Quantitative Analysis of Adulteration in Anoectochilus roxburghii Powder Using Hyperspectral Imaging and Multi-Channel Convolutional Neural Network
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Preparation
2.2. Hyperspectral Image System
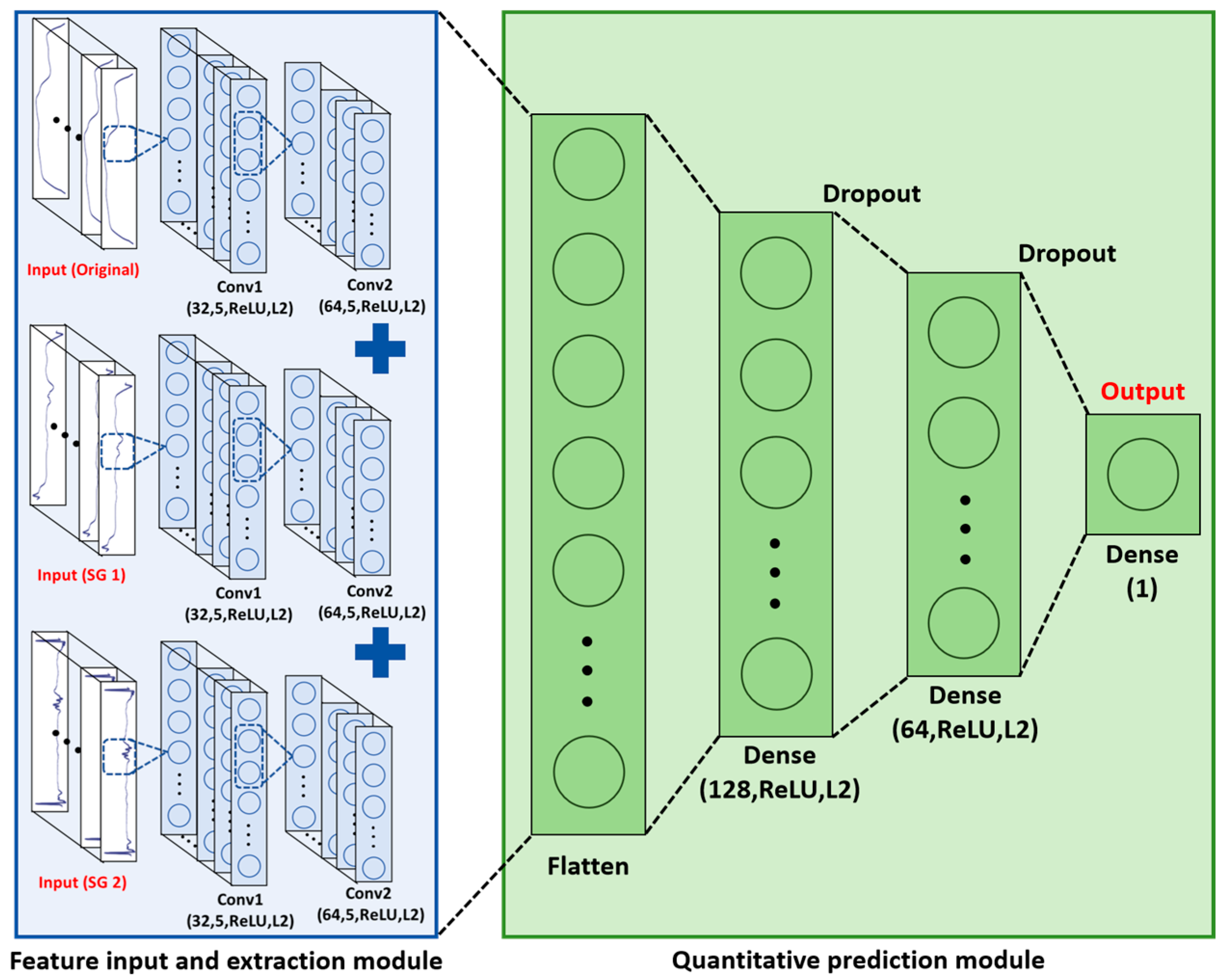
2.3. Spectral Data Analysis Models
3. Results and Discussion
3.1. Data Preprocessing
3.2. Traditional Machine Learning Models
3.3. Single-Channel Deep Learning Model
3.4. Multi-Channel Deep Learning Model
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Qu, A.; Wu, Q.; Su, J.; Li, C.; Yang, L.; Wang, Z.; Wang, Z.; Li, Z.; Ruan, X.; Zhao, Y.; et al. A Review on the Composition and Biosynthesis of Alkaloids and on the Taxonomy, Domestication, and Cultivation of Medicinal Fritillaria Species. Agronomy 2022, 12, 1844. [Google Scholar] [CrossRef]
- Bao, H.; Bao, H.; Wang, Y.; Wang, F.; Jiang, Q.; He, X.; Li, H.; Ding, Y.; Zhu, C. Challenges and Strategies in the Industrial Application of Dendrobium Officinale. Plants 2024, 13, 2961. [Google Scholar] [CrossRef]
- Luo, Q.; Meng, P.-H.; Jiang, D.-W.; Han, Z.-M.; Wang, Z.-H.; Tan, G.-F.; Zhang, J. Comprehensive Assessment of Houttuynia Cordata Thunb., an Important Medicinal Plant and Vegetable. Agronomy 2022, 12, 2582. [Google Scholar] [CrossRef]
- Zhu, S.; Liu, Q.; Qiu, S.; Dai, J.; Gao, X. DNA Barcoding: An Efficient Technology to Authenticate Plant Species of Traditional Chinese Medicine and Recent Advances. Chin. Med. 2022, 17, 112. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Yang, H.; Zheng, M.; Zhou, G.; Yang, Y.; Liu, S. Physiological and Transcriptomic Analyses Reveal the Regulatory Mechanisms of Anoectochilus Roxburghii in Response to High-Temperature Stress. BMC Plant Biol. 2024, 24, 584. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.; Shao, Q.; Xu, M.; Li, S.; Wu, M.; Tan, X.; Su, L. Effects of Light Quality on Morphology, Enzyme Activities, and Bioactive Compound Contents in Anoectochilus Roxburghii. Front. Plant Sci. 2017, 8, 857. [Google Scholar] [CrossRef] [PubMed]
- Han, T.; Xu, E.; Yao, L.; Zheng, B.; Younis, A.; Shao, Q. Regulation of Flowering Time Using Temperature, Photoperiod and Spermidine Treatments in Anoectochilus Roxburghii. Physiol. Mol. Biol. Plants 2020, 26, 247–260. [Google Scholar] [CrossRef]
- Xing, B.; Wan, S.; Su, L.; Riaz, M.W.; Li, L.; Ju, Y.; Zhang, W.; Zheng, Y.; Shao, Q. Two Polyamines -Responsive WRKY Transcription Factors from Anoectochilus Roxburghii Play Opposite Functions on Flower Development. Plant Sci. 2023, 327, 111566. [Google Scholar] [CrossRef]
