Genetic Dissection of Hypocotyl Elongation Responses to Light Quality in Brassica napus
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Plant Growth and Measurements of Hypocotyl Length
2.3. GWAS Analysis
2.4. Linkage Disequilibrium Analysis and Haplotype Analysis
2.5. Statistical Analysis
3. Results
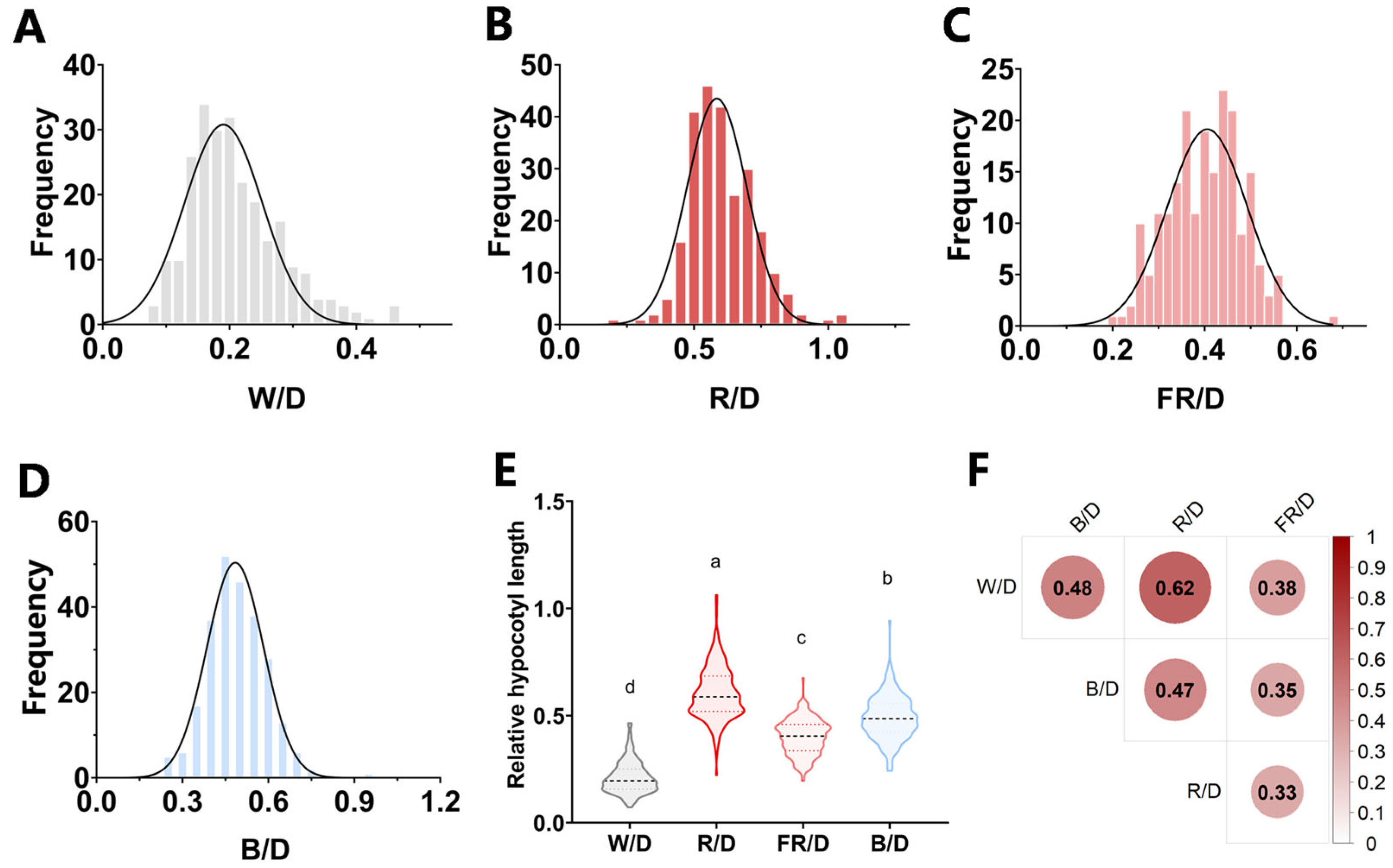
3.1. Phenotypic Variation in Hypocotyl Length Among 267 Rapeseed Accessions Exposed to Divergent Light Qualities
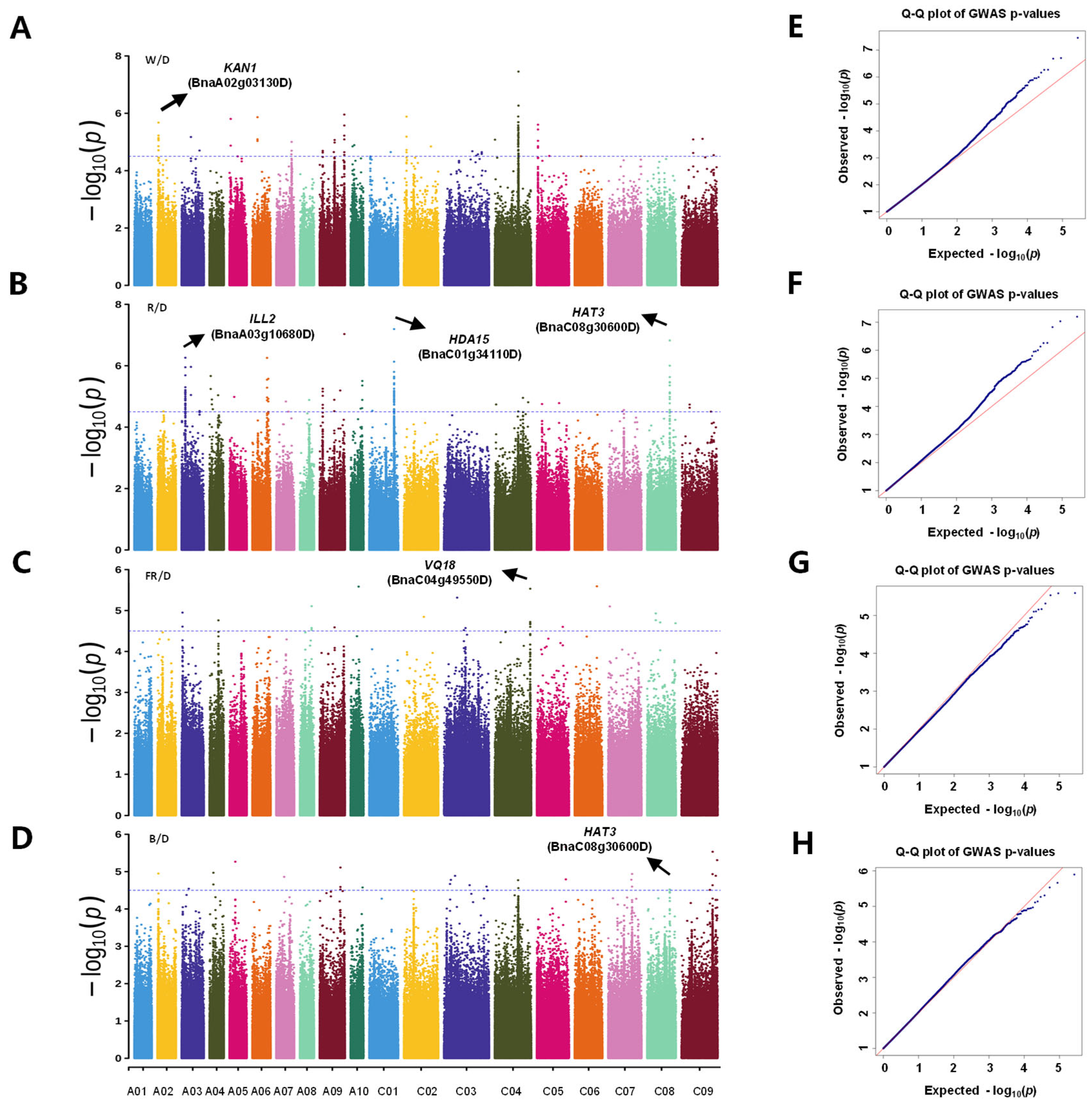
3.2. Genome-Wide Association Analysis of Relative Hypocotyl Length Under Different Light Qualities
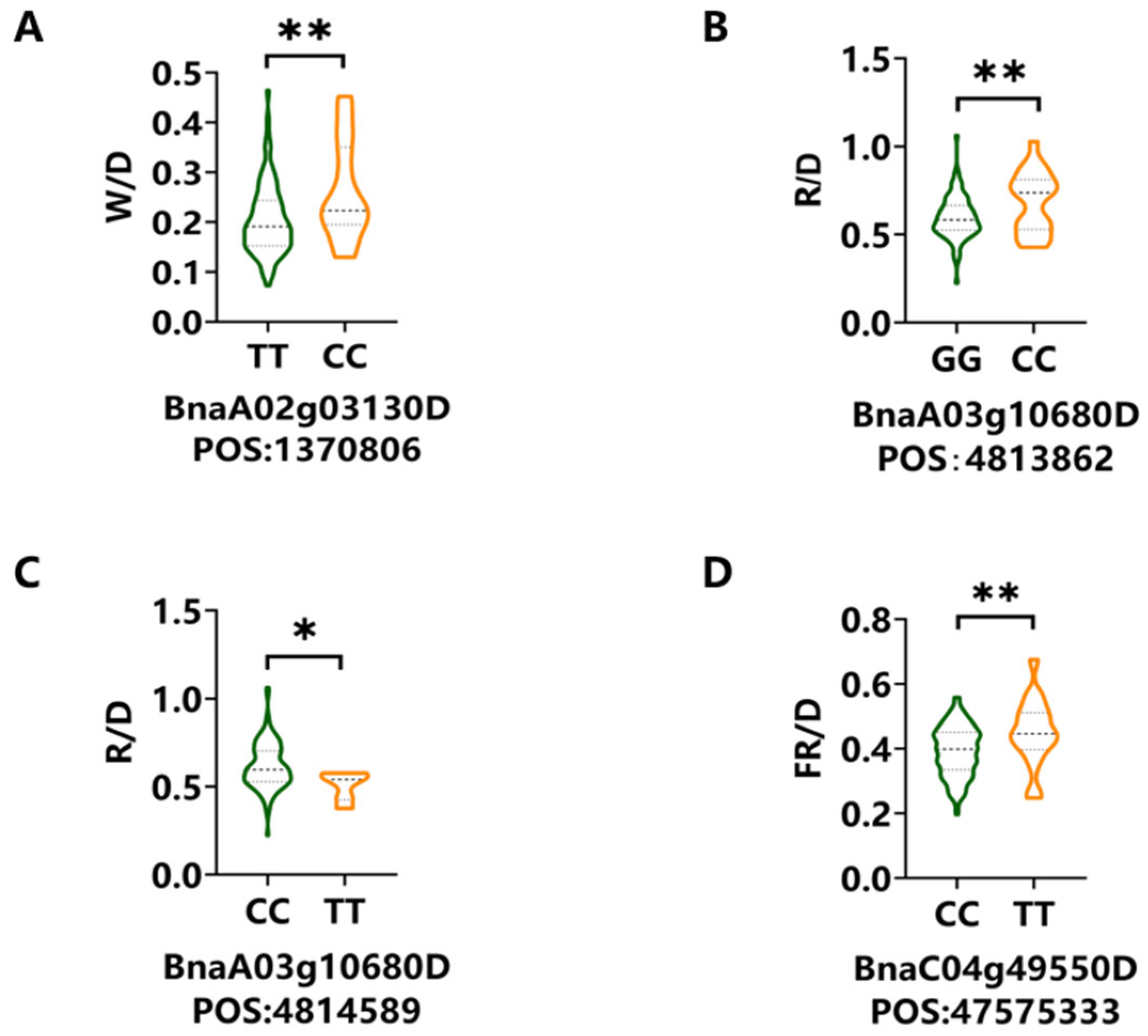
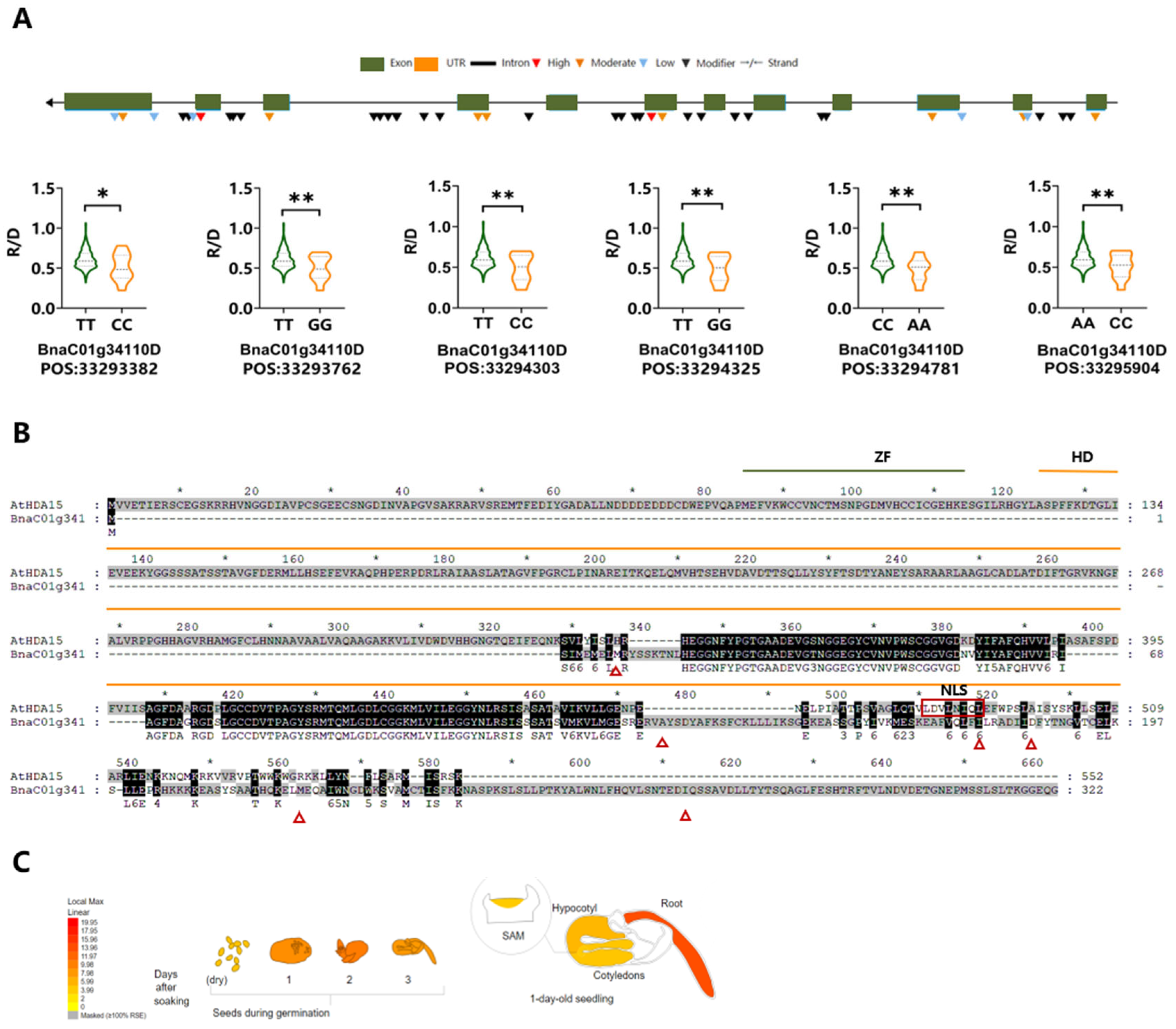
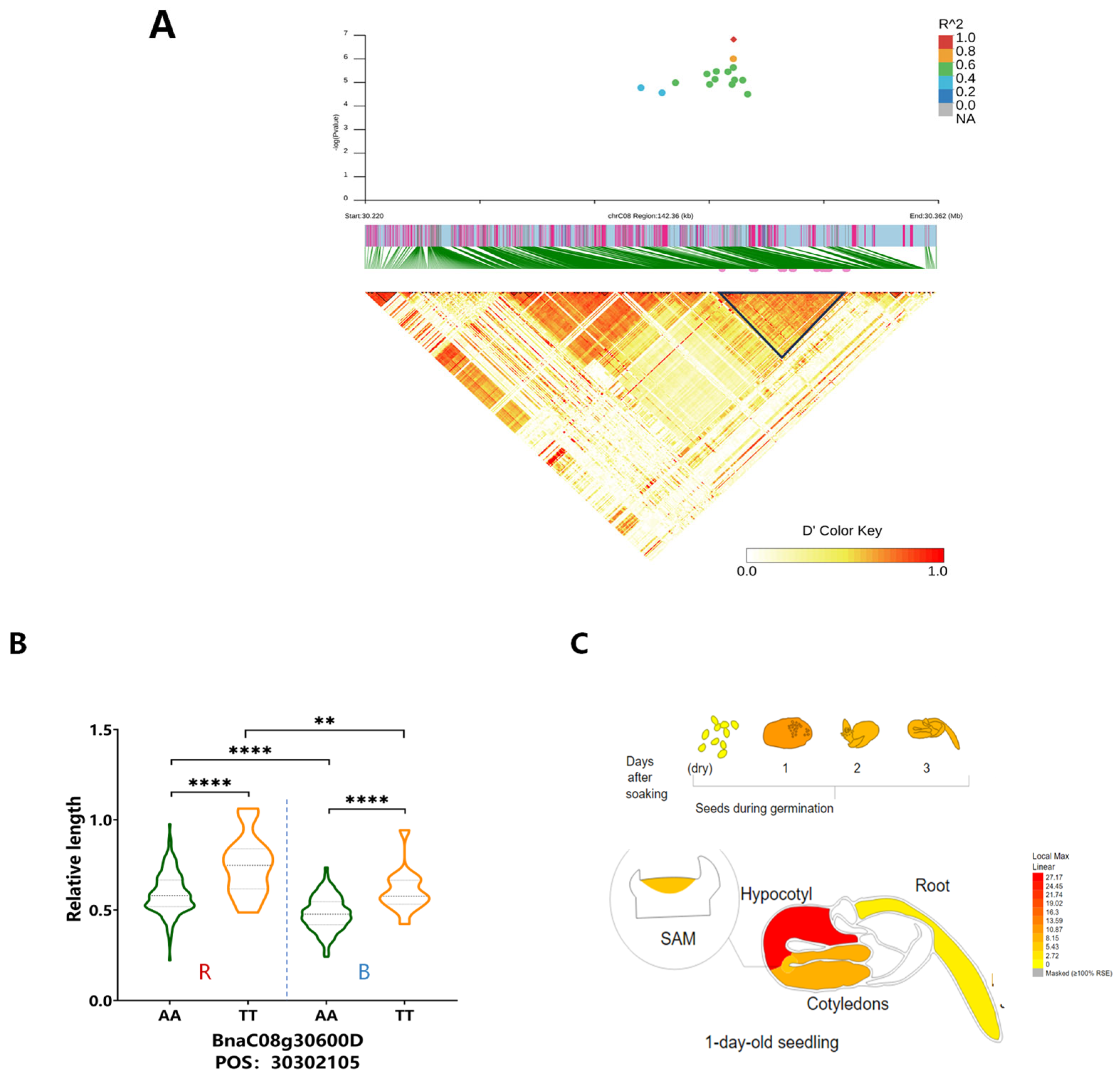
3.3. Key Candidate Genes Identified Under Different Light Conditions
4. Discussion
4.1. Light Quality Differentially Affects Plant Growth in B. napus
4.2. Identification of Genes Involved in Light-Mediated Inhibition of Hypocotyl Elongation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Nagaharu, U. Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. J. Exp. Bot. 1935, 7, 389–452. [Google Scholar]
- Smith, H.; Whitelam, G. The shade avoidance syndrome: Multiple responses mediated by multiple phytochromes. Plant Cell Environ. 1997, 20, 840–844. [Google Scholar] [CrossRef]
- Cheng, M.C.; Kathare, P.K.; Paik, I.; Huq, E. Phytochrome signaling networks. Annu. Rev. Plant Biol. 2021, 72, 217–244. [Google Scholar] [CrossRef] [PubMed]
