Keel Petal Fusion in Soybean: Anatomical Insights and Transcriptomic Identification of Candidate Regulators
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
2.2. Light and SEM
2.3. Histochemical Analysis
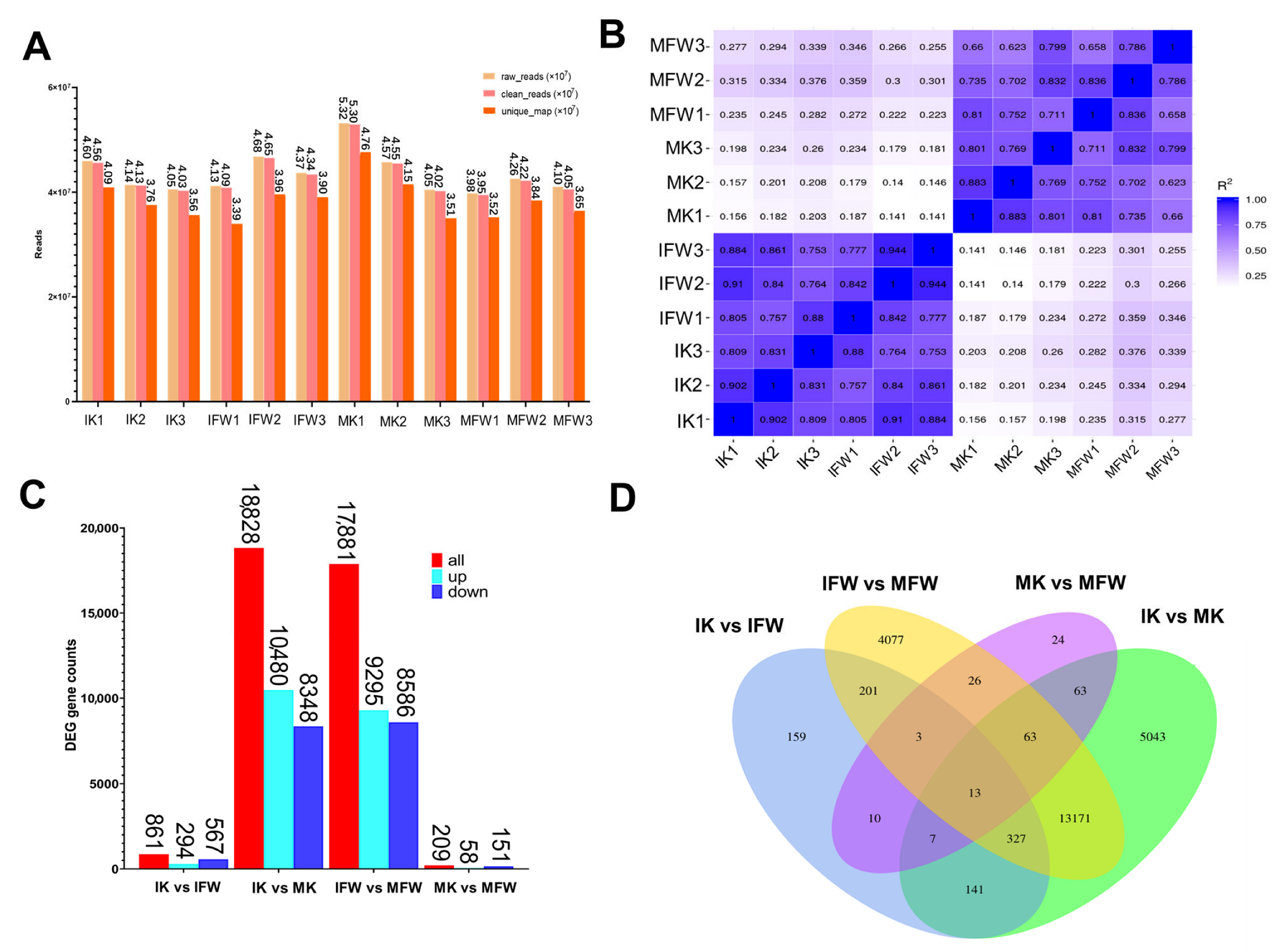
2.4. RNA Isolation, Library Construction, and Sequencing
2.5. Analysis of Differentially Expressed Genes (DEGs)
2.6. Validation and Functional Analysis of Candidate Gene Expression in Tissues
3. Results
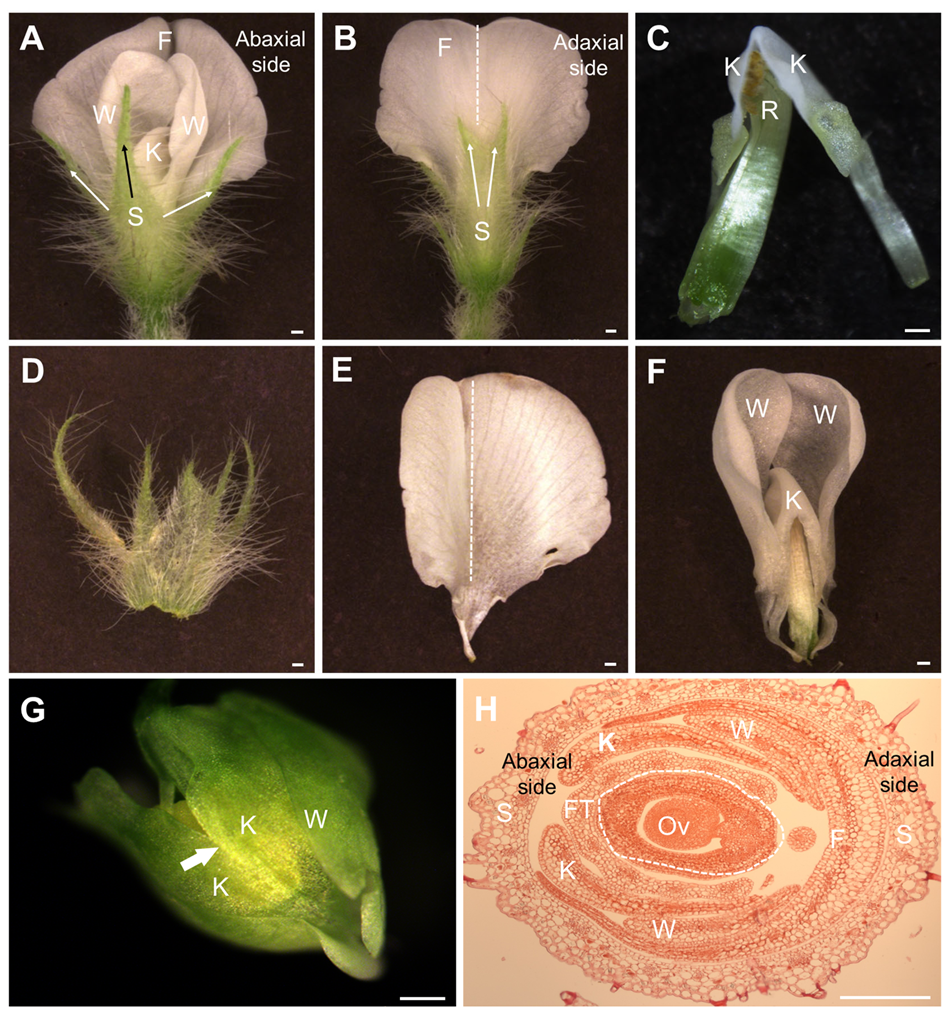
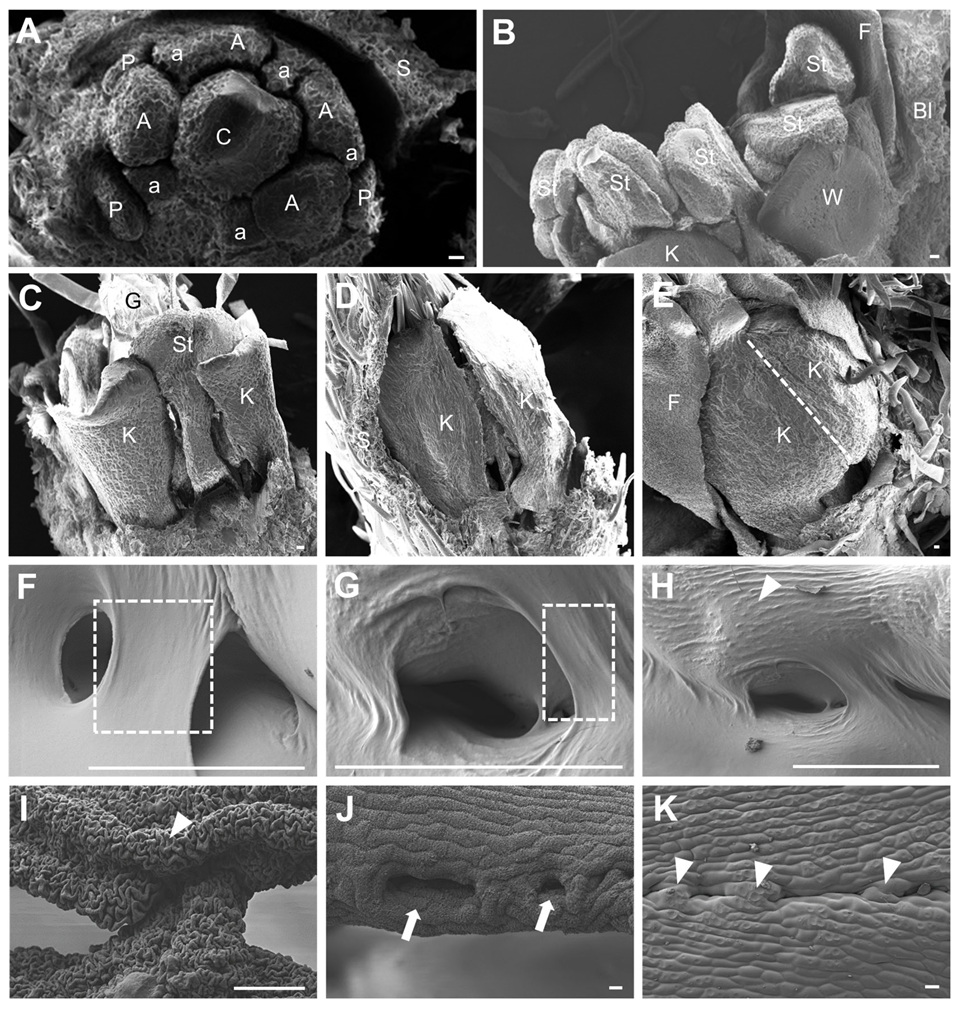
3.1. Morphology of the Papilionaceous Corolla of Soybean
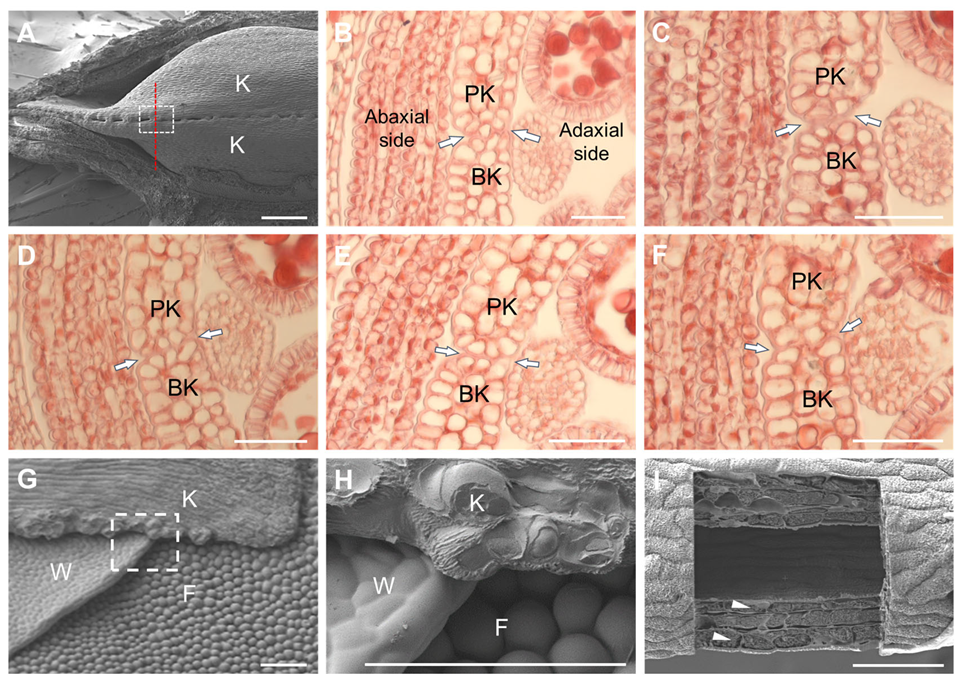
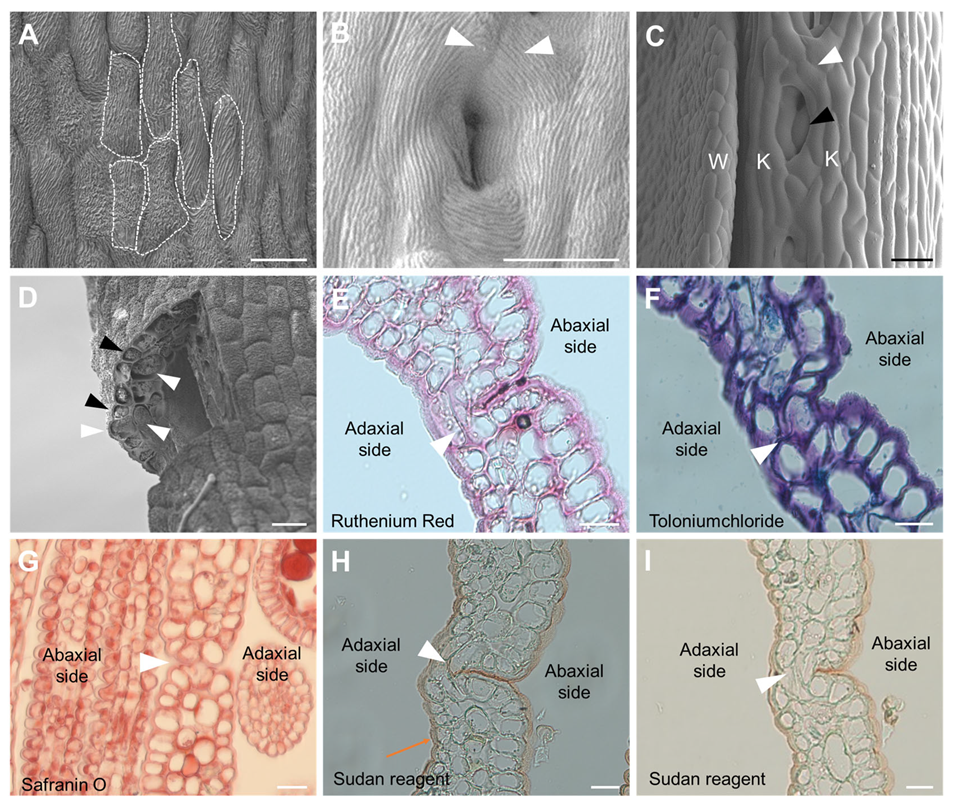
3.2. Different Morphology of the Opposite Marginal Cells at the Contact Site
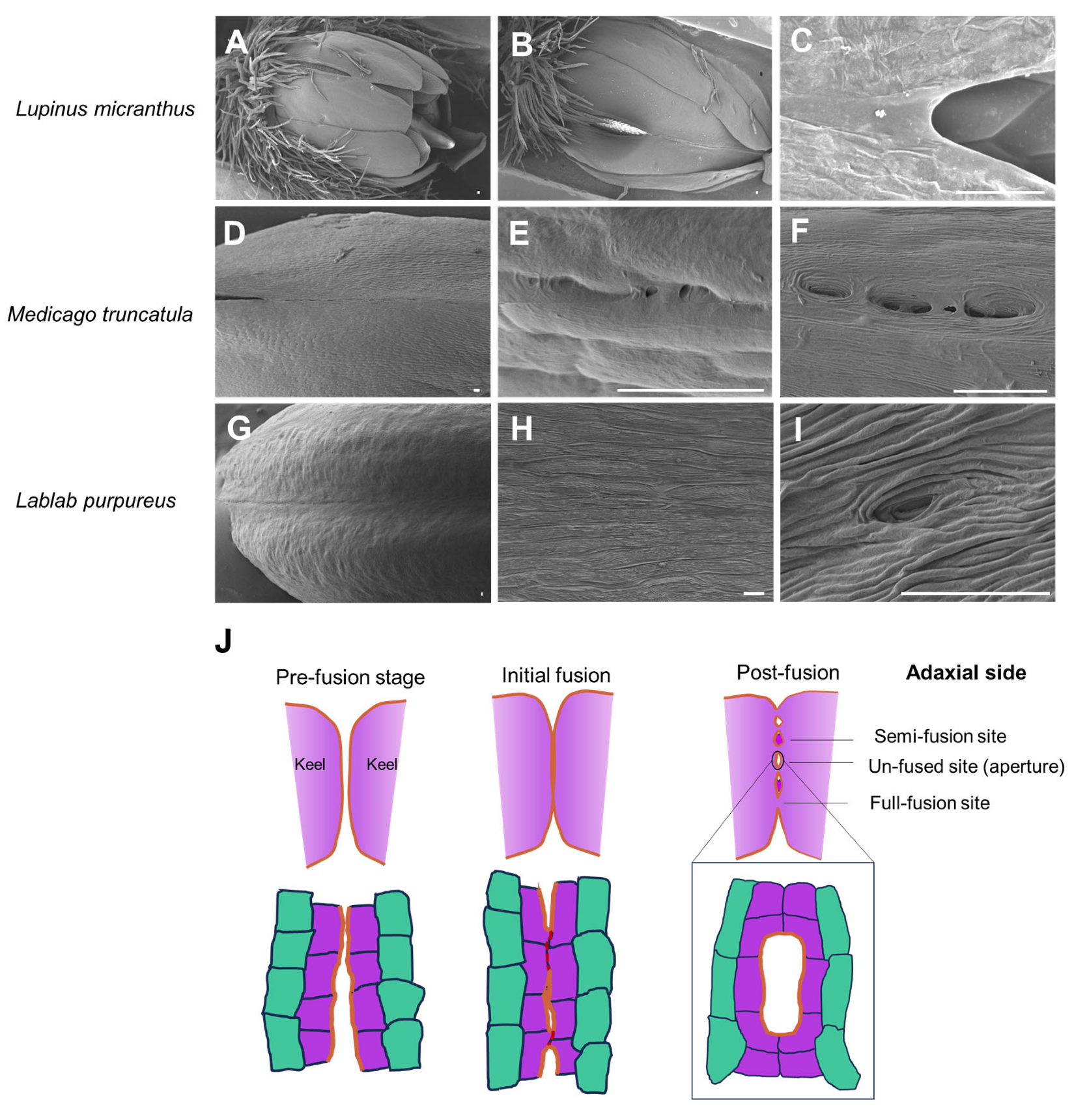
3.3. Cell Reshaping and Cuticle Changes on the Keel Fusion Site
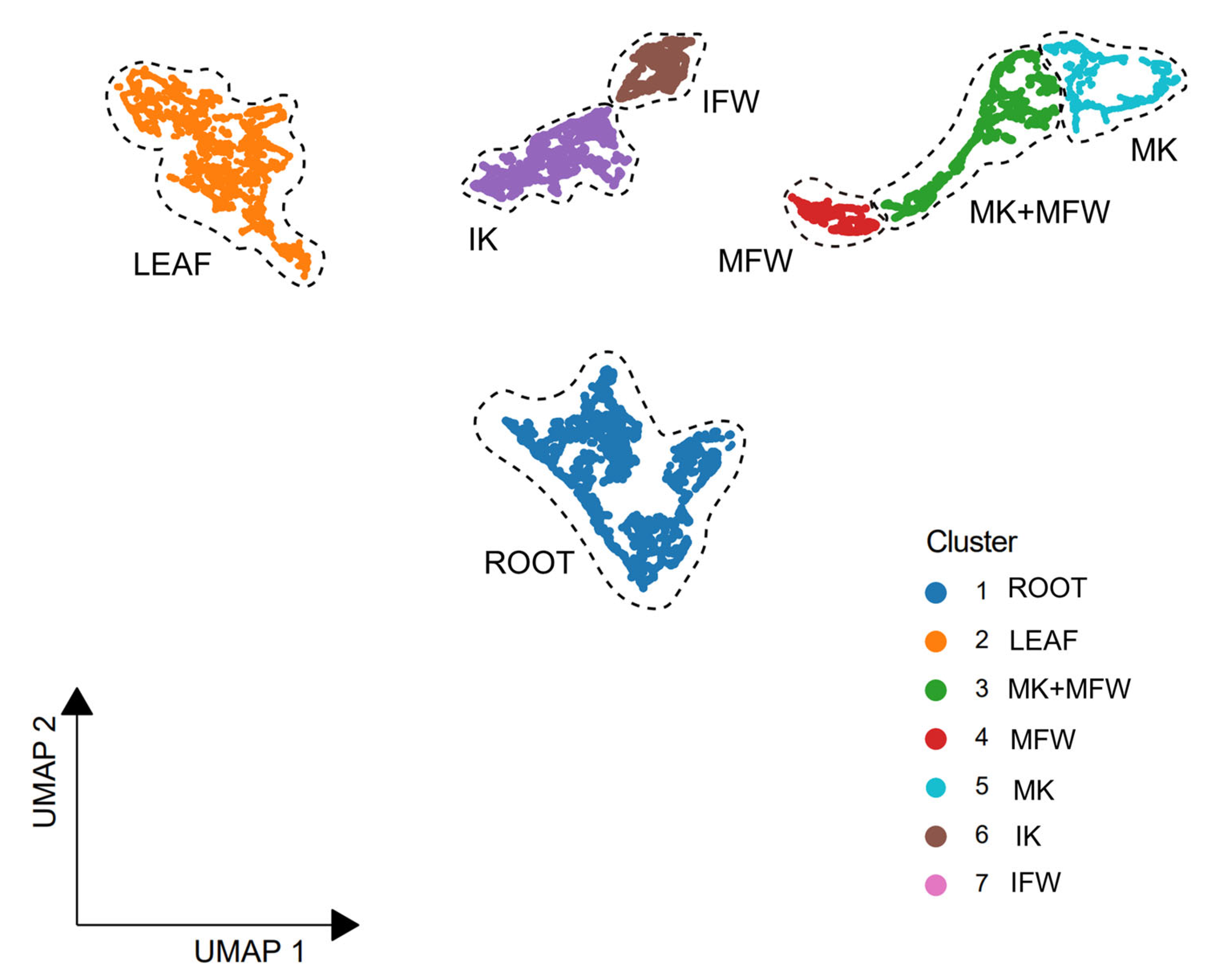
3.4. Distinct Transcriptomes Between the Petals
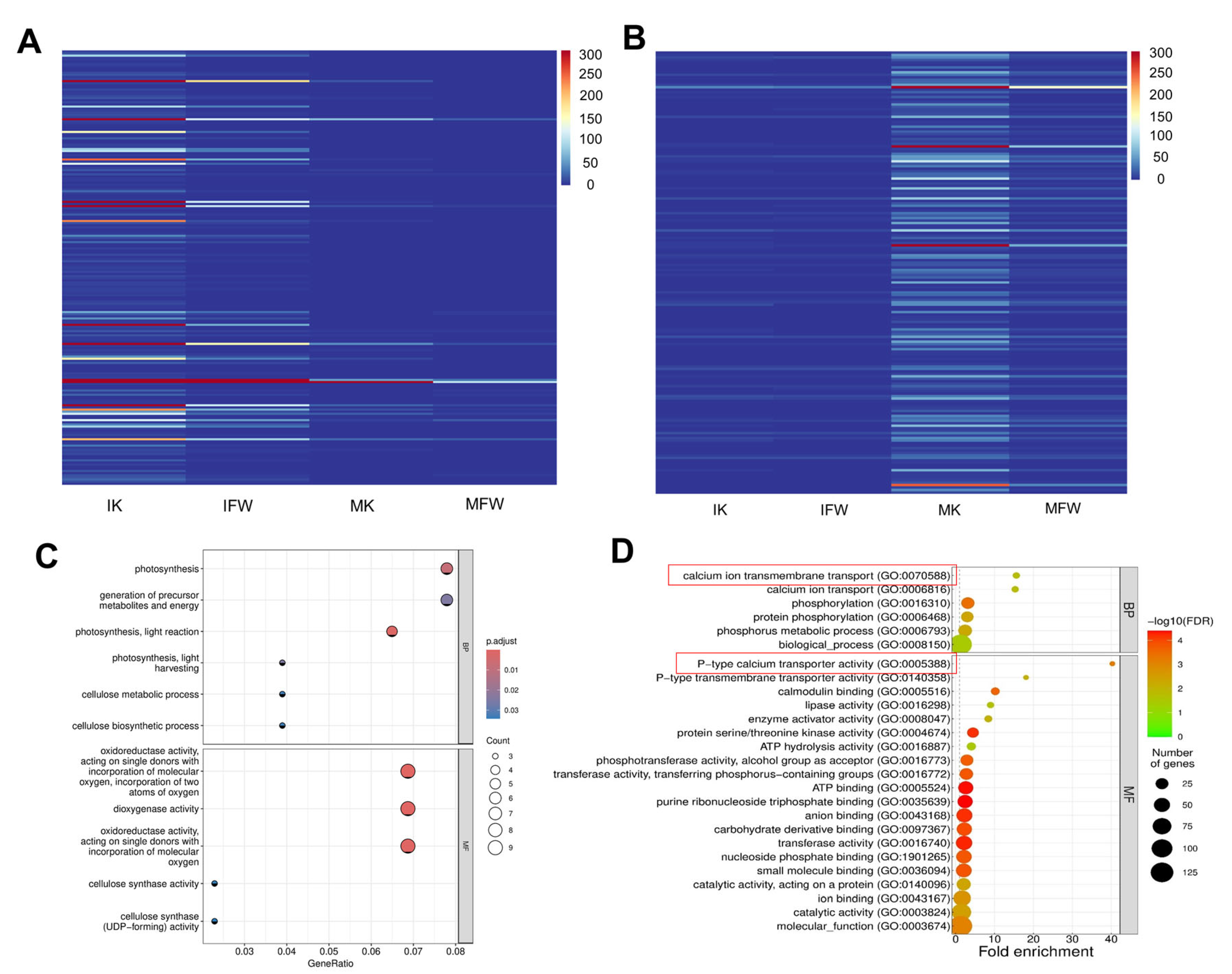
3.5. Analysis of Keel-Preferentially Expressed Genes and Screening of Candidate Regulators
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| BK | blunt keel petal |
| F | flag petal |
| IK | immature/unfused keel petal |
| IFW | immature/unfused mixed samples of flag and wing petals (keel petal stage) |
| K | keel petal |
| MK | mature/fused keel petal |
| MFW | mature/fused mixed samples of flag and wing petals (keel petal stage) |
| PK | pointed keel petal |
| W | wing petal |
References
- Zhao, Y.Y.; Zhang, R.; Jiang, K.W.; Qi, J.; Hu, Y.; Guo, J.; Zhu, R.B.; Zhang, T.K.; Egan, A.N.; Yi, T.S.; et al. Nuclear phylotranscriptomics and phylogenomics support numerous polyploidization events and hypotheses for the evolution of rhizobial nitrogen-fixing symbiosis in Fabaceae. Mol. Plant 2021, 14, 748–773. [Google Scholar] [CrossRef]
- Azani, N.; Babineau, M.; Bailey, C.D.; Banks, H.; Barbosa, A.; Barbosa Pinto, R.; Boatwright, J.; Borges, L.; Brown, G.; Bruneau, A.; et al. A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 2017, 66, 44–77. [Google Scholar] [CrossRef]
- Yahara, T.; Javadi, F.; Onoda, Y.; de Queiroz, L.P.; Faith, D.P.; Prado, D.E.; Akasaka, M.; Kadoya, T.; Ishihama, F.; Davies, S.; et al. Global legume diversity assessment: Concepts, key indicators, and strategies. Taxon 2013, 62, 249–266. [Google Scholar] [CrossRef]
- Lewis, G.; Schrire, B.; Mackinder, B.; Lock, M. Legumes of the World; Royal Botanic Gardens Kew: London, UK, 2005; pp. 1–577. [Google Scholar]
- Tucker, S.C. Floral development in legumes. Plant Physiol. 2003, 131, 911–926. [Google Scholar] [CrossRef] [PubMed]
- Saitoh, K.; Wakui, N.; Mahmood, T.; Kuroda, T. Differentiation and development of floral organs at each node and raceme order in an indeterminate type of soybean. Plant Prod. Sci. 1999, 2, 47–50. [Google Scholar] [CrossRef]
- Washburn, C.F.; Thomas, J.F. Reversion of flowering in Glycine Max (Fabaceae). Am. J. Bot. 2000, 87, 1425–1438. [Google Scholar] [CrossRef]
- Tilton, V.R.; Wilcox, L.W.; Palmer, R.G.; Albertsen, M.C. Stigma, style, and obturator of soybean, Glycine Max (L.) Merr. (Leguminosae) and their function in the reproductive process. Am. J. Bot. 1984, 71, 676–686. [Google Scholar] [CrossRef]