- Huang, T.; Wu, Y.; Huang, L.; Lin, R.; Li, Z.; Wang, X.; Wu, P.; Huang, L. Mechanism of the Effect of Compound Anoectochilus Roxburghii (Wall.) Lindl. Oral Liquid in Treating Alcoholic Rat Liver Injury by Metabolomics. Drug Des. Dev. Ther. 2023, 17, 3409–3428. [Google Scholar] [CrossRef]
- Jin, Q.-R.; Mao, J.-W.; Zhu, F. The Effects of Anoectochilus Roxburghii Polysaccharides on the Innate Immunity and Disease Resistance of Procambarus Clarkii. Aquaculture 2022, 555, 738210. [Google Scholar] [CrossRef]
- Xu, M.; Shao, Q.; Ye, S.; Li, S.; Wu, M.; Ding, M.; Li, Y. Simultaneous Extraction and Identification of Phenolic Compounds in Anoectochilus Roxburghii Using Microwave-Assisted Extraction Combined with UPLC-Q-TOF-MS/MS and Their Antioxidant Activities. Front. Plant Sci. 2017, 8, 1474. [Google Scholar] [CrossRef]
- Wang, P.; Yan, P.; Li, Z.; Jiang, J.; Lin, Y.; Ye, W. Transcriptomic and Metabolomic Insights into Key Genes Involved in Kinsenoside Biosynthesis in Anoectochilus Roxburghii. Plants 2025, 14, 688. [Google Scholar] [CrossRef]
- Xu, Y.; Ding, H.; Zhang, T.; Wang, Z.; Wang, H.; Zhou, L.; Dai, Y.; Liu, Z. Small-Sample Authenticity Identification and Variety Classification of Anoectochilus Roxburghii (Wall. ) Lindl. Using Hyperspectral Imaging and Machine Learning. Plants 2025, 14, 1177. [Google Scholar] [CrossRef]
- Vega-Castellote, M.; Sánchez, M.-T.; Torres-Rodríguez, I.; Entrenas, J.-A.; Pérez-Marín, D. NIR Sensing Technologies for the Detection of Fraud in Nuts and Nut Products: A Review. Foods 2024, 13, 1612. [Google Scholar] [CrossRef] [PubMed]
- Nallan Chakravartula, S.S.; Moscetti, R.; Bedini, G.; Nardella, M.; Massantini, R. Use of Convolutional Neural Network (CNN) Combined with FT-NIR Spectroscopy to Predict Food Adulteration: A Case Study on Coffee. Food Control 2022, 135, 108816. [Google Scholar] [CrossRef]
- Yu, D.; Guo, S.; Zhang, X.; Yan, H.; Zhang, Z.; Chen, X.; Chen, J.; Jin, S.; Yang, J.; Duan, J. Rapid Detection of Adulteration in Powder of Ginger (Zingiber Officinale Roscoe) by FT-NIR Spectroscopy Combined with Chemometrics. Food Chem. X 2022, 15, 100450. [Google Scholar] [CrossRef]
- Siche, R.; Vejarano, R.; Aredo, V.; Velasquez, L.; Saldaña, E.; Quevedo, R. Evaluation of Food Quality and Safety with Hyperspectral Imaging (HSI). Food Eng. Rev. 2016, 8, 306–322. [Google Scholar] [CrossRef]
- Li, W.; Huang, W.; Fan, D.; Gao, X.; Zhang, X.; Meng, Y.; Liu, T.C.Y. Rapid Quantification of Goat Milk Adulteration with Cow Milk Using Raman Spectroscopy and Chemometrics. Anal. Methods 2022, 15, 455–461. [Google Scholar] [CrossRef]
