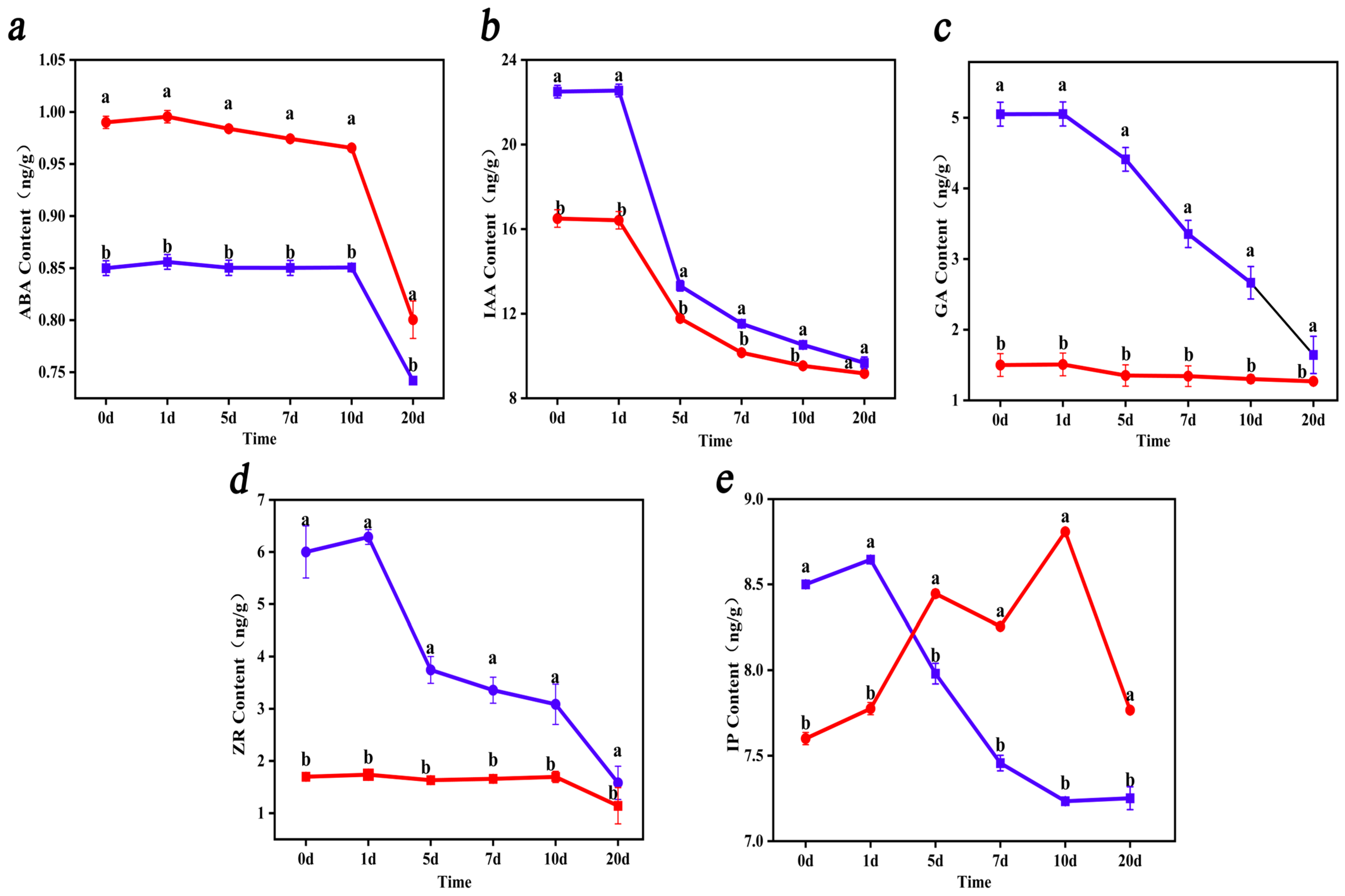
3.3. Analysis of the Endogenous Hormone Content of rolB-Transformed ‘Duli’
3.4. Transcriptomic Analysis During Adventitious Root Formation in rolB-Transformed ‘Duli’
3.4.1. Differentially Expressed Genes (DEGs) During Adventitious Root Development in rolB-Transformed ‘Duli’
3.4.2. GO Enrichment Analysis of DEGs
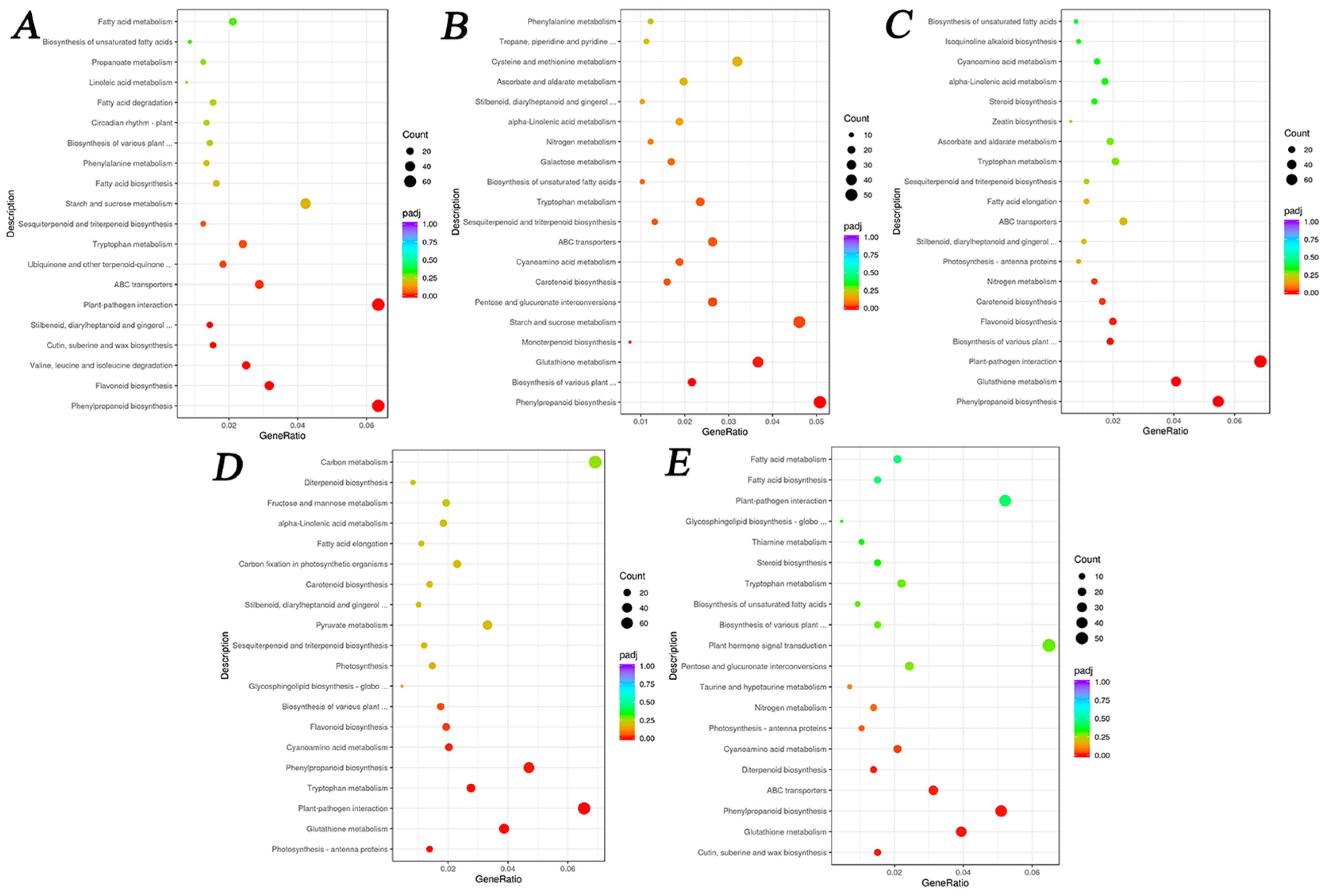
3.4.3. KEGG Pathway Enrichment Analysis of DEGs
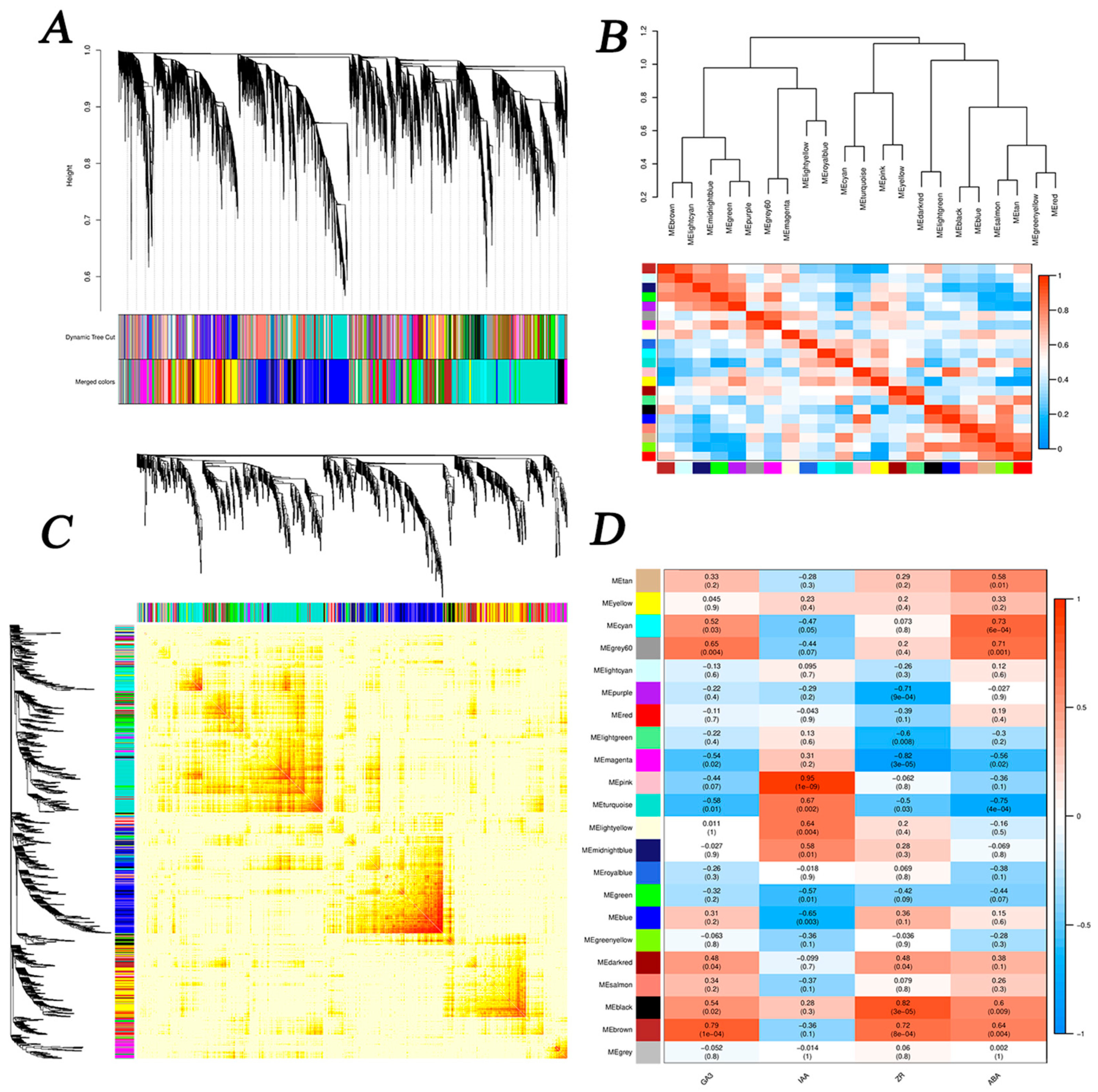
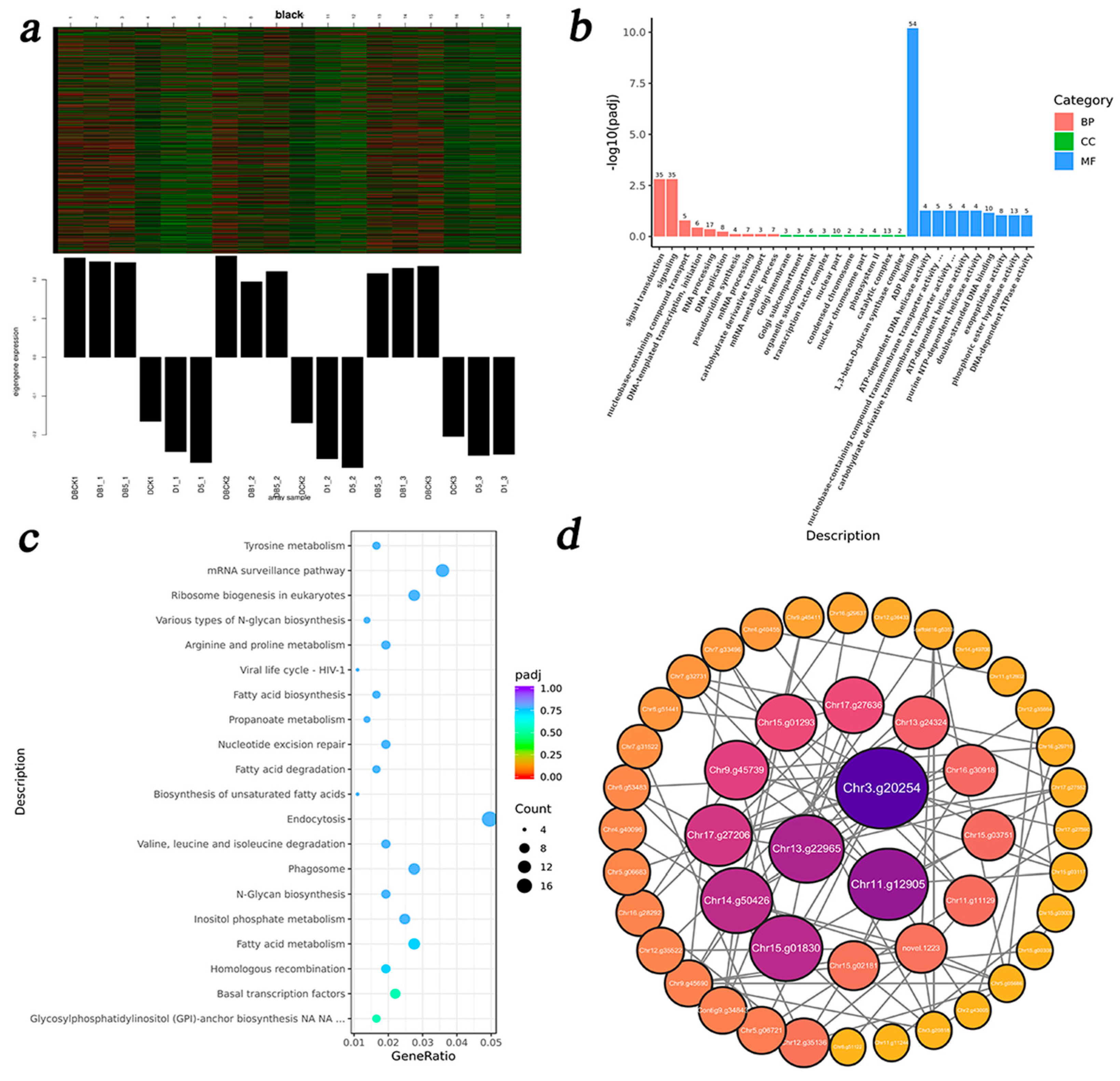
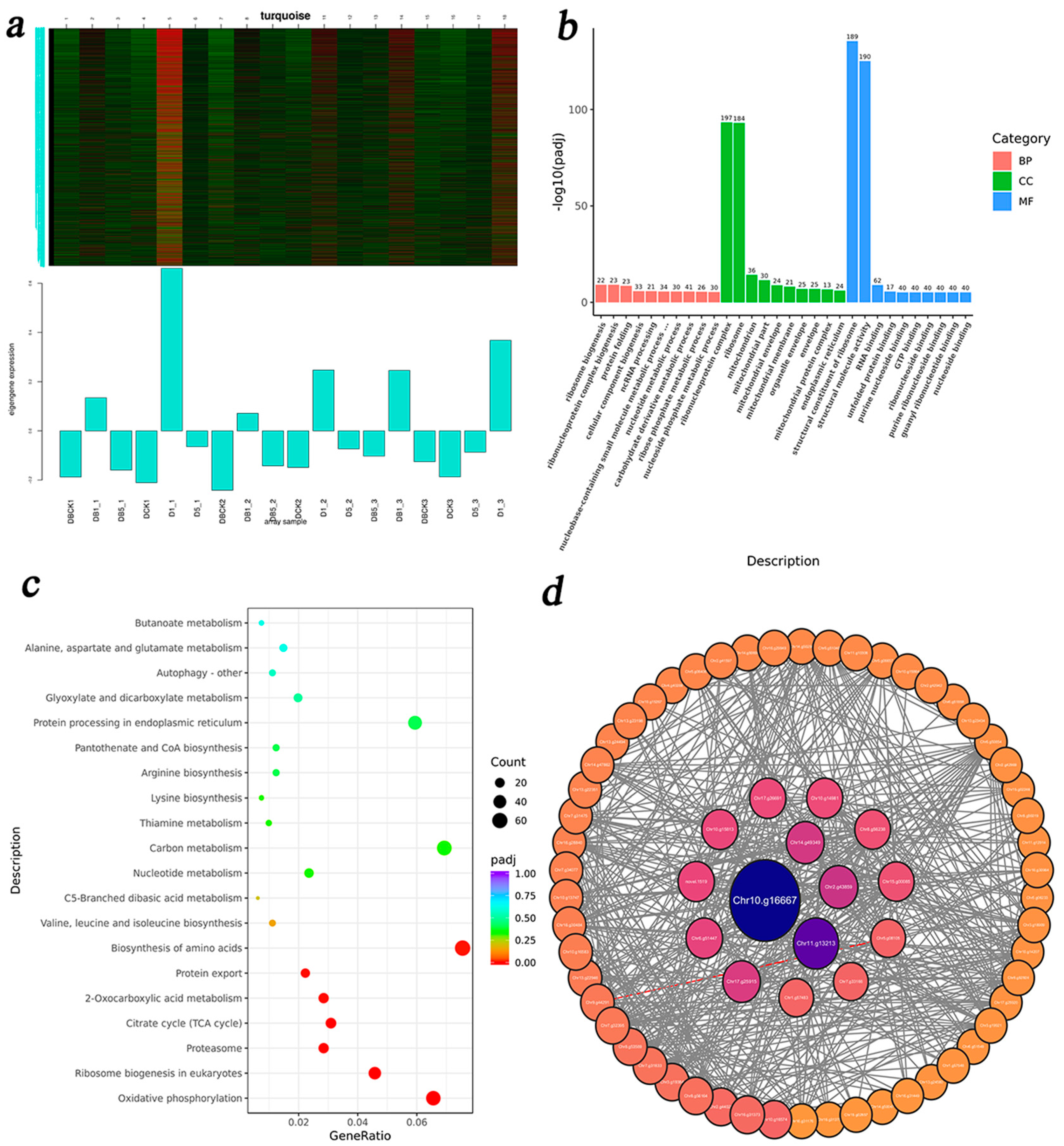
3.5. WGCNA
3.5.1. Identification of DEG Co-Expression Modules
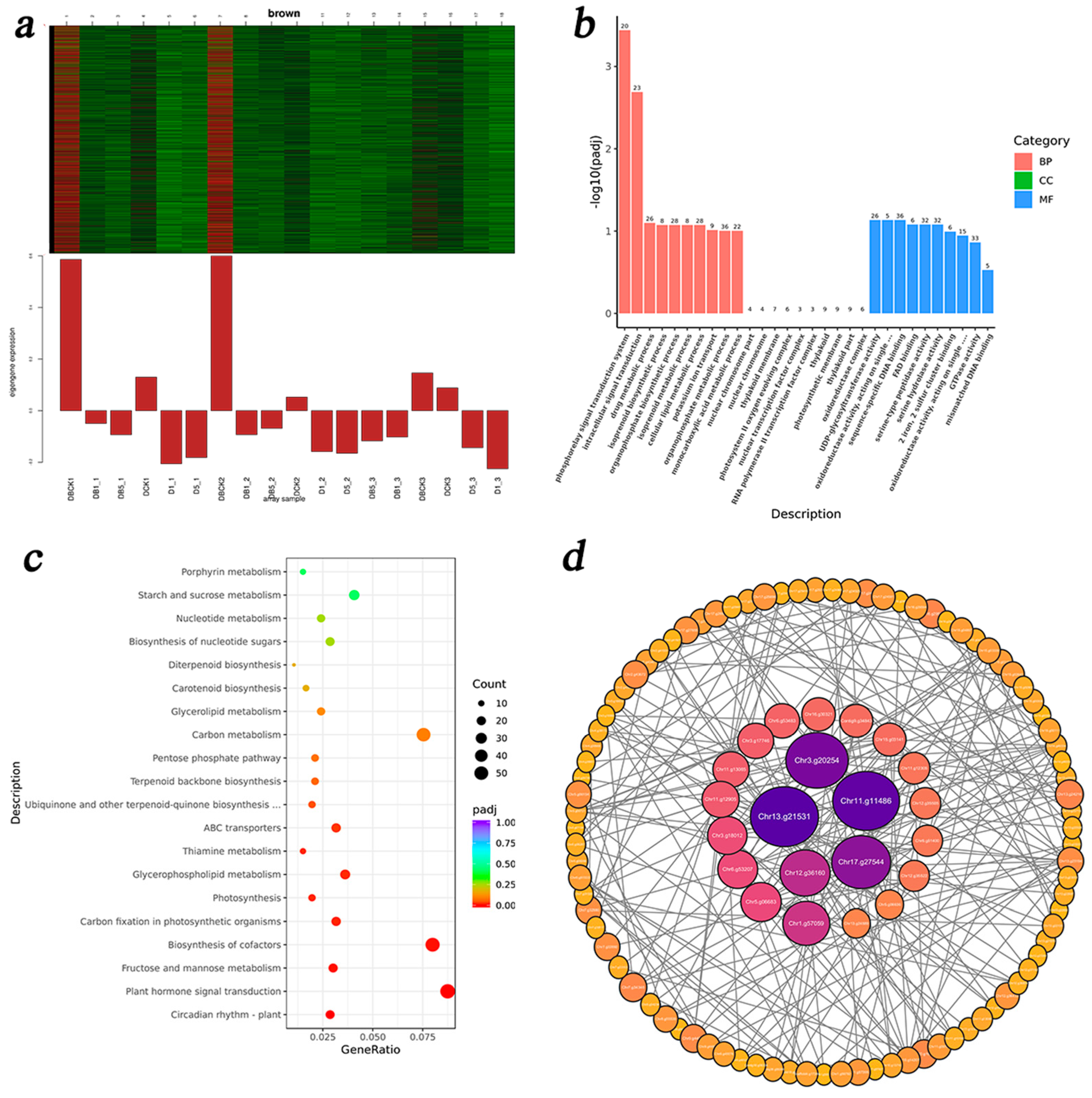
3.5.2. IAA-Related Modules and DEG Enrichment Analysis
3.5.3. DEG Enrichment Analysis of GA3-Related Modules
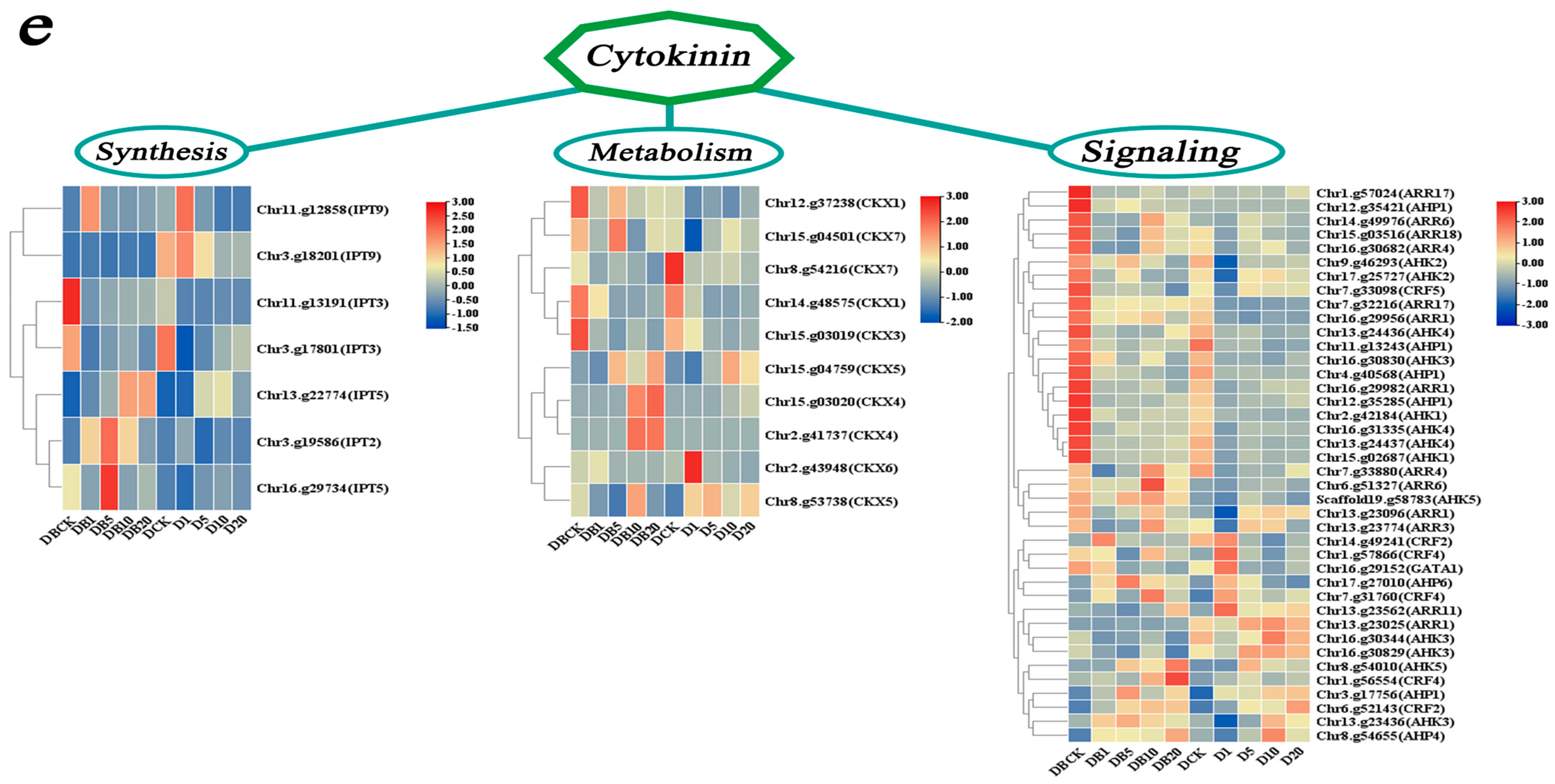
3.5.4. DEG Enrichment Analysis of ZR-Related Modules
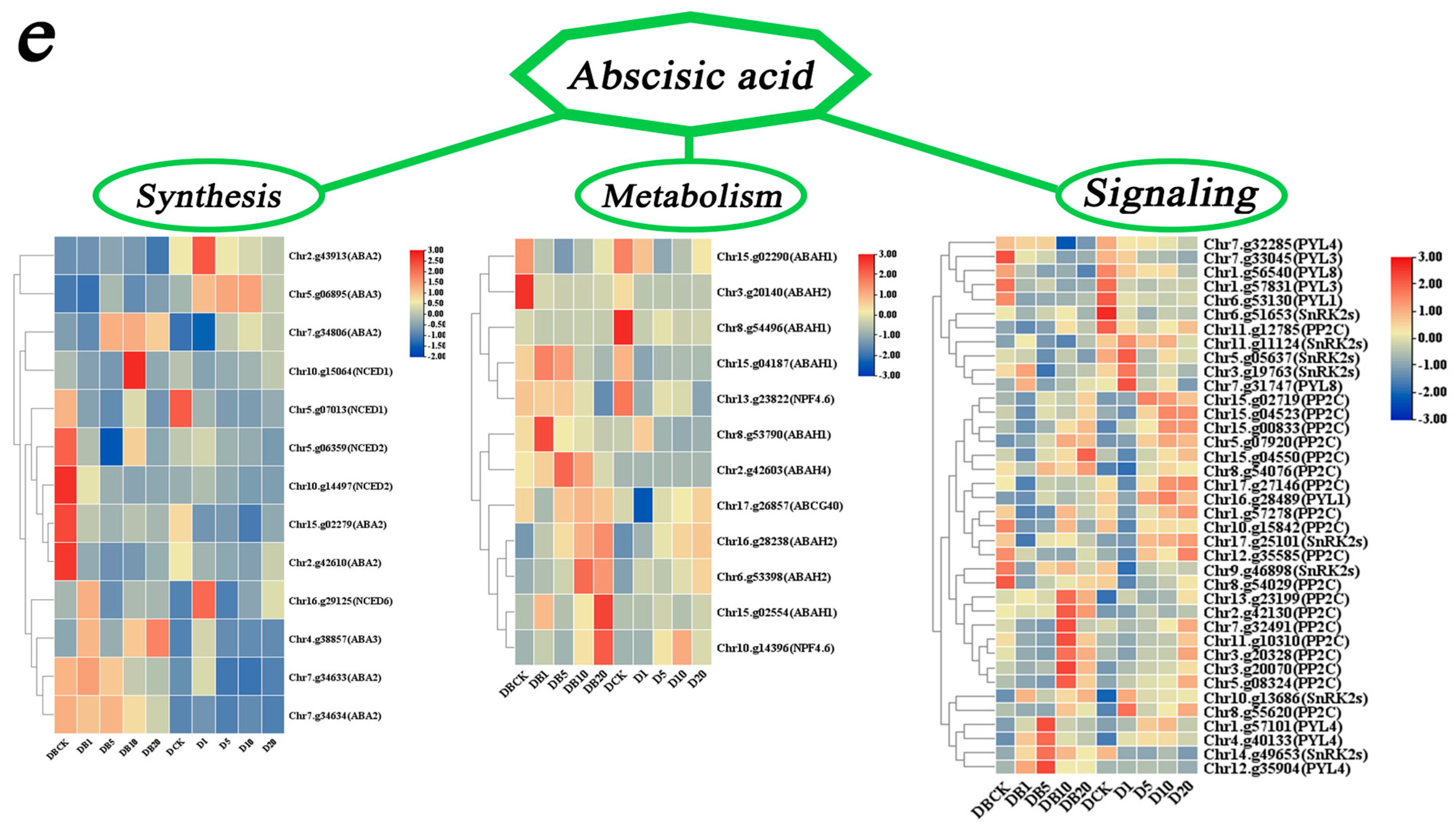
3.5.5. DEG Enrichment Analysis of ABA-Related Modules
3.6. Transcriptome Validation
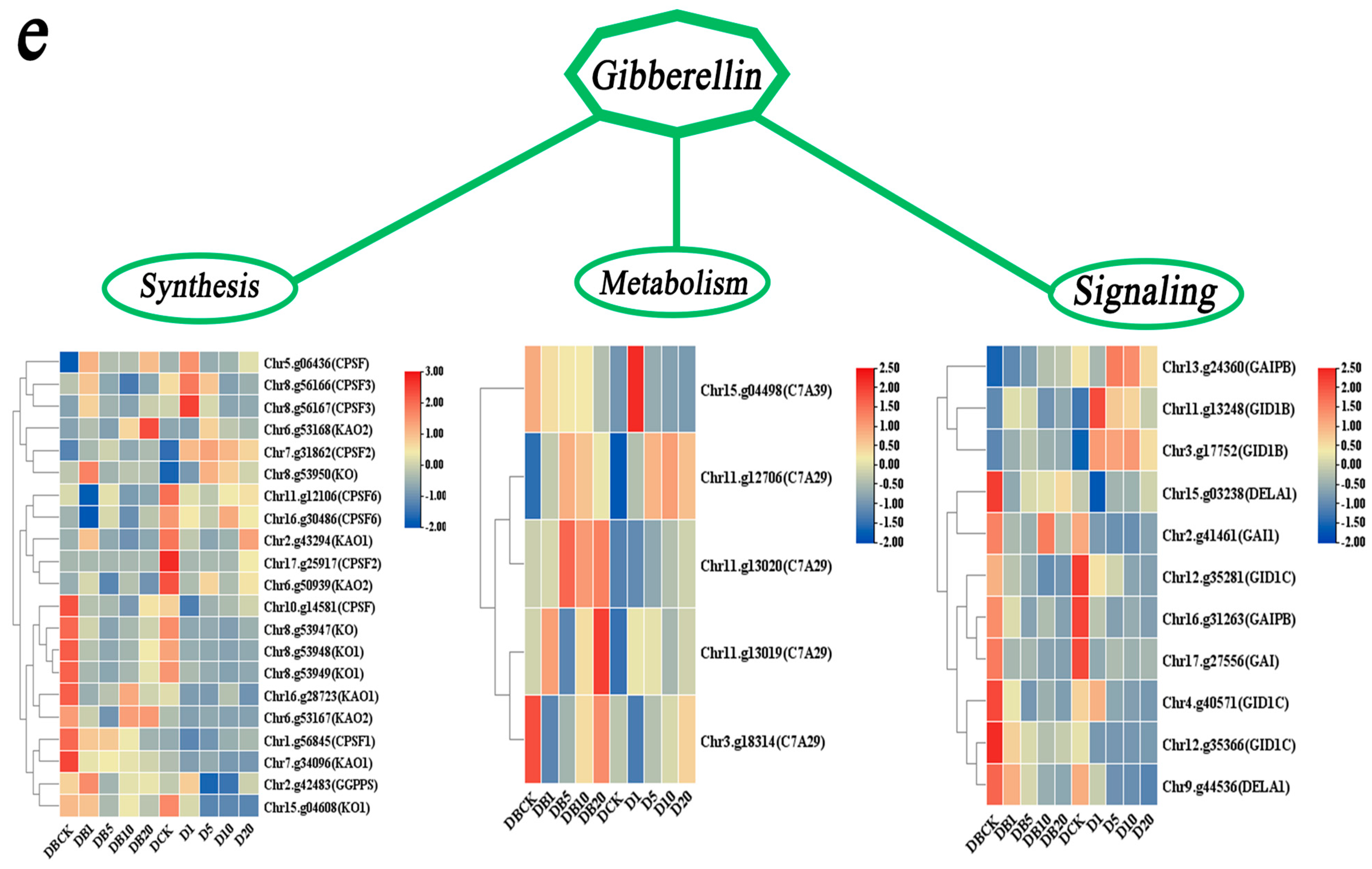
3.7. Analysis of the Molecular Mechanism by Which the rolB Gene Regulates Adventitious Root Growth Through Hormonal Control
4. Discussion
4.1. Phenotypic Analysis of Adventitious Root Development in rolB-Transformed ‘Duli’
4.2. Analysis of Key DEGs in the Adventitious Root Development of rolB-Transformed ‘Duli’
4.3. Hormonal Gene Analysis
4.3.1. IAA-Related Gene Analysis
4.3.2. Analysis of GA-Related Genes
4.3.3. Analysis of CTK-Related Genes
4.3.4. ABA-Related Gene Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bettini, P.P.; Marvasi, M.; Fani, F.; Lazzara, L.; Cosi, E.; Melani, L.; Mauro, M.L. Agrobacterium rhizogenes rolB gene affects photosynthesis and chlorophyll content in transgenic tomato (Solanum lycopersicum L.) plants. J. Plant Physiol. 2016, 204, 27–35. [Google Scholar] [CrossRef] [PubMed]