- Srinivasan, S.; Gaur, P.M. Genetics and characterization of an open flower mutant in chickpea. J. Hered. 2012, 103, 297–302. [Google Scholar] [CrossRef][Green Version]
- Feng, X.; Zhao, Z.; Tian, Z.; Xu, S.; Luo, Y.; Cai, Z.; Wang, Y.; Yang, J.; Wang, Z.; Weng, L.; et al. Control of petal shape and floral zygomorphy in Lotus japonicus. Proc. Natl. Acad. Sci. USA 2006, 103, 4970–4975. [Google Scholar] [CrossRef]
- Sauquet, H.; von Balthazar, M.; Magallón, S.; Doyle, J.A.; Endress, P.K.; Bailes, E.J.; Barroso de Morais, E.; Bull-Hereñu, K.; Carrive, L.; Chartier, M.; et al. The ancestral flower of angiosperms and its early diversification. Nat. Commun. 2017, 8, 16047. [Google Scholar] [CrossRef]
- Endress, P.K. Evolutionary diversification of the flowers in angiosperms. Am. J. Bot. 2011, 98, 370–396. [Google Scholar] [CrossRef]
- Bell, E.M.; Lin, W.C.; Husbands, A.Y.; Yu, L.F.; Jaganatha, V.; Jablonska, B.; Mangeon, A.; Neff, M.M.; Girke, T.; Springer, P.S. Arabidopsis lateral organ boundaries negatively regulates brassinosteroid accumulation to limit growth in organ boundaries. Proc. Natl. Acad. Sci. USA 2012, 109, 21146–21151. [Google Scholar] [CrossRef] [PubMed]
- Siegel, B.A.; Verbeke, J.A. Diffusible factors essential for epidermal cell redifferentiaion in Catharanthus roseus. Science 1989, 244, 580–582. [Google Scholar] [CrossRef] [PubMed]
- Phillips, H.R.; Landis, J.B.; Specht, C.D. Revisiting floral fusion: The evolution and molecular basis of a developmental innovation. J. Exp. Bot. 2020, 71, 3390–3404. [Google Scholar] [CrossRef]
- Verbeke, J.A. Fusion events during floral morphogenesis. Annu. Rev. Plant Physiol. Plant Mol. Biol 1992, 43, 583–598. [Google Scholar] [CrossRef]
- Li, F.; Shao, Y.P.; Ejaz, I.; Chen, Z.Y.; Wang, Z.W.; Wang, X.; Zhou, S.L. A morphological and anatomical study for tracking the growth and development of individual flowers and pods in soybean (Glycine max L.). Crop J. 2025, 13, 304–309. [Google Scholar] [CrossRef]
- Saitta, V.; Rebora, M.; Piersanti, S.; Gorb, E.; Gorb, S.; Salerno, G. Effect of leaf trichomes in different species of cucurbitaceae on attachment ability of the melon ladybird beetle Chnootriba elaterii. Insects 2022, 13, 1123. [Google Scholar] [CrossRef]
- Nogales, E.; Mahamid, J. Bridging structural and cell biology with cryo-electron microscopy. Nature 2024, 628, 47–56. [Google Scholar] [CrossRef]
- Schaffer, M.; Pfeffer, S.; Mahamid, J.; Kleindiek, S.; Laugks, T.; Albert, S.; Engel, B.D.; Rummel, A.; Smith, A.J.; Baumeister, W.; et al. A cryo-FIB lift-out technique enables molecular-resolution cryo-ET within native Caenorhabditis elegans tissue. Nat. Methods 2019, 16, 757–762. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, M.D.; Sun, Y.F.; Zhao, P.; Liu, C.; Qing, K.; Hu, X.T.; Zhong, Z.D.; Cheng, J.L.; Wang, H.J.; et al. Glycine max NNL1 restricts symbiotic compatibility with widely distributed bradyrhizobia via root hair infection. Nat. Plants 2021, 7, 73–86. [Google Scholar] [CrossRef]
- Lee, D.; Geisler, M.; Springer, P.S. LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 2009, 136, 2423–2432. [Google Scholar] [CrossRef]
