Progress of Acetylation Modification in Plants
Abstract
1. Introduction
2. Histone Acetylation Modifications Regulate Chromatin Structure and Gene Transcription
3. Histone Acetylation Modifications Regulate Plant Growth and Development
4. The Impact of Histone Acetyltransferases on Plant Root Development
5. Non-Histone Protein Acetylation Plays a Broader Role in Regulating Plant Growth and Development
6. Non-Histone Acetylation Modifications Are Present in Various Plant Cell Organelles
7. Non-Histone Protein Acetylation Is Involved in Diverse Physiological and Biochemical Processes
8. The Role of Non-Histone Protein Acetylation in Plant Responses to Biotic and Abiotic Stresses
9. Acetylation Modifications Interact with Other PTMs to Influence Gene Expression
10. Conclusions and Perspectives
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| ADA2 | alteration/deficiency in activation 2 |
| ATP | adenosine triphosphate |
| BPGA | glycerate 1,3-bisphosphate |
| CBB | Calvin–Benson–Bassham |
| CMS | cytoplasmic male sterility |
| CPC | CAPRICE |
| Cytb 6f | cytochrome b6f |
| DHAP | dihydroxyacetone phosphate |
| ETC1 | ENHANCER OF TRIPTYCHON AND CAPRICE1 |
| FBPase | fructose 1,6-bisphosphatase |
| FRAP | fluorescence recovery after photobleaching |
| F6-P | fructose 6-phosphate |
| GAPDH | glyceraldehyde 3-phosphate dehydrogenase |
| GAPC2 | glyceraldehyde-3-phosphate dehydrogenase cytosolic 2 |
| GCN5 | general control nonderepressible 5 |
| GL2 | GLABRA2 |
| GPI | glucose phosphate isomerase |
| HATs | histone acetyltransferases |
| HCT | HC-toxin |
| HDACs | histone deacetylases |
| HDT701 | histone deacetylase701 |
| JA | jasmonic acid |
| G3P | glyceraldehyde 3-phosphate |
| LysAc | lysine acetylation |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NSI | nuclear shuttle interacting |
| PDIL | protein disulfide isomerase |
| PGK | phosphoglycerate kinase |
| PGA | phosphoglycerate |
| PORA | protochlorophyllide oxidoreductase A |
| PSI | photosystem I |
| PSI-LHCI | photosystem I light-harvesting complex I |
| PSII | photosystem II |
| PSII-LHCII | photosystem II light-harvesting complex II |
| PTMs | protein post-translational modifications |
| RbcL | Rubisco large subunit |
| RCA | RuBisCO activase |
| RPE | ribulose phosphate epimerase; |
| RuBP | ribulose 1,5-bisphosphonate |
| Rubisco | ribulose 1,5-bisphosphate carboxylase/oxygenase |
| Ru5P | ribulose 5-phosphate |
| RPI | ribose-5-phosphate isomerase |
| R5P | ribose-5-phosphate |
| SBP | sedoheptulose-1,7-bisphosphate |
| SBPase | sedoheptulose-1,7-bisphosphatase |
| S7P | sedoheptulose-7-phosphate |
| TCA cycle | tricarboxylic acid cycle |
| TPL | TOPLESS |
| TPI | triose phosphate isomerase |
| WER | WEREWOLF |
| WOX11 | WUSCHEL-RELATED HOMEOBOX 11 |
| Xu5P | xylulose-5-phosphate |
| 2PG | 2-phosphoglycerate |
| 3-PGA | 3-phosphoglycerate |
References
- Allen, J.F. How does protein phosphorylation regulate photosynthesis? Trends Biochem. Sci. 1992, 17, 12–17. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Liu, M.; Zhu, L.; Luo, W.; Wang, Q.; Wang, M.; Chen, H.; Luo, Z.; Xiao, Y.; Zhang, Y.; et al. The E3 ubiquitin ligase CSIT1 regulates critical sterility-inducing temperature by ribosome-associated quality control to safeguard two-line hybrid breeding in rice. Mol. Plant 2023, 16, 1695–1709. [Google Scholar] [CrossRef]