- Casanova, E.; Trillas, M.I.; Moysset, L.; Vainstein, A. Influence of rol genes in floriculture. Biotechnol. Adv. 2005, 23, 3–39. [Google Scholar] [CrossRef]
- Altamura, M.M. Agrobacterium Rhizogenes rolB and rolD genes: Regulation and involvement in plant development. Plant Cell Rep. 2004, 77, 89–101. [Google Scholar] [CrossRef]
- Zhu, L.H.; Li, X.Y.; Ahlman, A.; Welander, M. The rooting ability of the dwarfing pear rootstock BP10030 (Pyrus communis) was significantly increased by introduction of the rolB gene. Plant Sci. 2003, 165, 829–835. [Google Scholar] [CrossRef]
- Kiyokawa, S.; Kobayashi, K.; Kikuchi, Y.; Kamada, H.; Harada, H. Root-inducing region of mikimopine type Ri plasmid pRi1724. Plant Physiol. 1994, 104, 801–808. [Google Scholar] [CrossRef]
- Mauro, M.L.; Bettini, P.P. Agrobacterium rhizogenes rolB oncogene: An intriguing player for many roles. Plant Physiol. Bioch. 2021, 165, 10–18. [Google Scholar] [CrossRef] [PubMed]
- Anwar, A.; Zafar, I.; Muzaffar, M.; Mirza, B.; Murtaza, I. Epigenetic modulation: An unexplored aspect of rolB gene in transgenic plants. Russ. J. Plant Physiol. 2023, 70, 28. [Google Scholar] [CrossRef]
- Kodahl, N.; Müller, R.; Lütken, H. The Agrobacterium rhizogenes oncogenes rolB and ORF13 increase formation of generative shoots and induce dwarfism in Arabidopsis thaliana (L.) Heynh. Plant Sci. 2016, 252, 22–29. [Google Scholar] [CrossRef]
- Delbarre, A.; Muller, P.; Imhoff, V.; Barbier-Brygoo, H.; Maurel, C.; Leblanc, N.; Perrot-Rechenmann, C.; Guern, J. The rolB gene of Agrobacterium rhizogenes does not increase the auxin sensitivity of tobacco protoplasts by modifying the intracellular auxin concentration. Plant Physiol. 1994, 105, 563–569. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Vereshchagina, Y.V.; Bulgakov, D.V.; Veremeichik, G.N.; Shkryl, Y.N. The rolB plant oncogene affects multiple signaling protein modules related to hormone signaling and plant defense. Sci. Rep. 2018, 8, 2285. [Google Scholar] [CrossRef]