- Wang, J.; Wang, X.F.; Yang, W.C.; Li, H.J. Loss of function of CENH3 causes genome instability in soybean. Seed Biol. 2023, 2, 24. [Google Scholar] [CrossRef]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, i884–i890. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. Feature counts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Matsuura, Y.; Ishiguro-Watanabe, M. KEGG: Biological systems database as a model of the real world. Nucleic Acids Res. 2024, 53, D672–D677. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudié, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinf. 2014, 15, 293. [Google Scholar] [CrossRef]
- He, Z.L.; Zhang, H.K.; Gao, S.H.; Lercher, M.J.; Chen, W.H.; Hu, S.N. Evolview v2: An online visualization and management tool for customized and annotated phylogenetic trees. Nucleic Acids Res. 2016, 44, W236–W241. [Google Scholar] [CrossRef]
- Nunes, A.C.; Vianna, G.R.; Cuneo, F.; Amaya-Farfán, J.; de Capdeville, G.; Rech, E.L.; Aragão, F.J. RNAi-mediated silencing of the myo-inositol-1-phosphate synthase gene (GmMIPS1) in transgenic soybean inhibited seed development and reduced phytate content. Planta 2006, 224, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Dang, X.; Yu, P.H.; Li, Y.J.; Yang, Y.Q.; Zhang, Y.; Ren, H.B.; Chen, B.; Lin, D.S. Reactive oxygen species mediate conical cell shaping in Arabidopsis thaliana petals. PLoS Genet. 2018, 14, e1007705. [Google Scholar] [CrossRef] [PubMed]
- Kozuka, T.; Kong, S.G.; Doi, M.; Shimazaki, K.; Nagatani, A. Tissue-autonomous promotion of palisade cell development by phototropin 2 in Arabidopsis. Plant Cell 2011, 23, 3684–3695. [Google Scholar] [CrossRef]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2019, 37, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.M.; Zhang, H.; Long, Y.P.; Shu, Y.; Zhai, J.X. Plant Public RNA-seq Database: A comprehensive online database for expression analysis of ~45,000 plant public RNA-Seq libraries. Plant Biotechnol. J. 2022, 20, 806–808. [Google Scholar] [CrossRef]
- Li-Beisson, Y.; Neunzig, J.; Lee, Y.; Philippar, K. Plant membrane-protein mediated intracellular traffic of fatty acids and acyl lipids. Curr. Opin. Plant Biol. 2017, 40, 138–146. [Google Scholar] [CrossRef]
- Zhu, L.; He, S.Y.; Liu, Y.Y.; Shi, J.X.; Xu, J. Arabidopsis FAX1 mediated fatty acid export is required for the transcriptional regulation of anther development and pollen wall formation. Plant Mol. Biol. 2020, 104, 187–201. [Google Scholar] [CrossRef] [PubMed]
- Bugaeva, W.; Könnel, A.; Peter, J.; Mees, J.; Hankofer, V.; Schick, C.; Schmidt, A.; Banguela-Castillo, A.; Philippar, K.; Philippar, K. Plastid fatty acid export (FAX) proteins in Arabidopsis thaliana-the role of FAX1 and FAX3 in growth and development. bioRxiv 2023. [Google Scholar] [CrossRef]