- Qi, H.; Huang, Z.; Sun, Z.; Tang, Q.; Zhao, G.; Zhu, X.; Zhang, C. Rice Seed Vigor Detection Based on Near-Infrared Hyperspectral Imaging and Deep Transfer Learning. Front. Plant Sci. 2023, 14, 1283921. [Google Scholar] [CrossRef]
- Zhou, Y.; Li, X.; Chen, C.; Zhou, L.; Zhao, Y.; Chen, J.; Tan, C.; Sun, J.; Zhang, L.; Hu, M.; et al. Coupling the PROSAIL Model and Machine Learning Approach for Canopy Parameter Estimation of Moso Bamboo Forests from UAV Hyperspectral Data. Forests 2024, 15, 946. [Google Scholar] [CrossRef]
- Zhao, D.; Pang, Y.; Liu, L.; Li, Z. Individual Tree Classification Using Airborne LiDAR and Hyperspectral Data in a Natural Mixed Forest of Northeast China. Forests 2020, 11, 303. [Google Scholar] [CrossRef]
- Ram, B.G.; Oduor, P.; Igathinathane, C.; Howatt, K.; Sun, X. A Systematic Review of Hyperspectral Imaging in Precision Agriculture: Analysis of Its Current State and Future Prospects. Comput. Electron. Agric. 2024, 222, 109037. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Wang, A. Comparison of Various Approaches for Estimating Leaf Water Content and Stomatal Conductance in Different Plant Species Using Hyperspectral Data. Ecol. Indic. 2022, 142, 109278. [Google Scholar] [CrossRef]
- Falcioni, R.; Santos, G.L.A.A.D.; Crusiol, L.G.T.; Antunes, W.C.; Chicati, M.L.; Oliveira, R.B.D.; Demattê, J.A.M.; Nanni, M.R. Non−Invasive Assessment, Classification, and Prediction of Biophysical Parameters Using Reflectance Hyperspectroscopy. Plants 2023, 12, 2526. [Google Scholar] [CrossRef] [PubMed]
- Lim, J.; Kim, G.; Mo, C.; Kim, M.S.; Chao, K.; Qin, J.; Fu, X.; Baek, I.; Cho, B.-K. Detection of Melamine in Milk Powders Using Near-Infrared Hyperspectral Imaging Combined with Regression Coefficient of Partial Least Square Regression Model. Talanta 2016, 151, 183–191. [Google Scholar] [CrossRef]
- Nargesi, M.H.; Amiriparian, J.; Bagherpour, H.; Kheiralipour, K. Detection of Different Adulteration in Cinnamon Powder Using Hyperspectral Imaging and Artificial Neural Network Method. Results Chem. 2024, 9, 101644. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, Q.; Zhang, T.; Zhao, S.; Gao, X.; Sun, T.; Dai, Y. Quantitative Modeling and Uncertainty Estimation for Small-Sample LIBS Using Gaussian Negative Log-Likelihood and Monte Carlo Dropout Methods. Opt. Laser Technol. 2025, 181, 111720. [Google Scholar] [CrossRef]
- Dai, Y.; Ma, Q.; Zhang, T.; Zhao, S.; Zhou, L.; Gao, X.; Liu, Z. Classification of Aluminum Alloy Using Laser-Induced Breakdown Spectroscopy Combined with Discriminative Restricted Boltzmann Machine. Chemom. Intell. Lab. Syst. 2025, 258, 105342. [Google Scholar] [CrossRef]