- Maurel, C.; Barbier-Brygoo, H.; Spena, A.; Tempé, J.; Guern, J. Single rol genes from the Agrobacterium rhizogenes TL-DNA alter some of the cellular responses to auxin in Nicotiana tabacum. Plant Physi. 1991, 97, 212–216. [Google Scholar] [CrossRef]
- Zhao, J.; Li, G.; Yi, G.X.; Wang, B.M.; Deng, A.X.; Nan, T.G.; Li, Z.H.; Li, Q.X. Comparison between conventional indirect competitive enzyme-linked immunosorbent assay (icELISA) and simplified icELISA for small molecules. Anal. Chim. Acat. 2006, 571, 79–85. [Google Scholar] [CrossRef]
- Yuan, S.; Zhang, W.L.; Zhang, Y.X. Characterization of SUPPRESSOR of MAX2 1-LIKE (SMXL) genes in ‘duli’ (Pyrus betulifolia L.) and expression analysis of PbSMXLs in response to plant growth regulators and salt stress. Agronomy 2024, 14, 2778. [Google Scholar] [CrossRef]
- Yuan, S.; Zhang, W.L.; Zhang, Y.X. Characterization of PHT genes in ‘duli’ (Pyrus betulifolia Bunge) and expression analysis of PbPHTs in response to plant growth regulators, P, and salt stress. Agriculture 2025, 15, 199. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.H.; Lin, H.H.; Liu, C.T.; Lin, T.C.; Liu, L.Y.D.; Lee, K.T. Transcriptomic analysis reveals that reactive oxygen species and genes encoding lipid transfer protein are associated with tobacco hairy root growth and branch development. Mol. Plant-Microbe Interact. 2014, 27, 678–687. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.H.; Yoon, E.S.; Jeong, J.H.; Choi, Y.E. Agrobacterium rhizogenes-mediated transformation of Taraxacum platycarpum and changes of morphological characters. Plant Cell Rep. 2004, 22, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Grigolon, S.; Bravi, B.; Martin, O.C. Plant responses to auxin signals: An operating principle for dynamical sensitivity yet high resilience. PLoS Comput. Biol. 2017, 13, e1005543. [Google Scholar]