- Lü, S.Y.; Song, T.; Kosma, D.K.; Parsons, E.P.; Rowland, O.; Jenks, M.A. Arabidopsis CER8 encodes long-chain acyl-coa synthetase 1 (LACS1) that has overlapping functions with LACS2 in plant wax and cutin synthesis. Plant J. 2009, 59, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Fiebig, A.; Mayfield, J.A.; Miley, N.L.; Chau, S.; Fischer, R.L.; Preuss, D. Alterations in CER6, a gene identical to CUT1, differentially affect long-chain lipid content on the surface of pollen and stems. Plant Cell 2000, 12, 2001–2008. [Google Scholar] [CrossRef]
- Beaudoin, F.; Wu, X.Z.; Li, F.L.; Haslam, R.P.; Markham, J.E.; Zheng, H.Q.; Napier, J.A.; Kunst, L. Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme A reductase candidates of the fatty acid elongase. Plant Physiol. 2009, 150, 1174–1191. [Google Scholar] [CrossRef]
- Bach, L.; Michaelson, L.V.; Haslam, R.; Bellec, Y.; Gissot, L.; Marion, J.; Da Costa, M.; Boutin, J.; Miquel, M.; Tellier, F.; et al. The very-long-chain hydroxy fatty acyl-CoA dehydratase pasticcino2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 2008, 105, 14727–14731. [Google Scholar] [CrossRef]
- Zheng, H.Q.; Rowland, O.; Kunst, L. Disruptions of the Arabidopsis enoyl-CoA reductase gene reveal an essential role for very-long-chain fatty acid synthesis in cell expansion during plant morphogenesis. Plant Cell 2005, 17, 1467–1481. [Google Scholar] [CrossRef]
- Takeda, S.; Iwasaki, A.; Matsumoto, N.; Uemura, T.; Tatematsu, K.; Okada, K. Physical interaction of floral organs controls petal morphogenesis in Arabidopsis. Plant Physiol. 2013, 161, 1242–1250. [Google Scholar] [CrossRef]
- Li, Y.H.; Beisson, F.; Koo, A.J.K.; Molina, I.; Pollard, M.; Ohlrogge, J. Identification of acyltransferases required for cutin biosynthesis and production of cutin with suberin-like monomers. Proc. Natl. Acad. Sci. USA 2007, 104, 18339–18344. [Google Scholar] [CrossRef]
- Panikashvili, D.; Shi, J.X.; Schreiber, L.; Aharoni, A. The Arabidopsis DCR encoding a soluble BAHD acyltransferase is required for cutin polyester formation and seed hydration properties. Plant Physiol. 2009, 151, 1773–1789. [Google Scholar] [CrossRef]
- Lee, E.J.; Kim, K.Y.; Zhang, J.; Yamaoka, Y.; Gao, P.; Kim, H.; Hwang, J.; Suh, M.C.; Kang, B.; Lee, Y. Arabidopsis seedling establishment under waterlogging requires ABCG5-mediated formation of a dense cuticle layer. New Phytol. 2021, 229, 156–172. [Google Scholar] [CrossRef]
- Kim, H.; Lee, S.B.; Kim, H.J.; Min, M.K.; Hwang, I.; Suh, M.C. Characterization of glycosylphosphatidylinositol-anchored lipid transfer protein 2 (LTPG2) and overlapping function between LTPG/LTPG1 and LTPG2 in cuticular wax export or accumulation in Arabidopsis thaliana. Plant Cell Physiol. 2012, 53, 1391–1403. [Google Scholar] [CrossRef]
- Hong, L.L.; Brown, J.; Segerson, N.A.; Rose, J.K.C.; Roeder, A.H.K. Cutin Synthase 2 maintains progressively developing cuticular ridges in Arabidopsis sepals. Mol. Plant. 2017, 10, 560–574. [Google Scholar] [CrossRef] [PubMed]
- Kannangara, R.; Branigan, C.; Liu, Y.; Penfield, T.; Rao, V.; Mouille, G.; Höfte, H.; Pauly, M.; Riechmann, J.L.; Broun, P. The transcription factor WIN1/SHN1 regulates cutin biosynthesis in Arabidopsis thaliana. Plant Cell 2007, 19, 1278–1294. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.B.; Kim, H.U.; Suh, M.C. MYB94 and MYB96 additively activate cuticular wax biosynthesis in Arabidopsis. Plant Cell Physiol. 2016, 57, 2300–2311. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.D.; Yang, X.P.; Zheng, M.L.; Lü, S.Y.; Zhao, H.Y. Fine-tuning the activities of β-ketoacyl-CoA synthase 3 (KCS3) and KCS12 in Arabidopsis is essential for maintaining cuticle integrity. J. Exp. Bot. 2023, 74, 6575–6587. [Google Scholar] [CrossRef]