- McNellis, T.W.; Deng, X.W. Light control of seedling morphogenetic pattern. Plant Cell 1995, 7, 1749–1761. [Google Scholar] [PubMed]
- Briggs, W.R. Editorial: In the light of day: Plant photomorphogenesis. Molecular Plant 2008, 1, 2–3. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Chory, J. Photomorphogenesis. Arab. Book. 2002, 1, e0054. [Google Scholar] [CrossRef]
- Nemhauser, J.L. Dawning of a new era: Photomorphogenesis as an integrated molecular network. Curr. Opin. Plant Biol. 2008, 11, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Lyu, M.; Luo, Y.; Liu, S.; Li, Y.; He, H.; Wei, N.; Deng, X.W.; Zhong, S. Genome-wide regulation of light-controlled seedling morphogenesis by three families of transcription factors. Proc. Natl. Acad. Sci. USA 2018, 115, 6482–6487. [Google Scholar] [CrossRef]
- Buti, S.; Hayes, S.; Pierik, R. The bHLH network underlying plant shade-avoidance. Physiol. Plant 2020, 169, 312–324. [Google Scholar] [CrossRef]
- Quail, P.H. Photosensory perception and signalling in plant cells: New paradigms? Curr. Opin. Cell Biol. 2002, 14, 180–188. [Google Scholar] [CrossRef]
- Izaguirre, M.M.; Mazza, C.A.; Biondini, M.; Baldwin, I.T.; Ballare, C.L. Remote sensing of future competitors: Impacts on plant defenses. Proc. Natl. Acad. Sci. USA 2006, 103, 7170–7174. [Google Scholar] [CrossRef]