- Yang, M.; Huang, H.; Ge, F. Lysine propionylation is a widespread post-translational modification involved in regulation of photosynthesis and metabolism in Cyanobacteria. Int. J. Mol. Sci. 2019, 20, 4792. [Google Scholar] [CrossRef]
- Finkemeier, I.; Laxa, M.; Miguet, L.; Howden, A.; Sweetlove, L. Proteins of diverse function and subcellular location are lysine acetylated in Arabidopsis. Plant Physiol. 2011, 155, 1779–1790. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.M.; Sprung, R.; Pei, J.M.; Tan, X.H.; Kim, S.; Zhu, H.; Liu, C.F.; Grishin, N.; Zhao, Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteom. 2009, 8, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Trefely, S.; Lovell, C.D.; Snyder, N.W.; Wellen, K.E. Compartmentalised acylCoA metabolism and roles in chromatin regulation. Mol. Metab. 2020, 38, 100941. [Google Scholar] [CrossRef]
- Shvedunova, M.; Akhtar, A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 329–349. [Google Scholar] [CrossRef]
- Verdin, E.; Ott, M. 50 years of protein acetylation: From gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 2015, 16, 258–264. [Google Scholar] [CrossRef]
- Rao, R.S.; Thelen, J.J.; Miernyk, J.A. Is Lys-Nε-acetylation the next big thing in post-translational modifications? Trends Plant Sci. 2014, 19, 550–553. [Google Scholar] [CrossRef]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversifcation of chromatin modifcation among multicellular eukaryotes. Nucleic. Acids. Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef]
- Hou, J.Q.; Ren, R.F.; Xiao, H.Z.; Chen, Z.F.; Yu, J.F.; Zhang, H.R.; Shi, Q.P.; Hou, H.L.; He, S.B.; Li, L.J. Characteristic and evolution of HAT and HDAC genes in Gramineae genomes and their expression analysis under diverse stress in Oryza sativa. Planta 2021, 253, 72. [Google Scholar] [CrossRef]
- Guo, J.; Wei, L.; Chen, S.S.; Cai, X.W.; Su, Y.N.; Li, L.; Chen, S.; He, X.J. The CBP/p300 histone acetyltransferases function as plant-specific MEDIATOR subunits in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 755–771. [Google Scholar] [CrossRef]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef]
- Kumar, V.; Thakur, J.K.; Prasad, M. Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 2021, 78, 4467–4486. [Google Scholar] [CrossRef]
- Zheng, Y.; Ge, J.Y.; Bao, C.; Chang, W.W.; Liu, J.J.; Shao, J.J.; Liu, X.Y.; Su, L.F.; Pan, L.; Zhou, D.X. Histone deacetylase HDA9 and WRKY53 transcription factor are mutual antagonists in regulation of plant stress response. Mol. Plant 2020, 13, 598–611. [Google Scholar] [CrossRef]
- Yang, L.Y.; Chen, X.S.; Wang, Z.X.; Sun, Q.; Hong, A.; Zhang, A.Q.; Zhong, X.H.; Hua, J. HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol. 2020, 226, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Schocken, J.; Kaldis, A.; Vlachonasios, K.E.; Hark, A.T.; McCain, E.R. The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in Arabidopsis. Planta 2009, 230, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.M.; Peng, T.; Chen, C.Y.; Ji, R.J.; Gu, D.C.; Li, T.T.; Zhang, D.D.; Tu, Y.T.; Wu, K.Q.; Liu, X.C. HY5 interacts with the histone deacetylase HDA15 to rrepress hypocotyl cell elongation in photomorphogenesis. Plant Physiol. 2019, 180, 1450–1466. [Google Scholar] [CrossRef]