- Lü, S.Y.; Zhao, H.Y.; Des Marais, D.L.; Parsons, E.P.; Wen, X.X.; Xu, X.J.; Bangarusamy, D.K.; Wang, G.C.; Rowland, O.; Juenger, T.; et al. Arabidopsis eceriferum9 involvement in cuticle formation and maintenance of plant water status. Plant Physiol. 2012, 159, 930–944. [Google Scholar] [CrossRef]
- Wu, R.H.; Li, S.B.; He, S.; Waßmann, F.; Yu, C.H.; Qin, G.J.; Schreiber, L.; Qu, L.J.; Gu, H.Y. CFL1, a WW domain protein, regulates cuticle development by modulating the function of HDG1, a class IV homeodomain transcription factor, in rice and Arabidopsis. Plant Cell 2011, 23, 3392–3411. [Google Scholar] [CrossRef]
- Huang, L.C.; Jin, L.; Li, J.; Zhang, X.; Yang, Y.; Wang, X. Floral morphology and its relationship with pollination systems in Papilionoideae. Acta Ecol. Sin. 2014, 34, 5360–5368. [Google Scholar] [CrossRef][Green Version]
- Walker, D.B. Postgenital carpel fusion in Catharanthus roseus (Apocynaceae). I. light and scanning electron microscopic study of gynoecial ontogeny. Am. J. Bot. 1975, 62, 457–467. [Google Scholar] [CrossRef]
- Verbeke, J.A.; Walker, D.B. Morphogenetic factors controlling differentiation and dedifferentiation of epidermal cells in the gynoecium of Catharanthus roseus. Planta 1986, 168, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Ingram, G.; Nawrath, C. The roles of the cuticle in plant development: Organ adhesions and beyond. J. Exp. Bot. 2017, 68, 5307–5321. [Google Scholar] [CrossRef] [PubMed]
- Yeats, T.H.; Rose, J.K.C. The formation and function of plant cuticles. Plant Physiol. 2013, 163, 5–20. [Google Scholar] [CrossRef] [PubMed]
- Delude, C.; Moussu, S.; Joubès, J.; Ingram, G.; Domergue, F. Plant surface lipids and epidermis development. Subcell. Biochem. 2016, 86, 287–313. [Google Scholar] [CrossRef]
- Bird, S.M.; Gray, J.E. Signals from the cuticle affect epidermal cell differentiation. New Phytol. 2003, 157, 9–23. [Google Scholar] [CrossRef]
- Bernard, A.; Joubès, J. Arabidopsis cuticular waxes: Advances in synthesis, export and regulation. Prog. Lipid Res. 2013, 52, 110–129. [Google Scholar] [CrossRef]
- Liu, X.; Bourgault, R.; Galli, M.; Strable, J.; Chen, Z.L.; Feng, F.; Dong, J.Q.; Molina, I.; Gallavotti, A. The fused leaves1-adherent1 regulatory module is required for maize cuticle development and organ separation. New Phytol. 2021, 229, 388–402. [Google Scholar] [CrossRef]
- Sinha, N.; Lynch, M. Fused organs in the adherent1 mutation in maize show altered epidermal walls with no perturbations in tissue identities. Planta 1998, 206, 184–195. [Google Scholar] [CrossRef]
- Yephremov, A.; Wisman, E.; Huijser, P.; Huijser, C.; Wellesen, K.; Saedler, H. Characterization of the fiddlehead gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell 1999, 11, 2187–2201. [Google Scholar] [CrossRef]