- Franklin, K.A. Shade avoidance. New Phytol. 2008, 179, 930–944. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Garcia, J.F.; Galstyan, A.; Salla-Martret, M.; Cifuentes-Esquivel, N.; Gallemi, M.; Bou-Torrent, J. Regulatory components of shade avoidance syndrome. Adv. Bot. Res. 2010, 53, 65–116. [Google Scholar]
- Fraser, D.P.; Panter, P.E.; Sharma, A.; Sharma, B.; Dodd, A.N.; Franklin, K.A. Phytochrome a elevates plant circadian-clock components to suppress shade avoidance in deep-canopy shade. Proc. Natl. Acad. Sci. USA 2021, 118, e2108176118. [Google Scholar] [CrossRef]
- Jang, I.T.; Lee, J.H.; Shin, E.J.; Nam, S.Y. Evaluation of growth, flowering, and chlorophyll fluorescence responses of viola cornuta cv. penny red wing according to spectral power distributions. J. People Plants Environ. 2023, 26, 335–349. [Google Scholar] [CrossRef]
- Li, L.; Ljung, K.; Breton, G.; Schmitz, R.J.; Pruneda-Paz, J.; Cowing-Zitron, C.; Cole, B.J.; Ivans, L.J.; Pedmale, U.V.; Jung, H.S.; et al. Linking photoreceptor excitation to changes in plant architecture. Genes Dev. 2012, 26, 785–790. [Google Scholar] [CrossRef]
- Roig-Villanova, I.; Bou-Torrent, J.; Galstyan, A.; Carretero-Paulet, L.; Portolés, S.; Rodríguez-Concepción, M.; Martínez-García, J.F. Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J. 2007, 26, 4756–4767. [Google Scholar] [CrossRef]