- Kong, L.Y.; Zhi, P.F.; Liu, J.; Li, H.Y.; Zhang, X.N.; Xu, J.; Zhou, J.Q.; Wang, X.Y.; Chang, C. Epigenetic activation of enoyl-CoA reductase by an acetyltransferase complex triggers wheat wax biosynthesis. Plant Physiol. 2020, 183, 1250–1267. [Google Scholar] [CrossRef]
- Ding, B.; Bellizzi, M.d.R.; Ning, Y.; Meyers, B.C.; Wang, G.L. HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 2012, 24, 3783–3794. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Yan, S.H.; Zhao, L.; Tan, J.J.; Zhang, Q.; Gao, F.; Wang, P.; Hou, H.L.; Li, L.J. Histone acetylation associated up-regulation of the cell wall related genes is involved in salt stress induced maize root swelling. BMC Plant Biol. 2014, 14, 105. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, X.B.; Lin, J.C.; Liu, X.Y.; Wang, Z.Y.; Xin, M.M.; Yao, Y.Y.; Peng, H.R.; Zhou, D.X.; Ni, Z.F.; et al. Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. 2019, 97, 587–602. [Google Scholar] [CrossRef]
- Wang, T.Y.; Xing, J.W.; Liu, Z.S.; Zheng, M.; Yao, Y.Y.; Hu, Z.R.; Peng, H.R.; Xin, M.M.; Zhou, D.X.; Ni, Z.F. Histone acetyltransferase GCN5-mediated regulation of long non-coding RNA At4 contributes to phosphate starvation response in Arabidopsis. J. Exp. Bot. 2019, 70, 6337–6348. [Google Scholar] [CrossRef]
- Yu, Y.; Zhao, F.; Yue, Y.P.; Zhao, Y.; Zhou, D.X. Lysine acetylation of histone acetyltransferase adaptor protein ADA2 is a mechanism of metabolic control of chromatin modification in plants. Nat. Plants 2024, 10, 439–452. [Google Scholar] [CrossRef]
- Hou, J.Q.; Xiao, H.Z.; Yao, P.; Ma, X.C.; Shi, Q.P.; Yang, J.; Hou, H.L.; Li, L.J. Unveiling the mechanism of broad-spectrum blast resistance in rice: The collaborative role of transcription factor OsGRAS30 and histone deacetylase OsHDAC1. Plant Biotechnol. J. 2024, 22, 1740–1756. [Google Scholar] [CrossRef]
- Giehl, R.F.; Gruber, B.D.; von Wirén, N. It’s time to make changes: Modulation of root system architecture by nutrient signals. J. Exp. Bot. 2014, 65, 769–778. [Google Scholar] [CrossRef]
- Czyzewicz, N.; Shi, C.L.; Vu, L.D.; Van De Cotte, B.; Hodgman, C.; Butenko, M.A.; De Smet, I. Modulation of Arabidopsis and monocot root architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 peptide. J. Exp. Bot. 2015, 66, 5229–5243. [Google Scholar] [CrossRef]
- Li, D.X.; Chen, W.Q.; Xu, Z.H.; Bai, S.N. HISTONE DEACETYLASE6-defective mutants show increased expression and acetylation of ENHANCER OF TRIPTYCHON AND CAPRICE1 and GLABRA2 with small but significant effects on root epidermis cellular pattern. Plant Physiol. 2015, 168, 1448–1458. [Google Scholar] [CrossRef]
- Otero, S.; Desvoyes, B.; Peiró, R.; Gutierrez, C. Histone H3 dynamics reveal domains with distinct proliferation potential in the Arabidopsis root. Plant Cell 2016, 28, 1361–1371. [Google Scholar] [CrossRef]
- Rosa, S.; Ntoukakis, V.; Ohmido, N.; Pendle, A.; Abranches, R.; Shaw, P. Cell differentiation and development in Arabidopsis are associated with changes in histone dynamics at the single-cell level. Plant Cell 2014, 26, 4821–4833. [Google Scholar] [CrossRef]
- Zhou, S.L.; Jiang, W.; Long, F.; Cheng, S.F.; Yang, W.J.; Zhao, Y.; Zhou, D.X. Rice homeodomain protein WOX11 recruits a histone acetyltransferase complex to establish programs of cell proliferation of crown root meristem. Plant Cell 2017, 29, 1088–1104. [Google Scholar] [CrossRef]