- Yeats, T.H.; Martin, L.B.B.; Viart, H.M.F.; Isaacson, T.; He, Y.H.; Zhao, L.X.; Matas, A.J.; Buda, G.J.; Domozych, D.S.; Clausen, M.H.; et al. The identification of cutin synthase: Formation of the plant polyester cutin. Nat. Chem. Biol. 2012, 8, 609–611. [Google Scholar] [CrossRef]
- Zhao, X.; Huang, L.; Kang, L.; Jetter, R.; Yao, L.H.; Li, Y.; Xiao, Y.; Wang, D.K.; Xiao, Q.L.; Ni, Y.; et al. Comparative analyses of cuticular waxes on various organs of faba bean (Vicia faba L.). Plant Physiol. Biochem. 2019, 139, 102–112. [Google Scholar] [CrossRef]
- Haslam, T.M.; Kunst, L. Extending the story of very-long-chain fatty acid elongation. Plant Sci. 2013, 210, 93–107. [Google Scholar] [CrossRef]
- de Silva, K.; Laska, B.; Brown, C.; Sederoff, H.W.; Khodakovskaya, M. Arabidopsis thaliana calcium-dependent lipid-binding protein (AtCLB): A novel repressor of abiotic stress response. J. Exp. Bot. 2011, 62, 2679–2689. [Google Scholar] [CrossRef]
- Gao, S.; Song, T.; Han, J.; He, M.; Zhang, Q.; Zhu, Y.; Zhu, Z. A calcium-dependent lipid binding protein, OsANN10, is a negative regulator of osmotic stress tolerance in rice. Plant Sci. 2020, 293, 110420. [Google Scholar] [CrossRef]
- Xiao, S.; Wu, C.; Zuo, D.; Cheng, H.; Zhang, Y.; Wang, Q.; Lv, L.; Song, G. Systematic analysis and comparison of CaLB genes reveal the functions of GhCaLB42 and GhCaLB123 in fiber development and abiotic stress in cotton. Ind. Crop Prod. 2022, 184, 115030. [Google Scholar] [CrossRef]
- Hepler, P.K.; Winship, L.J. Calcium at the cell wall-cytoplast interface. J. Integr. Plant Biol. 2010, 52, 147–160. [Google Scholar] [CrossRef]
- Hinderliter, A.K.; Almeida, P.F.F.; Biltonen, R.L.; Creutz, C.E. Membrane domain formation by calcium-dependent, lipid-binding proteins: Insights from the C2 motif. Biochim. Biophys. Acta 1998, 1448, 227–235. [Google Scholar] [CrossRef]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. Corrigendum to: The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2021, 33, 3743–3744. [Google Scholar] [CrossRef] [PubMed]
- Wagner, D.; Meyerowitz, E.M. SPLAYED, a novel SWI/SNF ATPase homolog, controls reproductive development in Arabidopsis. Curr. Biol. 2002, 12, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Kwon, C.S.; Chen, C.; Wagner, D. WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 2005, 19, 992–1003. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Chen, C.; Li, C.; Thapa, R.K.; Song, J.; Xie, X.; Nguyen, V.; Bian, S.; Liu, J.; Kohalmi, S.E.; et al. Genome-wide occupancy of Arabidopsis SWI/SNF chromatin remodeler SPLAYED provides insights into its interplay with its close homolog BRAHMA and Polycomb proteins. Plant J. 2021, 106, 200–213. [Google Scholar] [CrossRef]
- Caro, E.; Stroud, H.; Greenberg, M.V.; Bernatavichute, Y.V.; Feng, S.; Groth, M.; Vashisht, A.A.; Wohlschlegel, J.; Jacobsen, S.E. The SET-domain protein SUVR5 mediates H3K9me2 deposition and silencing at stimulus response genes in a DNA methylation-independent manner. PLoS Genet. 2012, 8, e1002995. [Google Scholar] [CrossRef]