- Hornitschek, P.; Kohnen, M.V.; Lorrain, S.; Rougemont, J.; Ljung, K.; López-Vidriero, I.; Franco-Zorrilla, J.M.; Solano, R.; Trevisan, M.; Pradervand, S.; et al. Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J. 2012, 71, 699–711. [Google Scholar] [CrossRef] [PubMed]
- Sharrock, R.A.; Clack, T. Patterns of expression and normalized levels of the five Arabidopsis phytochromes. Plant Physiol. 2002, 130, 442–456. [Google Scholar] [CrossRef]
- Castillon, A.; Shen, H.; Huq, E. Phytochrome Interacting Factors: Central players in phytochrome-mediated light signaling networks. Trends Plant Sci. 2007, 12, 514–521. [Google Scholar] [CrossRef] [PubMed]
- Duek, P.D.; Fankhauser, C. bHLH class transcription factors ake centre stage in phytochrome signalling. Trends Plant Sci. 2005, 10, 51–54. [Google Scholar] [CrossRef]
- Paik, I.; Yang, S.; Choi, G. Phytochrome regulates translation of mRNA in the cytosol. Proc. Natl. Acad. Sci. USA 2012, 109, 1335–1340. [Google Scholar] [CrossRef]
- Gu, D.; Chen, C.Y.; Zhao, M.; Zhao, L.; Duan, X.; Duan, J.; Wu, K.; Liu, X. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 2017, 45, 7137–7150. [Google Scholar] [CrossRef]
- Paulišić, S.; Qin, W.; Arora Verasztó, H.; Then, C.; Alary, B.; Nogue, F.; Tsiantis, M.; Hothorn, M.; Martínez-García, J.F. Adjustment of the PIF7-HFR1 transcriptional module activity controls plant shade adaptation. EMBO J. 2021, 40, e104273. [Google Scholar] [CrossRef]