- Xu, C.R.; Liu, C.; Wang, Y.L.; Li, L.C.; Chen, W.Q.; Xu, Z.H.; Bai, S.N. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proc. Natl. Acad. Sci. USA 2005, 102, 14469–14474. [Google Scholar] [CrossRef]
- Tersenidis, C.; Poulios, S.; Komis, G.; Panteris, E.; Vlachonasios, K. Roles of histone acetylation and deacetylation in root development. Plants 2024, 13, 2760. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specifc DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606. [Google Scholar] [CrossRef]
- Fang, X.; Chen, W.; Zhao, Y.L.; Ruan, S.; Zhang, H.; Yan, C.; Jin, L.; Cao, L.; Zhu, J.; Ma, H.; et al. Global analysis of lysine acetylation in strawberry leaves. Front. Plant Sci. 2015, 6, 739. [Google Scholar] [CrossRef]
- Xiong, Q.; Chen, Z.; Ge, F. Proteomic analysis of post translational modifications in cyanobacteria. J. Proteom. 2016, 134, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Bienvenut, W.V.; Espagne, C.; Martinez, A.; Majeran, W.; Valot, B.; Zivy, M.; Vallon, O.; Adam, Z.; Meinnel, T.; Giglione, C. Dynamics of post-translational modifications and protein stability in the stroma of Chlamydomonas reinhardtii chloroplasts. Proteomics 2011, 11, 1734–1750. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Peng, X.; Cheng, Z.; Liu, W.; Wang, G.L. A comprehensive catalog of the lysine-acetylation targets in rice (Oryza sativa) based on proteomic analyses. J. Proteom. 2016, 138, 20–29. [Google Scholar] [CrossRef]
- Smith-Hammond, C.L.; Swatek, K.N.; Johnston, M.L.; Thelen, J.J.; Miernyk, J.A. Initial description of the developing soybean seed protein Lys-Nε-acetylome. J. Proteom. 2014, 96, 56–66. [Google Scholar] [CrossRef]
- Walley, J.W.; Shen, Z.; McReynolds, M.R.; Schmelz, E.A.; Briggs, S.P. Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl. Acad. Sci. USA 2018, 115, 210–215. [Google Scholar] [CrossRef]
- Mulligan, R.; Houtz, R.; Tolbert, N. Reaction-intermediate analogue binding by ribulose bisphosphate carboxylase/oxygenase causes specific changes in proteolytic sensitivity: The amino-terminal residue of the large subunit is acetylated proline. Proc. Natl. Acad. Sci. USA 1988, 85, 1513–1517. [Google Scholar] [CrossRef]
- Zybailov, B.; Rutschow, H.; Friso, G.; Rudella, A.; Emanuelsson, O.; Sun, Q.; van Wijk, K. Sorting signals, n-terminal modifications and abundance of the chloroplast proteome. PLoS ONE 2008, 3, e1994. [Google Scholar] [CrossRef]
- Wang, Y.; Hou, Y.; Qiu, J.; Li, Z.; Zhao, J.; Tong, X.; Zhang, J. A quantitative acetylomic analysis of early seed development in rice (Oryza sativa L.). Int. J. Mol. Sci. 2017, 18, 1376. [Google Scholar] [CrossRef]
- Meng, X.; Lv, Y.; Mujahid, H.; Edelmann, M.J.; Zhao, H.; Peng, X.; Peng, Z. Proteome-wide lysine acetylation identification in developing rice (Oryza sativa) seeds and protein co-modification by acetylation, succinylation, ubiquitination, and phosphorylation. Biochim. Biophys. Acta (BBA)-Proteins Proteom. 2018, 1866, 451–463. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Bello, B.K.; Ajadi, A.A.; Tong, X.; Chang, Y.; Zhang, J. Construction of a quantitative acetylomic tissue atlas in rice (Oryza sativa L.). Molecules 2018, 23, 2843. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Liu, Q.; Chen, Z.; Yue, Y.; Liu, Y.; Zhao, Y.; Zhou, D.X. Histone deacetylases control lysine acetylation of ribosomal proteins in rice. Nucleic Acids Res. 2021, 49, 4613–4628. [Google Scholar] [CrossRef] [PubMed]