- De Klerk, G.J.; Hanecakova, J.; Jasik, J. The role of cytokinins in rooting of stem slices cut from apple microcuttings. Plant Biosyst. 2001, 135, 79–84. [Google Scholar] [CrossRef]
- Guo, D.; Liang, J.; Li, L. Abscisic acid (ABA) inhibition of lateral root formation involves endogenous ABA biosynthesis in Arachis hypogaea L. Plant Growth Regul. 2009, 58, 173–179. [Google Scholar] [CrossRef]
- Sun, T.; Rao, S.; Zhou, X.; Li, L. Plant carotenoids: Recent advances and future perspectives. Mol. Hortic. 2002, 2, 3. [Google Scholar]
- Brunetti, C.; Fini, A.; Sebastiani, F.; Gori, A.; Tattini, M. Modulation of phytohormone signaling: A primary function of flavonoids in plant-environment interactions. Front. Plant Sci. 2018, 9, 1042. [Google Scholar] [CrossRef]
- Yoda, H.; Yamaguchi, Y.; Sano, H. Induction of hypersensitive cell death by hydrogen peroxide produced through polyamine degradation in tobacco plants. Plant Physiol. 2003, 132, 1973–1981. [Google Scholar] [CrossRef]
- Noctor, G.; Cohen, M.; Trémulot, L.; Châtel-Innocenti, G.; Van Breusegem, F.; Mhamdi, A. Glutathione: A key modulator of plant defence and metabolism through multiple mechanisms. J. Exp. Bot. 2024, 75, 4549–4572. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA gene family in plants: Molecular structure, regulation, and function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Cheng, W.; Zhang, M.; Cheng, T.; Wang, J.; Zhang, Q. Genome-wide identification of Aux/IAA gene family and their expression analysis in Prunus mume. Front. Genet. 2022, 13, 1013822. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Jung, J.H.; Han, D.Y.; Seo, P.J.; Park, W.J.; Park, C.M. Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 2012, 235, 923–938. [Google Scholar] [PubMed]
- Lange, T. Molecular biology of gibberellin synthesis. Planta 1998, 204, 409–419. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Huang, Y.; Yang, W.; Pei, Z.; Guo, X.; Liu, D.; Sun, J.; Zhang, A. The genes for gibberellin biosynthesis in wheat. Funct. Integr. Genom. 2012, 12, 199–206. [Google Scholar]