- Jiao, Y.; Lau, O.S.; Deng, X.W. Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 2007, 8, 217–230. [Google Scholar] [CrossRef] [PubMed]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef]
- Luo, X.; Xue, Z.; Ma, C.; Hu, K.; Zeng, Z.; Dou, S.; Tu, J.; Shen, J.; Yi, B.; Fu, T. Joint genome-wide association and transcriptome sequencing reveals a complex polygenic network underlying hypocotyl elongation in rapeseed (Brassica napus L.). Sci. Rep. 2017, 7, 41561. [Google Scholar] [CrossRef]
- Wu, D.; Liang, Z.; Yan, T.; Xu, Y.; Xuan, L.; Tang, J.; Zhou, G.; Lohwasser, U.; Hua, S.; Wang, H.; et al. Whole-genome resequencing of a worldwide collection of rapeseed accessions reveals the genetic basis of ecotype divergence. Mol. Plant 2019, 12, 30–43. [Google Scholar] [CrossRef]
- Wang, H.; Wang, Q.; Pak, H.; Yan, T.; Chen, M.; Chen, X.; Wu, D.; Jiang, L. Genome-wide association study reveals a patatin-like lipase relating to the reduction of seed oil content in Brassica napus. BMC Plant Biol. 2021, 21, 6. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wu, G.; Zhang, J.; Hu, W.; Yao, X.; Jiang, L.; Zhu, Y. Integration of GWAS and transcriptome analysis to identify temperature-dependent genes involved in germination of rapeseed (Brassica napus L.). Front. Plant Sci. 2025, 16, 1551317. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.S.; He, W.M.; Ji, J.J.; Zhang, C.; Guo, Y.; Yang, T.L. LDBlockShow: A fast and convenient tool for visualizing linkage disequilibrium and haplotype blocks based on variant call format files. Brief. Bioinform. 2021, 22, bbaa227. [Google Scholar] [CrossRef]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Klepikova, A.V.; Kasianov, A.S.; Gerasimov, E.S.; Logacheva, M.D.; Penin, A.A. A high resolution map of the Arabidopsis thaliana developmental transcriptome based on RNA-seq profiling. Plant J. 2016, 88, 1058–1070. [Google Scholar] [CrossRef]
- Zhang, X.; Fang, T.M.; Huang, Y.Q.; Sun, W.Y.; Cai, S.G. Transcriptional regulation of photomorphogenesis in seedlings of Brassica napus under different light qualities. Planta 2022, 256, 77. [Google Scholar] [CrossRef]
- Berger, S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. 2002, 12, 142–148. [Google Scholar] [CrossRef]
- Liu, X.; Chen, C.Y.; Wang, K.C.; Luo, M.; Tai, R.; Yuan, L.; Zhao, M.; Yang, S.; Tian, G.; Cui, Y.; et al. PHYTOCHROME INTERACTING FACTOR3 associates with the histone deacetylase HDA15 in repression of chlorophyll biosynthesis and photosynthesis in etiolated Arabidopsis seedlings. Plant Cell 2013, 25, 1258–1273. [Google Scholar] [CrossRef]