- Smith-Hammond, C.L.; Hoyos, E.; Miernyk, J.A. The pea seedling mitochondrial Nε-lysine acetylome. Mitochondrion 2014, 19, 154–165. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Shen, Z.; Gao, Z.F.; Chao, Q.; Qian, C.R.; Zheng, H.; Wang, B.C. A comprehensive analysis of the lysine acetylome revealsdiverse functions of acetylated proteins during de-etiolation in Zea mays. J. Plant Physiol. 2020, 248, 153158. [Google Scholar] [CrossRef]
- König, A.C.; Hartl, M.; Pham, P.A.; Laxa, M.; Boersema, P.J.; Orwat, A.; Kalitventseva, I.; Plöchinger, M.; Braun, H.P.; Leister, D.; et al. The Arabidopsis class II sirtuin is a lysine deacetylase and interacts with mitochondrial energy metabolism. Plant Physiol. 2014, 164, 1401–1414. [Google Scholar] [CrossRef]
- Hartl, M.; Füßl, M.; Boersema, P.J.; Jost, J.O.; Kramer, K.; Bakirbas, A.; Sindlinger, J.; Plöchinger, M.; Leister, D.; Uhrig, G.; et al. Lysine acetylome profiling uncovers novel histone deacetylase substrate proteins in Arabidopsis. Mol. Syst. Biol. 2017, 13, 949. [Google Scholar] [CrossRef]
- Wu, X.; Oh, M.H.; Schwarz, E.M.; Larue, C.T.; Sivaguru, M.; Imai, B.S.; Yau, P.M.; Ort, D.R.; Huber, S.C. Lysine acetylation is a widespread protein modification for diverse proteins in Arabidopsis. Plant Physiol. 2011, 155, 1769–1778. [Google Scholar] [CrossRef]
- Zhou, H.; Finkemeier, I.; Guan, W.; Tossounian, M.A.; Wei, B.; Young, D.; Huang, J.; Messens, J.; Yang, X.; Zhu, J.; et al. Oxidative stress-triggered interactions between the succinyl- and acetyl-proteomes of rice leaves. Plant Cell Environ. 2018, 41, 1139–1153. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Yang, C.; Xiong, H.; Lin, Y.; Yao, J.; Li, H.; Xie, L.; Zhao, W.; Yao, Y.; et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science 2010, 327, 1004–1007. [Google Scholar] [CrossRef]
- Narita, T.; Weinert, B.T.; Choudhary, C. Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 2019, 20, 156–174. [Google Scholar] [CrossRef]
- Liu, Z.; Song, J.; Miao, W.; Yang, B.; Zhang, Z.; Chen, W.; Tan, F.; Suo, H.; Dai, X.; Zou, X.; et al. Comprehensive proteome and lysine acetylome analysis reveals the widespread involvement of acetylation in cold resistance of pepper (Capsicum annuum L.). Front. Plant Sci. 2021, 12, 730489. [Google Scholar] [CrossRef] [PubMed]
- Uhrig, R.G.; Schläpfer, P.; Roschitzki, B.; Hirsch-Hoffmann, M.; Gruissem, W. Diurnal changes in concerted plant protein phosphorylation and acetylation in Arabidopsis organs and seedlings. Plant J. 2019, 99, 176–194. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Chai, X.; Mei, Y.; Du, J.; Du, H.; Shi, H.; Zhu, J.K.; Zhang, H. Acetylproteomics analyses reveal critical features of lysine-ε-acetylation in Arabidopsis and a role of 14-3-3 protein acetylation in alkaline response. Stress Biol. 2022, 2, 1. [Google Scholar] [CrossRef]
- Tilak, P.; Kotnik, F.; Née, G.; Seidel, J.; Sindlinger, J.; Heinkow, P.; Eirich, J.; Schwarzer, D.; Finkemeier, I. Proteome-wide lysine acetylation profiling to investigate the involvement of histone deacetylase HDA5 in the salt stress response of Arabidopsis leaves. Plant J. 2023, 115, 275–292. [Google Scholar] [CrossRef]