- Linder, S.J.; Bernasocchi, T.; Martínez-Pastor, B.; Sullivan, K.D.; Galbraith, M.D.; Lewis, C.A.; Ferrer, C.M.; Boon, R.; Silveira, G.G.; Cho, H.M.; et al. Inhibition of the proline metabolism rate-limiting enzyme P5CS allows proliferation of glutamine-restricted cancer cells. Nat. Metab. 2023, 5, 2131–2147. [Google Scholar] [CrossRef]
- Rai, A.N.; Penna, S. Molecular evolution of plant P5CS gene involved in proline biosynthesis. Mol. Biol. Rep. 2013, 40, 6429–6435. [Google Scholar] [CrossRef] [PubMed]
- Slugina, M.A.; Efremov, G.I.; Shchennikova, A.V.; Kochieva, E.Z. Characterization of RIN isoforms and their expression in tomato fruit ripening. Cells 2021, 10, 1739. [Google Scholar] [CrossRef] [PubMed]
- Meekins, D.A.; Vander Kooi, C.W.; Gentry, M.S. Structural mechanisms of plant glucan phosphatases in starch metabolism. FEBS J. 2016, 283, 2427–2447. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.M.; Sun, H.; Xia, H.F.; Wu, T.T.; Li, P.C.; Xu, C.W.; Yang, Z.F. Natural variation and domestication selection of ZmCKX5 with root morphological traits at the seedling stage in maize. Plants 2020, 10, 1. [Google Scholar] [CrossRef]
- Galuszka, P.; Frébort, I.; Šebela, M.; Peč, P. Degradation of cytokinins by cytokinin oxidases in plants. Plant Growth Regul. 2000, 32, 315–327. [Google Scholar] [CrossRef]
- McGaw, B.A.; Horgan, R. Cytokinin catabolism and cytokinin oxidase. Phytochemistry 1983, 22, 1103–1105. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef]
- Antoniadi, I.; Mateo-Bonmatí, E.; Pernisová, M.; Brunoni, F.; Antoniadi, M.; Villalonga, M.G.; Ament, A.; Karády, M.; Turnbull, C.; Doležal, K.; et al. IPT9, a cis-zeatin cytokinin biosynthesis gene, promotes root growth. Front. Plant Sci. 2002, 13, 932008. [Google Scholar] [CrossRef]