- Zhao, L.; Peng, T.; Chen, C.Y.; Ji, R.; Gu, D.; Li, T.; Zhang, D.; Tu, Y.T.; Wu, K.; Liu, X. HY5 Interacts with the Histone Deacetylase HDA15 to Repress Hypocotyl Cell Elongation in Photomorphogenesis. Plant Physiol. 2019, 180, 1450–1466. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Chang, C.H.; Wu, C.H.; Tu, Y.T.; Wu, K. Arabidopsis cyclin-dependent kinase C2 interacts with HDA15 and is involved in far-red light-mediated hypocotyl cell elongation. Plant J. 2022, 112, 1462–1472. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Jing, Y.; Gao, Y.; Liu, Y.; Fang, X.; Lin, R. The PIF1/PIF3-MED25-HDA19 transcriptional repression complex regulates phytochrome signaling in Arabidopsis. New Phytol. 2023, 240, 1097–1115. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Wang, H.; Hu, Y.; Yu, D. Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J. 2018, 95, 529–544. [Google Scholar] [CrossRef]
- Neha, B.; Henrik, Å.; Henrik, J.; Marcus, G.H. Quantitative analysis of auxin sensing in leaf primordia argues against proposed role in regulating leaf dorsoventrality. ELife 2019, 8, e39298. [Google Scholar]
- Preciado, J.; Begcy, K.; Liu, T. The Arabidopsis HDZIP class II transcription factor ABA INSENSITIVE TO GROWTH 1 functions in leaf development. J. Exp. Bot. 2022, 73, 1978–1991. [Google Scholar] [CrossRef] [PubMed]
- Ohgishi, M.; Oka, A.; Morelli, G.; Ruberti, I.; Aoyama, T. Negative autoregulation of the Arabidopsis homeobox gene ATHB-2. Plant J. 2001, 25, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S.; Ohgishi, M.; Goda, H.; Higuchi, K.; Shimada, Y.; Yoshida, S.; Koshiba, T. The