- Ahmad, T.; Aziz, M.N. Data Preprocessing and Feature Selection for Machine Learning Intrusion Detection Systems. ICIC Express Lett. 2019, 13, 93–101. [Google Scholar] [CrossRef]
- Rochac, J.F.R.; Zhang, N.; Thompson, L.; Oladunni, T. A Data Augmentation-Assisted Deep Learning Model for High Dimensional and Highly Imbalanced Hyperspectral Imaging Data. In Proceedings of the 2019 9th International Conference on Information Science and Technology (ICIST), Hulunbuir, China, 2–5 August 2019; pp. 362–367. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral–Spatial Feature Extraction for Hyperspectral Image Classification: A Dimension Reduction and Deep Learning Approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]
- Gao, W.; Xiao, Z.; Bao, T. Detection and Identification of Potato-Typical Diseases Based on Multidimensional Fusion Atrous-CNN and Hyperspectral Data. Appl. Sci. 2023, 13, 5023. [Google Scholar] [CrossRef]
- Yan, T.; Xu, W.; Lin, J.; Duan, L.; Gao, P.; Zhang, C.; Lv, X. Combining Multi-Dimensional Convolutional Neural Network (CNN) With Visualization Method for Detection of Aphis Gossypii Glover Infection in Cotton Leaves Using Hyperspectral Imaging. Front. Plant Sci. 2021, 12, 604510. [Google Scholar] [CrossRef]
- Chen, X.; Lv, X.; Ma, L.; Chen, A.; Zhang, Q.; Zhang, Z. Optimization and Validation of Hyperspectral Estimation Capability of Cotton Leaf Nitrogen Based on SPA and RF. Remote Sens. 2022, 14, 5201. [Google Scholar] [CrossRef]
- Gong, J.; Chen, G.; Deng, Y.; Li, C.; Fang, K. Non-Destructive Detection of Tea Polyphenols in Fu Brick Tea Based on Hyperspectral Imaging and Improved PKO-SVR Method. Agriculture 2024, 14, 1701. [Google Scholar] [CrossRef]
- Burnett, A.C.; Anderson, J.; Davidson, K.J.; Ely, K.S.; Lamour, J.; Li, Q.; Morrison, B.D.; Yang, D.; Rogers, A.; Serbin, S.P. A Best-Practice Guide to Predicting Plant Traits from Leaf-Level Hyperspectral Data Using Partial Least Squares Regression. J. Exp. Bot. 2021, 72, 6175–6189. [Google Scholar] [CrossRef]
- Feng, G.; Li, Z.; Zhang, J.; Wang, M. Multi-Scale Spatial Attention-Based Multi-Channel 2D Convolutional Network for Soil Property Prediction. Sensors 2024, 24, 4728. [Google Scholar] [CrossRef] [PubMed]
- Alsakar, Y.M.; Sakr, N.A.; Elmogy, M. An Enhanced Classification System of Various Rice Plant Diseases Based on Multi-Level Handcrafted Feature Extraction Technique. Sci. Rep. 2024, 14, 30601. [Google Scholar] [CrossRef] [PubMed]