- Jiang, D.; Kong, N.C.; Gu, X.; Li, Z.; He, Y. Arabidopsis COMPASS-like complexes mediate histone H3 lysine-4 trimethylation to control floral transition and plant development. PLoS Genet. 2011, 7, e1001330. [Google Scholar] [CrossRef]

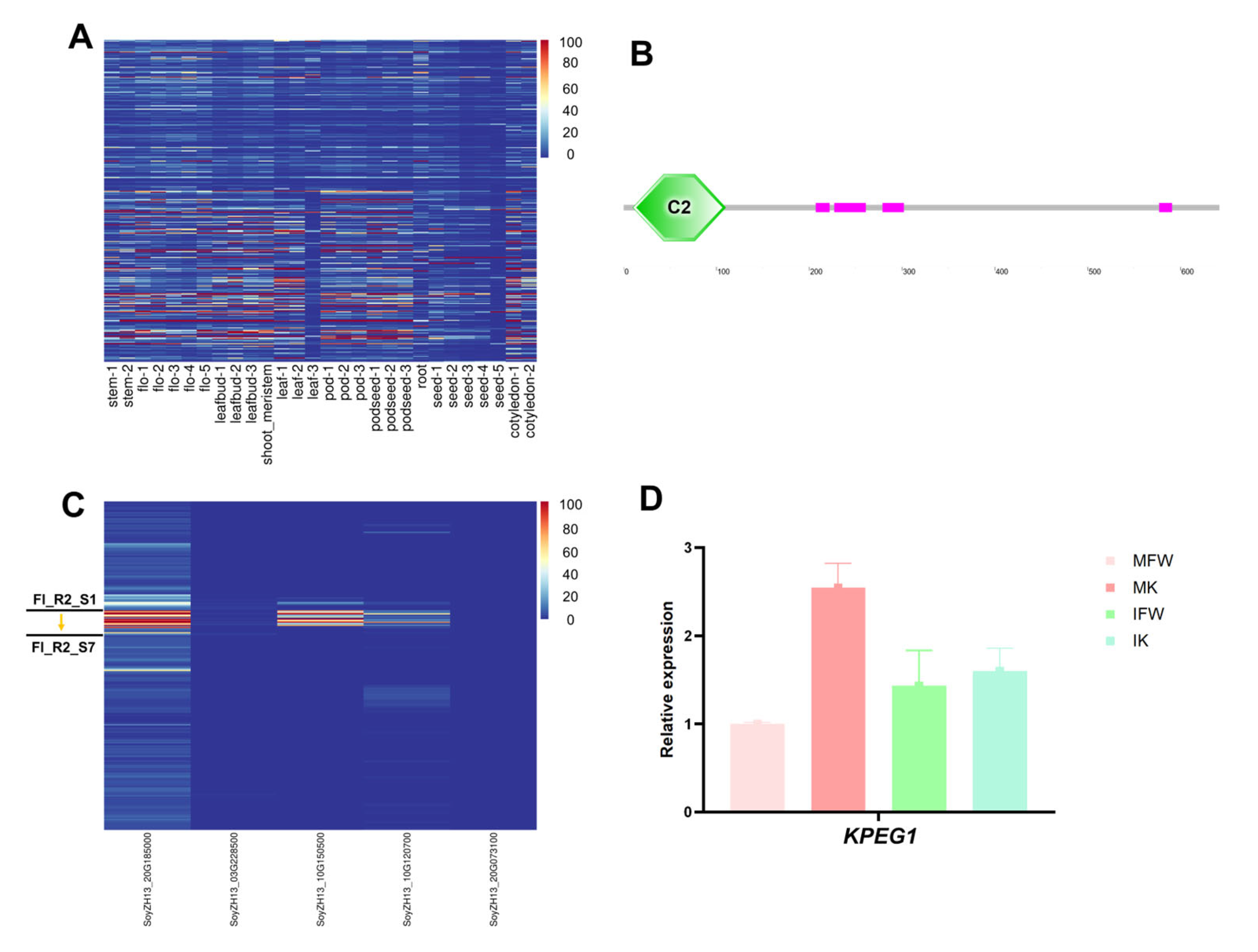
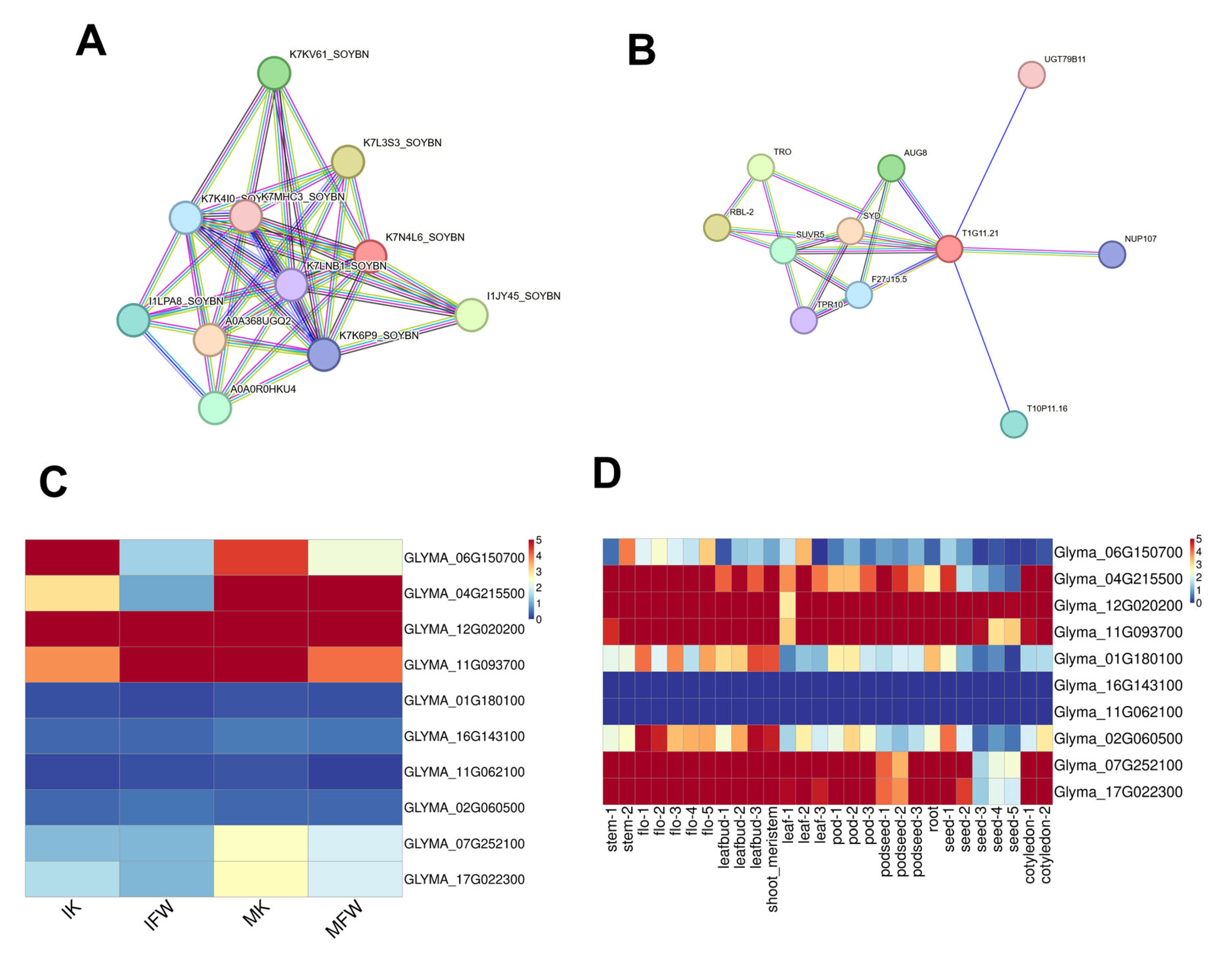
| Gene Name | Gene ID | Soybean Ortholog | Biological Function | Types of Regulators | Description in Mutant | Reference |
|---|---|---|---|---|---|---|
| FAX1 | At3g57280 | Glyma.03G133200, Glyma.19G135000 | Plastidial FAs export protein | positive | Abnormal wax accumulation in stems, abnormal tapetum development, and pollen wall formation | [42,43] |
| LACS1 | At2g47240 | Glyma.02G010300, Glyma.03G092900, Glyma.03G221400, Glyma.10G010800, Glyma.19G218300 | Long-chain acyl-coenzyme A synthetase | positive | Reduced the amount of wax and cutin on the stem and leaf | [44] |
| CER6 | At1g68530 | Glyma.08G261100, Glyma.10G241700, Glyma.10G274400, Glyma.20G115500 | Keto-acyl-CoA Synthase | positive | Reduced wax in stem and lipid contents in the pollen coats | [45] |
| KCR1 | At1g67730 | Glyma.11G245600, Glyma.11G245700, Glyma.18G011500, Glyma.18G011600 | Keto-acyl-CoA reductase | positive | Reduced wax in stem, fused rosette leaves, and embryo-lethal | [46] |
| PAS2 | At5g10480 | Glyma.01G023400, Glyma.08G279700, Glyma.18G146900 | Hydroxy-acyl-CoA dehydratase | positive | Reduced wax deposition, fused flower buds, embryo lethal | [47] |
| CER10 | At3g55360 | Glyma.02G273300, Glyma.14G043500 | Enoyl-CoA Reductase | positive | Reduced wax deposition in stem | [48] |
| FOP1 | At5g53390 | Glyma.06G291200, Glyma.06G291300, Glyma.07G000300, Glyma.09G196400 | Bifunctional wax ester synthase/diacylglycerol acyltransferase | positive | Flattened surface of the epidermal cells in petal, folded petal | [49] |
| GPAT8 | At4g00400 | Glyma.03G008300, Glyma.07G069700 | Glycerol-3-phosphate acyltransferases | positive | Reduced cutin on the stem and leaves, increased water loss, susceptibility to pathogens, and altered stomata structure | [50] |
| DCR | At5g23940 | Glyma.09G134600, Glyma.16G180500 | BAHD family of acyltransferases | positive | Reduced cutin monomer and postgenital fusions between the rosette leaves and flower buds | [51] |
| ABCG5 | At2g13610 | Glyma.04G211800, Glyma.05G192700, Glyma.06G154500, Glyma.08G000800 | ATP-binding cassette transporter subfamily G proteins | positive | Reduced wax contents in cotyledons, and seedling failed to develop true leaves under waterlogged conditions | [52] |
| LTPG1 | At1g27950 | Glyma.05G147700, Glyma.11G251100, Glyma.18G005800 | Glycosylphosphatidylinositol-anchored lipid transfer protein | positive | Reduced wax loads and altered wax composition in stem and silique | [53] |
| CUS2 | At5g33370 | Glyma.01G106900, Glyma.03G252600, Glyma.03G252700, Glyma.03G252800, Glyma.04G109900, Glyma.05G116300, Glyma.09G241400, Glyma.10G168400, Glyma.10G168500, Glyma.13G044500, Glyma.13G045100, Glyma.18G254800, Glyma.19G050000, Glyma.19G050800, Glyma.19G050900, Glyma.19G051000, Glyma.19G051100, Glyma.19G250100, Glyma.19G250400, Glyma.20G221200 | Glycine-aspartic acid-serine-leucine-motif lipase/hydrolase | positive | Reduced cuticular ridges on mature sepals | [54] |