HAT2 gene, a member of the HD-Zip gene family, isolated as an auxin inducible gene by DNA microarray screening, affects auxin response in Arabidopsis. Plant J. 2002, 32, 1011–1022. [Google Scholar] [CrossRef] [PubMed]
- Sorin, C.; Salla-Martret, M.; Bou-Torrent, J.; Roig-Villanova, I.; Martínez-García, J.F. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J. 2009, 59, 266–277. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhou, Y.; Wan, Q.; Huang, T.; Hu, Z.; Zhang, X.; Cai, S.; Zhao, H. Genetic Dissection of Hypocotyl Elongation Responses to Light Quality in Brassica napus. Agronomy 2025, 15, 2047. https://doi.org/10.3390/agronomy15092047
Zhou Y, Wan Q, Huang T, Hu Z, Zhang X, Cai S, Zhao H. Genetic Dissection of Hypocotyl Elongation Responses to Light Quality in Brassica napus. Agronomy. 2025; 15(9):2047. https://doi.org/10.3390/agronomy15092047
Chicago/Turabian StyleZhou, Yichen, Qi Wan, Tonghao Huang, Zengjie Hu, Xin Zhang, Shengguan Cai, and Huifang Zhao. 2025. "Genetic Dissection of Hypocotyl Elongation Responses to Light Quality in Brassica napus" Agronomy 15, no. 9: 2047. https://doi.org/10.3390/agronomy15092047
APA StyleZhou, Y., Wan, Q., Huang, T., Hu, Z., Zhang, X., Cai, S., & Zhao, H. (2025). Genetic Dissection of Hypocotyl Elongation Responses to Light Quality in Brassica napus. Agronomy, 15(9), 2047. https://doi.org/10.3390/agronomy15092047