- Rantala, M.; Ivanauskaite, A.; Laihonen, L.; Kanna, S.; Ughy, B.; Mulo, P. Chloroplast acetyltransferase GNAT2 is involved in the organization and dyna mics of thylakoid structure. Plant Cell Physiol. 2022, 63, 1205–1214. [Google Scholar] [CrossRef]
- Koskela, M.M.; Brünje, A.; Ivanauskaite, A.; Grabsztunowicz, M.; Lassowskat, I.; Neumann, U.; Dinh, T.V.; Sindlinger, J.; Schwarzer, D.; Wirtz, M.; et al. Chloroplast acetyltransferase NSI is required for state transitions in Arabidopsis thaliana. Plant Cell 2018, 30, 1695–1709. [Google Scholar] [CrossRef] [PubMed]
- Gao, X.; Hong, H.; Li, W.C.; Yang, L.; Huang, J.; Xiao, Y.L.; Chen, X.Y.; Chen, G.Y. Downregulation of Rubisco activity by non-enzymatic acetylation of RbcL. Mol. Plant 2016, 9, 1018–1027. [Google Scholar] [CrossRef]
- Liang, M.T.; Gu, D.C.; Lie, Z.Y.; Yang, Y.Y.; Lu, L.X.; Dai, G.Y.; Peng, T.; Deng, L.; Zheng, F.; Liu, X.C. Regulation of chlorophyll biosynthesis by light-dependent acetylation of NADPH: Protochlorophyll oxidoreductase A in Arabidopsis. Plant Sci. 2023, 330, 111641. [Google Scholar] [CrossRef]
- Lehtimäki, N.; Koskela, M.M.; Dahlström, K.M.; Pakula, E.; Lintala, M.; Scholz, M.; Hippler, M.; Hanke, G.T.; Rokka, A.; Battchikova, N.; et al. Posttranslational modifications of FERREDOXIN-NADP+ OXIDOREDUCTASE in Arabidopsis chloroplasts. Plant Physiol. 2014, 166, 1764–1776. [Google Scholar] [CrossRef]
- Angelovici, R.; Fait, A.; Fernie, A.R.; Galili, G.A. seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol. 2011, 189, 148–159. [Google Scholar] [CrossRef]
- Kim, S.C.; Sprung, R.; Chen, Y.; Xu, Y.; Ball, H.; Pei, J.; Cheng, T.; Kho, Y.; Xiao, H.; Xiao, L.; et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell 2006, 23, 607–618. [Google Scholar] [CrossRef]
- Guarente, L. The logic linking protein acetylation and metabolism. Cell Metab. 2011, 14, 151–153. [Google Scholar] [CrossRef][Green Version]
- König, A.C.; Hartl, M.; Boersema, P.J.; Mann, M.; Finkemeier, I. The mitochondrial lysine acetylome of Arabidopsis. Mitochondrion 2014, 19, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Back, K. Overexpression of rice serotonin N-acetyltransferase 1 in transgenic rice plants confers resistance to cadmium and senescence and increases grain yield. J. Pineal Res. 2017, 62, e12392. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.; Back, K. Cloning and characterization of the serotonin N-acetyltransferase-2 gene (SNAT2) in rice (Oryza sativa). J. Pineal. Res. 2016, 61, 198–207. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; He, J.; Velanis, C.N.; Zhu, Y.; He, Y.; Tang, K.; Zhu, M.; Graser, L.; de Leau, E.; Wang, X.; et al. A domesticated Harbinger transposase forms a complex with HDA6 and promotes histone H3 deacetylation at genes but not TEs in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 1462–1474. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.Q.; Zheng, X.K.; Ren, R.F.; Shi, Q.P.; Xiao, H.Z.; Chen, Z.F.; Yue, M.X.; Wu, Y.Q.; Hou, H.L.; Li, L.J. The histone deacetylase 1/GSK3/SHAGGY-like kinase 2/BRASSINAZOLE-RESISTANT 1 module controls lateral root formation in rice. Plant Physiol. 2022, 189, 858–873. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.X.; Chen, W.; Ma, C.L.; Shen, S.Y.; Zhou, Y.Y.; Zhou, L.Q.; Chen, L. Proteome and acetyl-proteome profiling of camellia sinensis cv. ‘Anjin Baicha’ during periodic albinism reveals alterations in photosynthetic and secondary metabolite biosynthetic pathways. Front. Plant Sci. 2017, 11, 2104. [Google Scholar]