- Schmülling, T.; Werner, T.; Riefler, M.; Krupková, E.; Manns, I.B. Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 2003, 116, 241–252. [Google Scholar] [CrossRef] [PubMed]
- Müller, B.; Sheen, J. Arabidopsis cytokinin signaling pathway. Sci. Signal 2007, 2007, cm5. [Google Scholar]
- Higuchi, M.; Pischke, M.S.; Mähönen, A.P.; Miyawaki, K.; Hashimoto, Y.; Seki, M.; Kobayashi, M.; Shinozaki, K.; Kato, T.; Tabata, S.; et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl. Acad. Sci. USA 2004, 101, 8821–8826. [Google Scholar] [CrossRef] [PubMed]
- Argyros, R.D.; Mathews, D.E.; Chiang, Y.H.; Palmer, C.M.; Thibault, D.M.; Etheridge, N.; Argyros, D.A.; Mason, M.G.; Kieber, J.J.; Schaller, G.E. Type B response regulators of Arabidopsis play key roles in cytokinin signaling and plant development. Plant Cell 2008, 20, 2102–2116. [Google Scholar] [CrossRef]
- Willems, A.; De Veylder, L. The plant anaphase-promoting complex/cyclosome. Annu. Rev. Cell Dev. Biol. 2022, 38, 25–48. [Google Scholar] [CrossRef]
- Kwee, H.S.; Sundaresan, V. The NOMEGA gene required for female gametophyte development encodes the putative APC6/CDC16 component of the anaphase promoting complex in Arabidopsis. Plant J. 2003, 36, 853–866. [Google Scholar] [CrossRef]
- Zhao, M.; Ma, L.; Song, N.; Cheng, J.; Zhao, Z.; Wu, J. The regulation of Alternaria alternata resistance by LRR-RK4 through ERF109, defensin19, and phytoalexin scopoletin in Nicotiana attenuata. Plant Sci. 2022, 323, 111414. [Google Scholar] [CrossRef]
- Jwa, M.; Kim, J.H.; Chan, C.S. Regulation of Sli15/INCENP, kinetochore, and Cdc14 phosphatase functions by the ribosome biogenesis protein Utp7. J. Cell Biol. 2008, 182, 1099–1111. [Google Scholar] [CrossRef]
- Gao, Q.; Xu, S.; Zhu, X.; Wang, L.; Yang, Z.; Zhao, X. Genome-wide identification and characterization of the RIO atypical kinase family in plants. Genes Genom. 2018, 40, 669–683. [Google Scholar] [CrossRef]