- Signoroni, A.; Savardi, M.; Baronio, A.; Benini, S. Deep Learning Meets Hyperspectral Image Analysis: A Multidisciplinary Review. J. Imaging 2019, 5, 52. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Islam, M.T.; Uddin, M.P.; Ulhaq, A. Improving Hyperspectral Image Classification with Compact Multi-Branch Deep Learning. Remote Sens. 2024, 16, 2069. [Google Scholar] [CrossRef]
- Li, P.; Tang, S.; Chen, S.; Tian, X.; Zhong, N. Hyperspectral Imaging Combined with Convolutional Neural Network for Accurately Detecting Adulteration in Atlantic Salmon. Food Control 2023, 147, 109573. [Google Scholar] [CrossRef]
- Wang, X.; Feng, Y.; Wang, Y.; Zhu, H.; Song, D.; Shen, C.; Luo, Y. Enhancing Optical Non-Destructive Methods for Food Quality and Safety Assessments with Machine Learning Techniques: A Survey. J. Agric. Food Res. 2025, 19, 101734. [Google Scholar] [CrossRef]
- Jia, W.; Georgouli, K.; Martinez-Del Rincon, J.; Koidis, A. Challenges in the Use of AI-Driven Non-Destructive Spectroscopic Tools for Rapid Food Analysis. Foods 2024, 13, 846. [Google Scholar] [CrossRef]
- Lun, Z.; Wu, X.; Dong, J.; Wu, B. Deep Learning-Enhanced Spectroscopic Technologies for Food Quality Assessment: Convergence and Emerging Frontiers. Foods 2025, 14, 2350. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Z.; Zhang, T.; Ding, H.; Wang, Z.; Wang, H.; Zhou, L.; Dai, Y.; Xu, Y. Quantitative Analysis of Adulteration in Anoectochilus roxburghii Powder Using Hyperspectral Imaging and Multi-Channel Convolutional Neural Network. Agronomy 2025, 15, 1894. https://doi.org/10.3390/agronomy15081894
Liu Z, Zhang T, Ding H, Wang Z, Wang H, Zhou L, Dai Y, Xu Y. Quantitative Analysis of Adulteration in Anoectochilus roxburghii Powder Using Hyperspectral Imaging and Multi-Channel Convolutional Neural Network. Agronomy. 2025; 15(8):1894. https://doi.org/10.3390/agronomy15081894
Chicago/Turabian StyleLiu, Ziyuan, Tingsong Zhang, Haoyuan Ding, Zhangting Wang, Hongzhen Wang, Lu Zhou, Yujia Dai, and Yiqing Xu. 2025. "Quantitative Analysis of Adulteration in Anoectochilus roxburghii Powder Using Hyperspectral Imaging and Multi-Channel Convolutional Neural Network" Agronomy 15, no. 8: 1894. https://doi.org/10.3390/agronomy15081894
APA StyleLiu, Z., Zhang, T., Ding, H., Wang, Z., Wang, H., Zhou, L., Dai, Y., & Xu, Y. (2025). Quantitative Analysis of Adulteration in Anoectochilus roxburghii Powder Using Hyperspectral Imaging and Multi-Channel Convolutional Neural Network. Agronomy, 15(8), 1894. https://doi.org/10.3390/agronomy15081894