- Fujii, S.; Kazama, T.; Yamada, M.; Toriyama, K. Discovery of global genomic re-organization based on comparison of two newly sequenced rice mitochondrial genomes with cytoplasmic male sterility-related genes. BMC Genom. 2010, 11, 209–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Boavida, L.C.; Ron, M.; McCormick, S. Truncation of a protein disulfide isomerase, PDIL2-1, delays embryosac maturation and disrupts pollen tube guidance in Arabidopsis thaliana. Plant Cell 2019, 20, 3300–3311. [Google Scholar] [CrossRef]
- Chen, P.; Wei, F.; Li, R.; Li, Z.Q.; Kashif, M.H.; Zhou, R.Y. Comparative acetylomic analysis reveals differentially acetylatedproteins regulating anther and pollen development in kenaf cytoplasmic male sterility line. Physiol. Plant 2019, 166, 960–978. [Google Scholar] [CrossRef]
- Lämke, J.; Bäurle, I. Epigenetic and chromatin-based mechanisms in environmental stress adaptation and stress memory in plants. Genome Biol. 2017, 18, 124. [Google Scholar] [CrossRef]
- Gururani, M.A.; Venkatesh, J.; Tran, L.S. Regulation of photosynthesis during abiotic stress-induced photoinhibition. Mol. Plant 2015, 8, 1304–1320. [Google Scholar] [CrossRef]
- An, C.; Deng, L.; Zhai, H.; You, Y.; Wu, F.; Zhai, Q.; Goossens, A.; Li, C. Regulation of jasmonate signaling by reversible acetylation of TOPLESS in Arabidopsis. Mol. Plant 2022, 15, 1329–1346. [Google Scholar] [CrossRef]
- Yang, Z.H.; Du, J.; Tan, X.H.; Zhang, H.H.; Li, L.L.; Li, Y.J.; Wei, Z.Y.; Xu, Z.T.; Lu, Y.W.; Chen, J.P.; et al. Histone deacetylase OsHDA706 orchestrates rice broad-spectrum antiviral immunity and is impeded by a viral effector. Cell Rep. 2024, 43, 113838. [Google Scholar] [CrossRef]
- Zhang, N.; Lv, F.; Qiu, F.; Han, D.; Xu, Y.; Liang, W. Pathogenic fungi neutralize plant-derived ROS via Srpk1 deacetylation. EMBO J. 2023, 42, e112634. [Google Scholar] [CrossRef]
- Zhang, N.; Hu, J.; Liu, Z.; Liang, W.; Song, L. Sir2-mediated cytoplasmic deacetylation facilitates pathogenic fungi infection in host plants. New Phytol. 2024, 241, 1732–1746. [Google Scholar] [CrossRef]
- Sun, Y.; Xie, Z.Z.; Jin, L.; Qin, T.; Zhan, C.H.; Huang, J.L. Histone deacetylase OsHDA716 represses rice chilling tolerance by deacetylating OsbZIP46 to reduce its transactivation function and protein stability. Plant Cell 2024, 36, 1913–1936. [Google Scholar] [CrossRef]
- Liu, Z.; Yang, Q.; Liu, X.; Li, J.; Zhang, L.; Chu, W.; Lin, J.; Liu, D.; Zhao, D.; Peng, X.; et al. Suppression of TaHDA8-mediated lysine deacetylation of TaAREB3 acts as a drought adaptive mechanism in wheat root development. Mol. Plant 2025, 18, 1222–1240. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.Y.; Tu, Y.T.; Hsu, J.C.; Hung, H.C.; Liu, T.C.; Lee, Y.H.; Chou, C.C.; Cheng, Y.S.; Wu, K.Q. Structure of Arabidopsis HISTONE DEACETYLASE15. Plant Physiol. 2020, 184, 1585–1600. [Google Scholar] [CrossRef]