- Shimizu, T.; Kanno, Y.; Suzuki, H.; Watanabe, S.; Seo, M. Arabidopsis NPF4.6 and NPF5.1 control leaf stomatal aperture by regulating abscisic acid transport. Genes 2021, 12, 885. [Google Scholar] [CrossRef]
- Borghi, L.; Kang, J.; Ko, D.; Lee, Y.; Martinoia, E. The role of ABCG-type ABC transporters in phytohormone transport. Biochem. Soc. Trans. 2015, 43, 924–930. [Google Scholar] [CrossRef]
- Orman-Ligeza, B.; Morris, E.C.; Parizot, B.; Lavigne, T.; Babé, A.; Ligeza, A.; Klein, S.; Sturrock, C.; Xuan, W.; Novák, O.; et al. The xerobranching response represses lateral root formation when roots are not in contact with water. Curr. Biol. 2018, 28, 3165–3173. [Google Scholar] [CrossRef]
- Zhao, Y.; Xing, L.; Wang, X.; Hou, Y.J.; Gao, J.; Wang, P.; Duan, C.G.; Zhu, X.; Zhu, J.K. The ABA receptor PYL8 promotes lateral root growth by enhancing MYB77-dependent transcription of auxin-responsive genes. Sci. Signal. 2014, 7, ra53. [Google Scholar] [CrossRef]
- Zeng, Y.; Verstraeten, I.; Trinh, H.K.; Heugebaert, T.; Stevens, C.V.; Garcia-Maquilon, I.; Rodriguez, P.L.; Vanneste, S.; Geelen, D. Arabidopsis hypocotyl adventitious root formation is suppressed by ABA signaling. Genes 2021, 12, 1141. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.; Xiong, L. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 2012, 586, 1742–1747. [Google Scholar] [CrossRef]



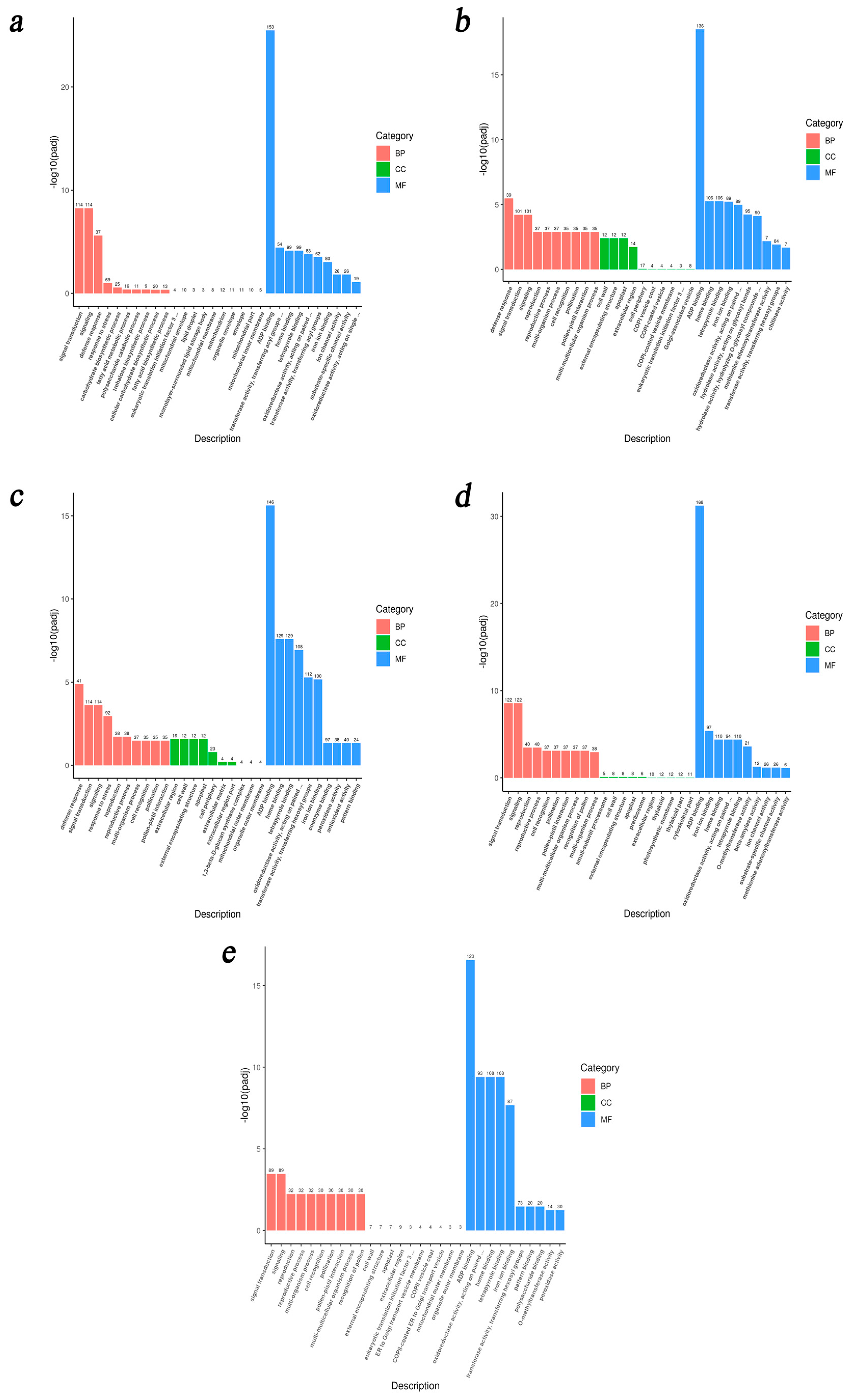

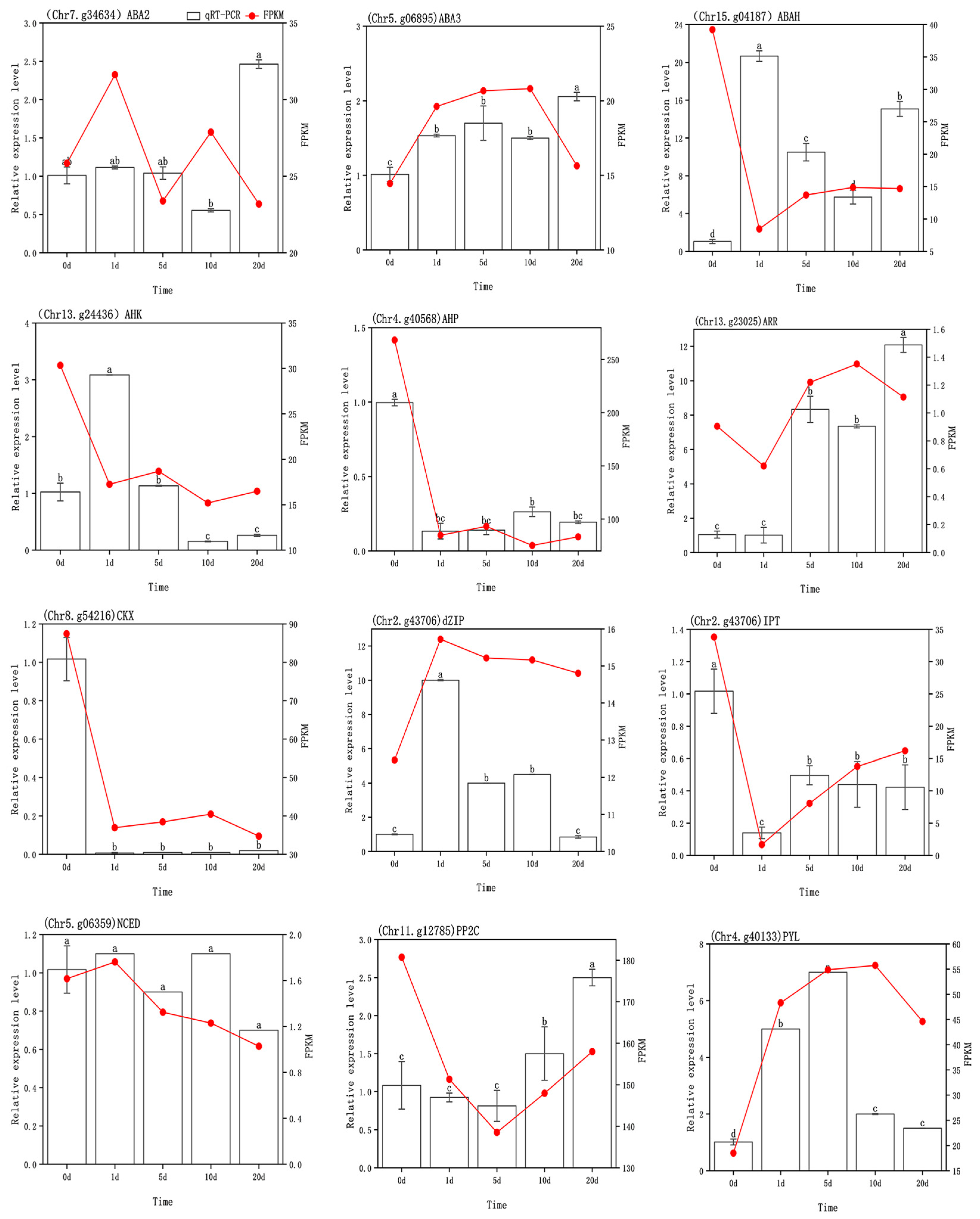
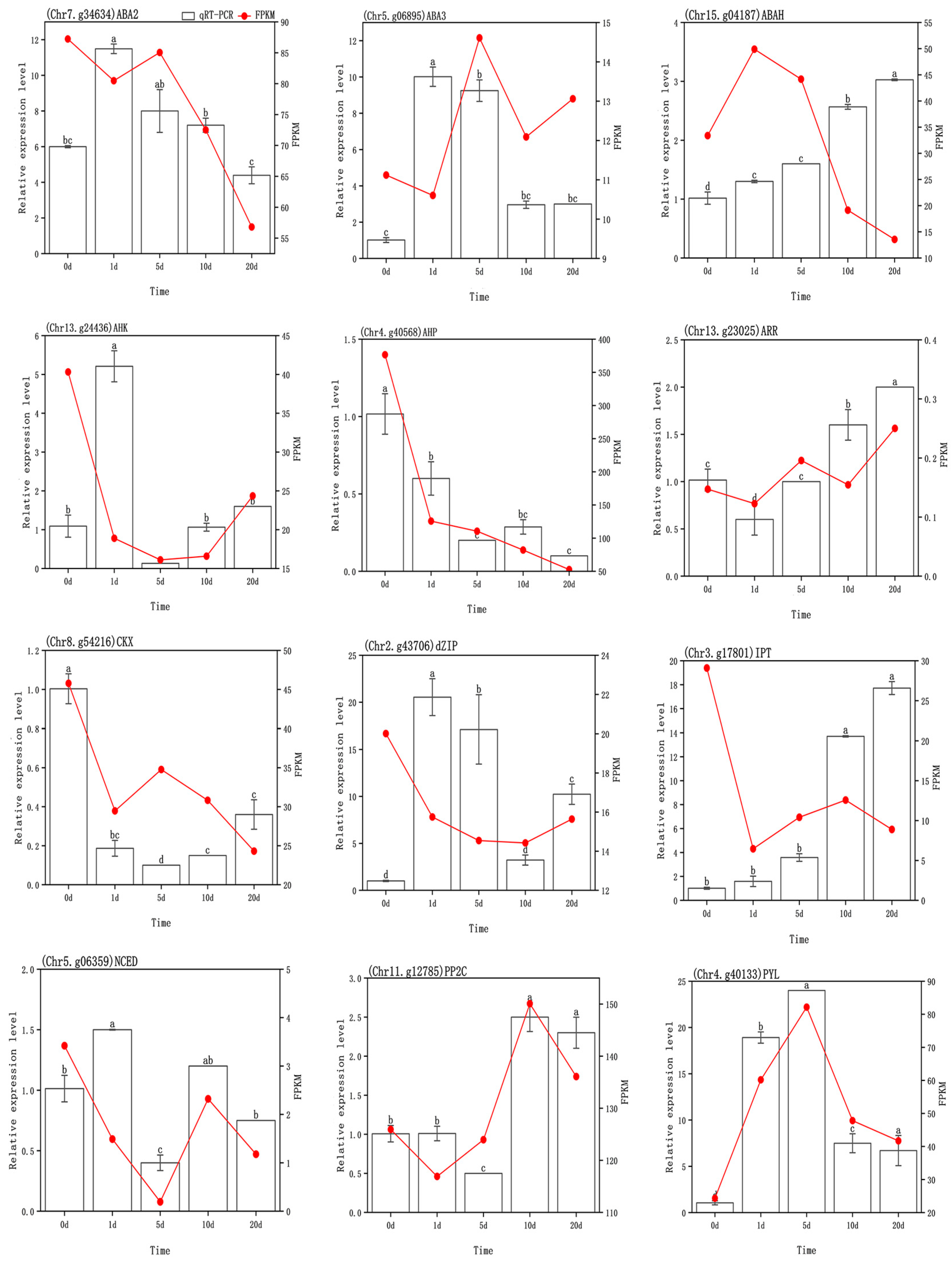
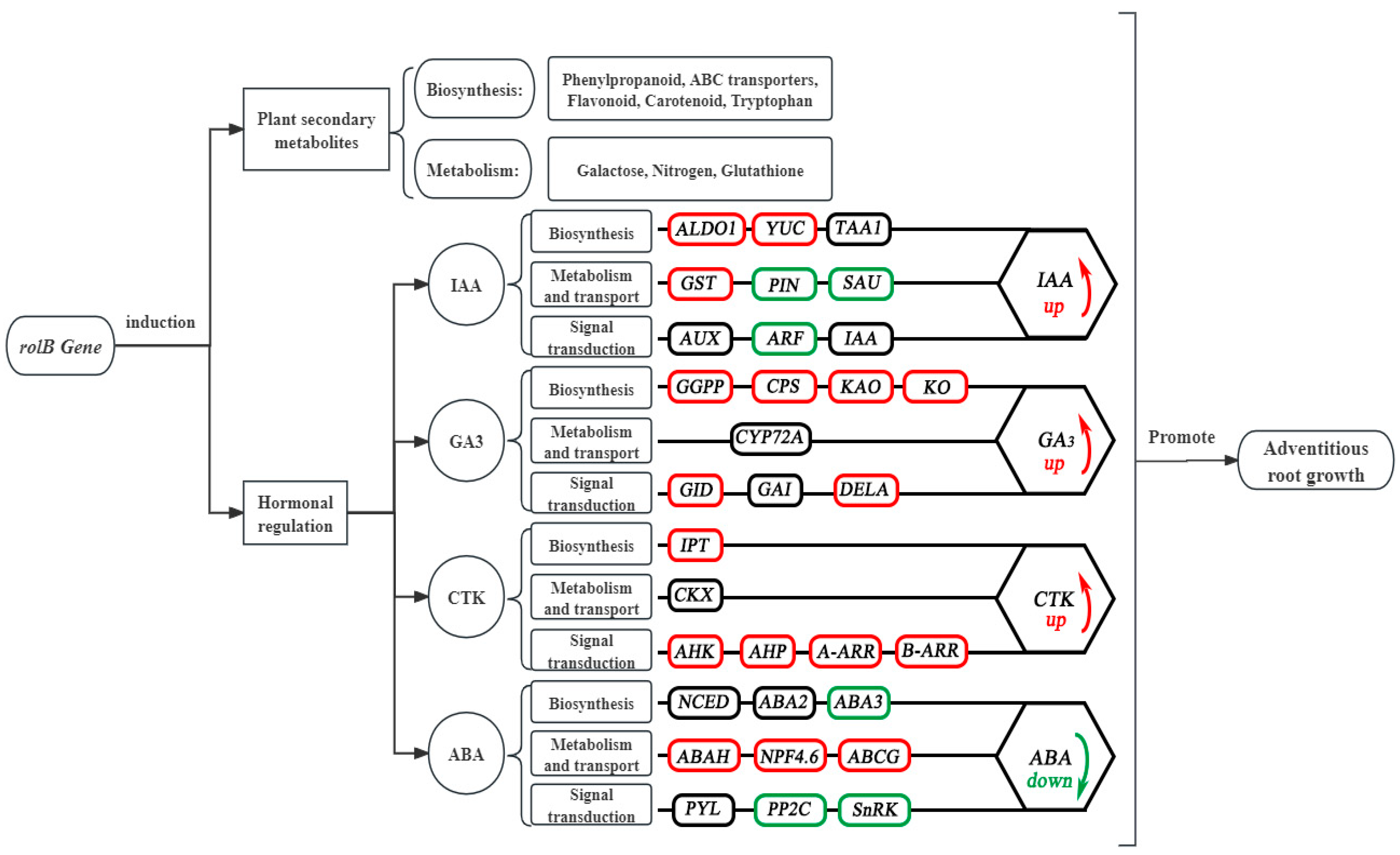
| Comparison | Biological Process (BP) | Cellular Component (CC) | Molecular Function (MF) |
|---|---|---|---|
| DB0vsD0 | 1533 | 409 | 2323 |
| DB1vsD1 | 1587 | 394 | 2288 |
| DB5vsD5 | 1853 | 478 | 2721 |
| DB10vsD10 | 1674 | 431 | 2467 |
| DB20vsD20 | 1353 | 345 | 2007 |
| Gene ID | Gene Name | Forward Primer 5′−3′ | Reverse Primer 5′−3′ |
|---|---|---|---|