| WIN1 | At1g15360 | Glyma.04G147500, Glyma.06G221800, Glyma.07G031200, Glyma.08G211600, Glyma.13G166700, Glyma.15G008600, Glyma.17G114500 | Transcription factor of the ethylene response factor (ERF) family | positive | Reduced cutin composition on flower | [55] |
| MYB94 | At3g47600 | Glyma.03G090800, Glyma.04G170100, Glyma.05G027000, Glyma.06G193600, Glyma.17G099800 | Abscisic acid (ABA)-responsive R2R3-type MYB transcription factor | positive | Reduced wax on the stem and leaves, more permeable cuticle, sensitive to droughts | [56] |
| KCS12 | AT2G28630 | Glyma.12G075100, Glyma.13G331600, Glyma.15G042500 | Ketoacyl-CoA synthases | negative | Increased wax and cutin contents in flower and leaves | [57] |
| CER9 | At4g34100 | Glyma.02G103800, Glyma.07G215200 | E3 ubiquitin ligase | negative | Increased cutin monomers and cuticle membrane thickness in leaves and stems | [58] |
| CFL1 | At2g33510 | Glyma.18G302000 | WW domain protein | negative | Increased epicuticular wax on the surface of trichomes on the inflorescence stem | [59] |
| Arabidopsis Gene ID | Wm82 Gene ID | ZH13 Gene ID | UniProt ID |
|---|---|---|---|
| AT1G04540 | Glyma.20G199700 (KPEG1) | SoyZH13_20G185000 | K7N4L6_SOYBN |
| Glyma.03G249700 | SoyZH13_03G228500 | I1JRR2_SOYBN | |
| Glyma.10G165300 | SoyZH13_10G150500 | I1LBP5_SOYBN | |
| Glyma.10G130100 | SoyZH13_10G120700 | K7LJ48_SOYBN | |
| Glyma.20G081900 | SoyZH13_20G073100 | I1NEL6_SOYBN |
| Gene Name | Arabidopsis Gene ID | UniProt ID | Soybean Gene ID | UniProt ID |
|---|---|---|---|---|
| ATG1T | AT1G49180 | F27J15.5 | Glyma.06G150700 | K7KV61_SOYBN |
| Glyma.04G215500 | I1JY45_SOYBN | |||
| TRO | AT1G51450 | Q9C8J7 | Glyma.12G020200 | I1LPA8_SOYBN |
| Glyma.11G093700 | A0A0R0HKU4 | |||
| SUVR5 | AT2G23740 | O64827 | Glyma.01G180100 | K7K4I0_SOYBN |
| Glyma.16G143100 | K7MHC3_SOYBN | |||
| Glyma.11G062100 | K7LNB1_SOYBN | |||
| Glyma.02G060500 | K7K6P9_SOYBN | |||
| SYD | AT2G28290 | F4IHS2 | Glyma.07G252100 | K7L3S3_SOYBN |
| Glyma.17G022300 | A0A368UGQ2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jia, S.-G.; Guo, L.-N.; Wang, X.-F.; Wang, D.-L.; Chen, D.; Yang, W.-C.; Li, H.-J. Keel Petal Fusion in Soybean: Anatomical Insights and Transcriptomic Identification of Candidate Regulators. Agronomy 2025, 15, 1971. https://doi.org/10.3390/agronomy15081971
Jia S-G, Guo L-N, Wang X-F, Wang D-L, Chen D, Yang W-C, Li H-J. Keel Petal Fusion in Soybean: Anatomical Insights and Transcriptomic Identification of Candidate Regulators. Agronomy. 2025; 15(8):1971. https://doi.org/10.3390/agronomy15081971
Chicago/Turabian StyleJia, Shun-Geng, Li-Na Guo, Xiao-Fei Wang, De-Li Wang, Dan Chen, Wei-Cai Yang, and Hong-Ju Li. 2025. "Keel Petal Fusion in Soybean: Anatomical Insights and Transcriptomic Identification of Candidate Regulators" Agronomy 15, no. 8: 1971. https://doi.org/10.3390/agronomy15081971
APA StyleJia, S.-G., Guo, L.-N., Wang, X.-F., Wang, D.-L., Chen, D., Yang, W.-C., & Li, H.-J. (2025). Keel Petal Fusion in Soybean: Anatomical Insights and Transcriptomic Identification of Candidate Regulators. Agronomy, 15(8), 1971. https://doi.org/10.3390/agronomy15081971