- Yu, C.W.; Tai, R.; Wang, S.C.; Yang, P.; Luo, M.; Yang, S.; Cheng, K.; Wang, W.C.; Cheng, Y.S.; Wu, K.Q. HISTONE DEACETYLASE6 Acts in Concert with Histone Methyltransferases SUVH4, SUVH5, and SUVH6 to Regulate Transposon Silencing. Plant Cell 2017, 29, 1970–1983. [Google Scholar] [CrossRef]
- Hao, Y.H.; Wang, H.J.; Qiao, S.L.; Leng, L.N.; Wang, X.L. Histone deacetylase HDA6 enhances brassinosteroid signaling by inhibiting the BIN2 kinase. Proc. Natl. Acad. Sci. USA 2016, 113, 10418–10423. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.W.; Guan, S.H.; Sun, Y.; Deng, Z.P.; Tang, W.; Shang, J.X.; Sun, Y.; Burlingame, A.L.; Wang, Z.Y. Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 2009, 11, 1254–1260. [Google Scholar] [CrossRef] [PubMed]
- Senior, A.W.; Evans, R.; Jumper, J.; Kirkpatrick, J.; Sifre, L.; Green, T.; Qin, C.; Žídek, A.; Nelson, A.W.R.; Bridgland, A.; et al. Improved protein structure prediction using potentials from deep learning. Nature 2020, 577, 706–710. [Google Scholar] [CrossRef]
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444. [Google Scholar] [CrossRef]

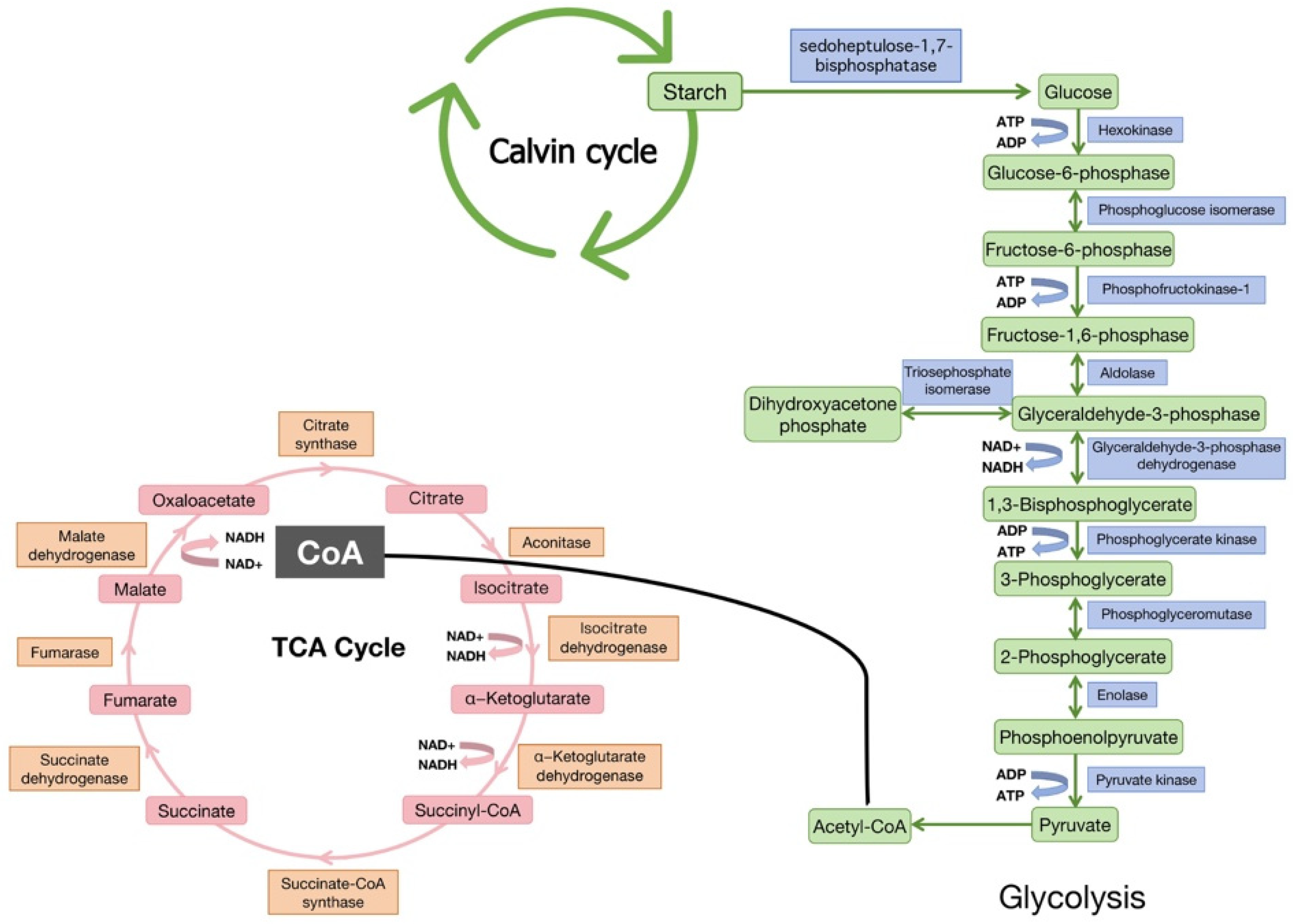
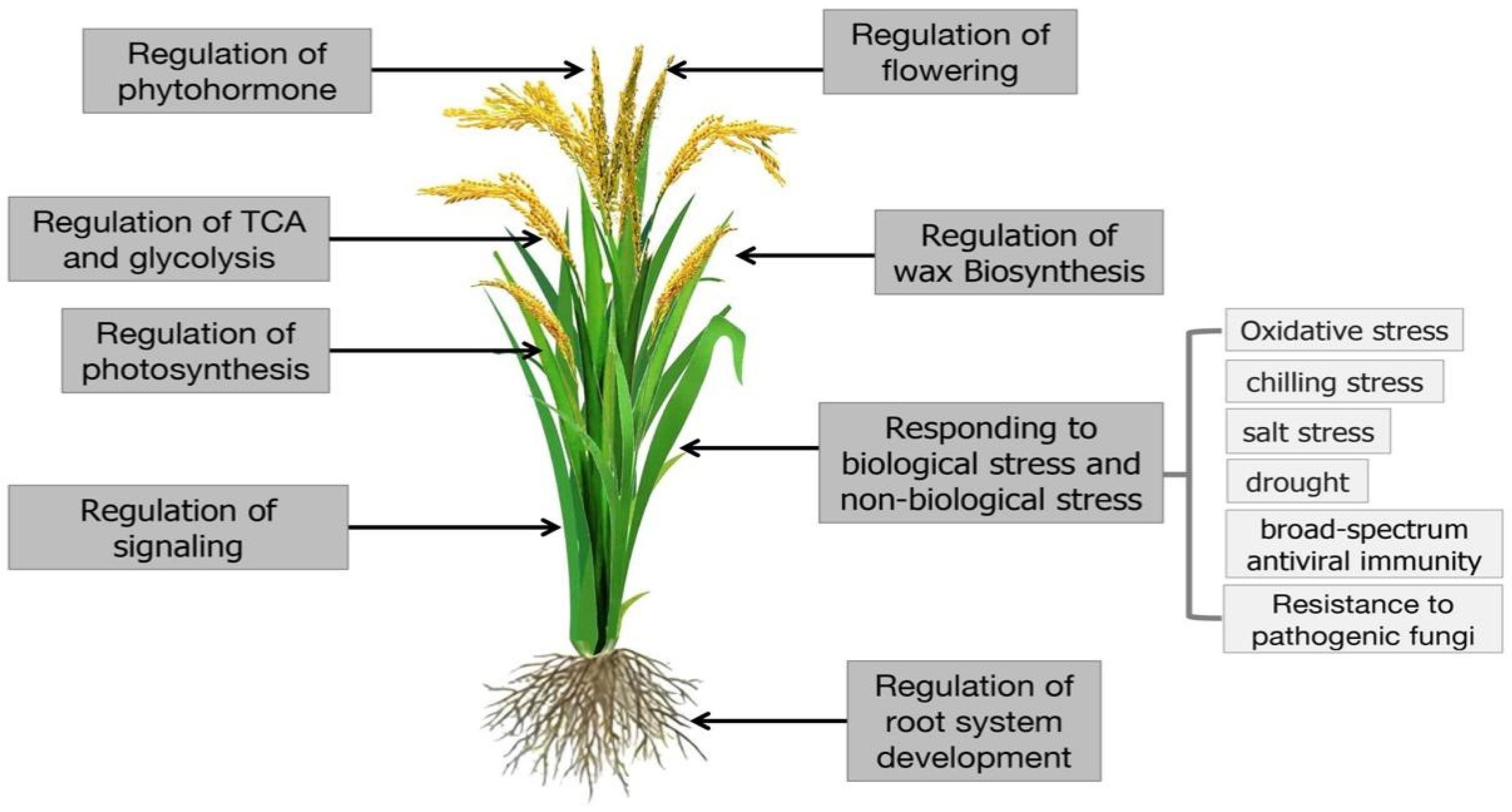
| Species | Number of Kac Sites | Number of Kac Proteins | Tissues/Organs | Main Location | References |
|---|---|---|---|---|---|
| Arabidopsis thaliana | 91 | 74 | Leaves | Chloroplast | [4] |
| Arabidopsis thaliana | 348 | 204 | Mitochondria | Mitochondria | [49] |
| Arabidopsis thaliana | 2152 | 1022 | Leaves | Chloroplast, mitochondria, cytoplasm, nucleus | [50] |
| Arabidopsis thaliana | 64 | 57 | Leaves | Chloroplast, mitochondria, cytoplasm, nucleus | [51] |
| Fragaria ananassa | 1392 | 684 | Leaves | Chloroplast, mitochondria, cytoplasm, nucleus | [35] |
| Oryza sativa | 1337 | 716 | Leaves, stems, roots | Chloroplast, mitochondria, cytoplasm, nucleus | [36] |
| Oryza sativa | 1003 | 692 | Seeds | Vacuole, mitochondria, cytoplasm, nucleus | [44] |
| Oryza sativa | 1536 | 890 | Callus, root, leaves, panicle | Chloroplast, mitochondria, cytoplasm, nucleus | [45] |
| Oryza sativa | 1669 | 1024 | Leaves | Ribosome, chloroplast, mitochondria, nucleus | [52] |
| Oryza sativa | 4868 | 1952 | Seeds | Ribosome | [46] |
| Glycine max | 245 | 400+ | Seeds | Mitochondria, cytoplasm, nucleus | [39] |
| Pisum sativum | 358 | 664 | Mitochondria | Mitochondria | [47] |
| Zea mays | 912 | 2791 | Leaves | Chloroplast, mitochondria, cytoplasm, nucleus | [40] |
| Zea mays | 462 | 814 | Leaves | Chloroplast, mitochondria, cytoplasm, nucleus | [48] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Li, X.; He, Y.; Chen, X.; Li, J.; Zhuang, C. Progress of Acetylation Modification in Plants. Agronomy 2025, 15, 1910. https://doi.org/10.3390/agronomy15081910
Li R, Li X, He Y, Chen X, Li J, Zhuang C. Progress of Acetylation Modification in Plants. Agronomy. 2025; 15(8):1910. https://doi.org/10.3390/agronomy15081910
Chicago/Turabian StyleLi, Ruiqi, Xuezhong Li, Ying He, Xiaoyuan Chen, Jing Li, and Chuxiong Zhuang. 2025. "Progress of Acetylation Modification in Plants" Agronomy 15, no. 8: 1910. https://doi.org/10.3390/agronomy15081910
APA StyleLi, R., Li, X., He, Y., Chen, X., Li, J., & Zhuang, C. (2025). Progress of Acetylation Modification in Plants. Agronomy, 15(8), 1910. https://doi.org/10.3390/agronomy15081910