| Chr7.g34634 | ABA2 | AGGACAACCTCGGCTTAC | TACCGTGACATCACAATGGA |
| Chr5.g06895 | ABA3 | TTGGCTACTCTAGCATCG | GGCTCTACATCTTGTCCC |
| Chr5.g06359 | NCED | AAAATGTACGGCGGTGAG | AGTTCTTCTCGTCGTGAACGAACGT |
| Chr15.g04187 | ABA8′-H | CCTGGGATGTCCAAGTGT | TGACGGACCAATCAAACG |
| Chr4.g40133 | PYL | CCGCCCAAATCATCAATC | ACTGTGGCCCGCTTCTTA |
| Chr11.g12785 | PP2C | TTTAATTGACTGCCAGAAG | TTTGATACGAAACCACCT |
| Chr2.g43706 | dZIP | GGTTGATGCGAATGTGG | TTGGCGGTGGTAAAGG |
| Chr8.g54216 | CKX | CGTTGGCGGAGCATTT | ACGCACCGTTTCAGCAC |
| Chr3.g17801 | IPT | ACTCGTGCTCGGTTACTA | CAGATTCCACCCTTTGATA |
| Chr13.g23025 | ARR | ATGTTCCTGGGCTTACTA | TCAAATTATTCTGGTGCTG |
| Chr13.g24436 | AHK | CTATGACGGCGGATGT | CGAAGGGCTTTGAGAC |
| Chr4.g40568 | AHP | TGCGATGAGCAGAACG | ACCTTGGAAGCGAACC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, T.; Wang, W.; Nie, K.; He, Z.; He, J.; Zhang, Y.; Liu, N.; Li, Y. rolB Promotes Adventitious Root Development in Pyrus betulaefolia by Modulating Endogenous Hormones and Gene Expression. Agronomy 2025, 15, 2165. https://doi.org/10.3390/agronomy15092165
Xie T, Wang W, Nie K, He Z, He J, Zhang Y, Liu N, Li Y. rolB Promotes Adventitious Root Development in Pyrus betulaefolia by Modulating Endogenous Hormones and Gene Expression. Agronomy. 2025; 15(9):2165. https://doi.org/10.3390/agronomy15092165
Chicago/Turabian StyleXie, Ting, Weimin Wang, Kuozhen Nie, Zijuan He, Jiaojiao He, Yuxing Zhang, Na Liu, and Yingli Li. 2025. "rolB Promotes Adventitious Root Development in Pyrus betulaefolia by Modulating Endogenous Hormones and Gene Expression" Agronomy 15, no. 9: 2165. https://doi.org/10.3390/agronomy15092165
APA StyleXie, T., Wang, W., Nie, K., He, Z., He, J., Zhang, Y., Liu, N., & Li, Y. (2025). rolB Promotes Adventitious Root Development in Pyrus betulaefolia by Modulating Endogenous Hormones and Gene Expression. Agronomy, 15(9), 2165. https://doi.org/10.3390/agronomy15092165





