Assessment of Genetic Diversity in Elite Stevia Genotypes Utilizing Distinguishability, Homogeneity and Stability (DHS) Through Morphological Descriptors
Abstract
1. Introduction
2. Materials and Methods
3. Results
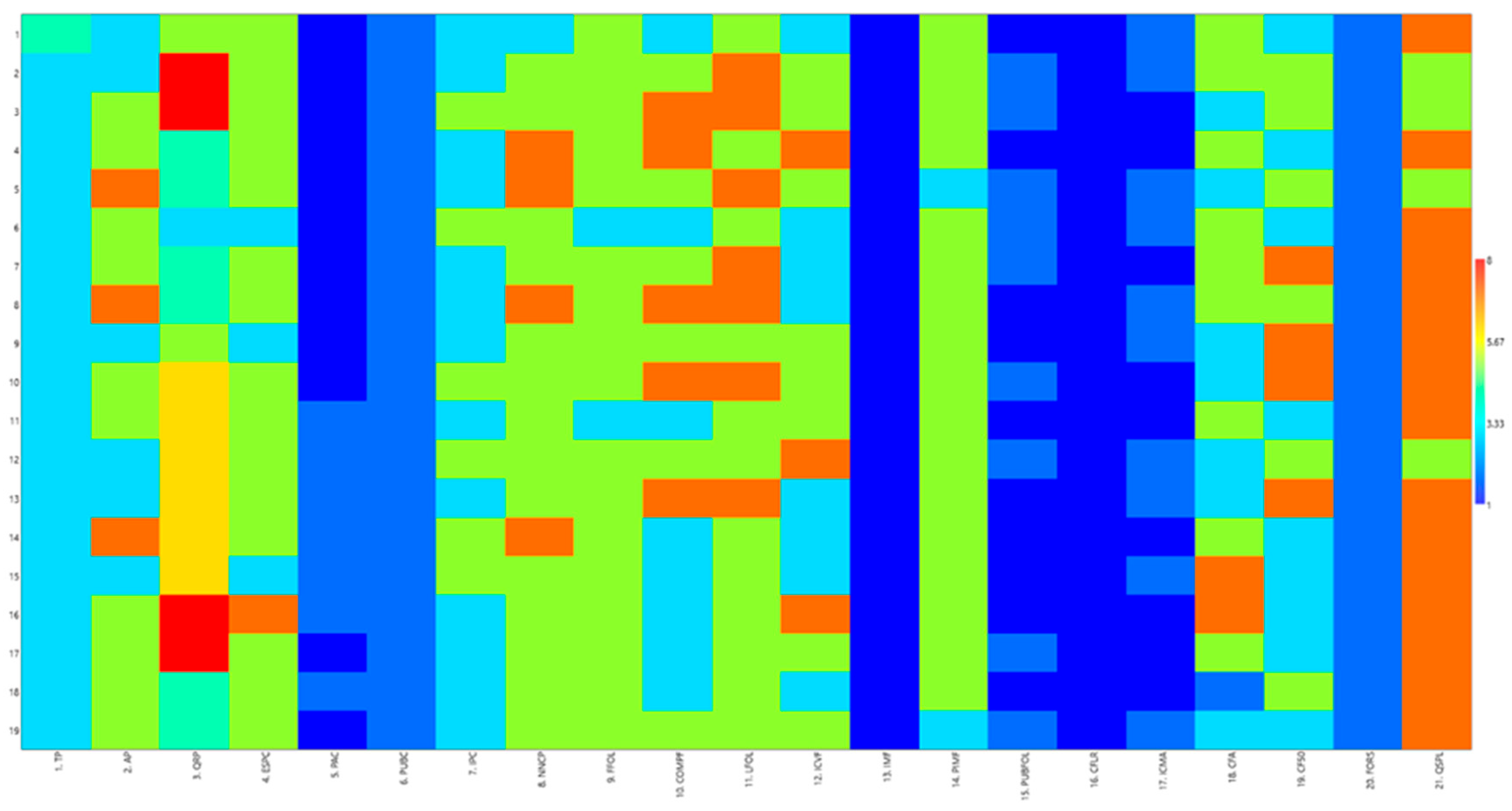
3.1. Characterization of the Descriptors
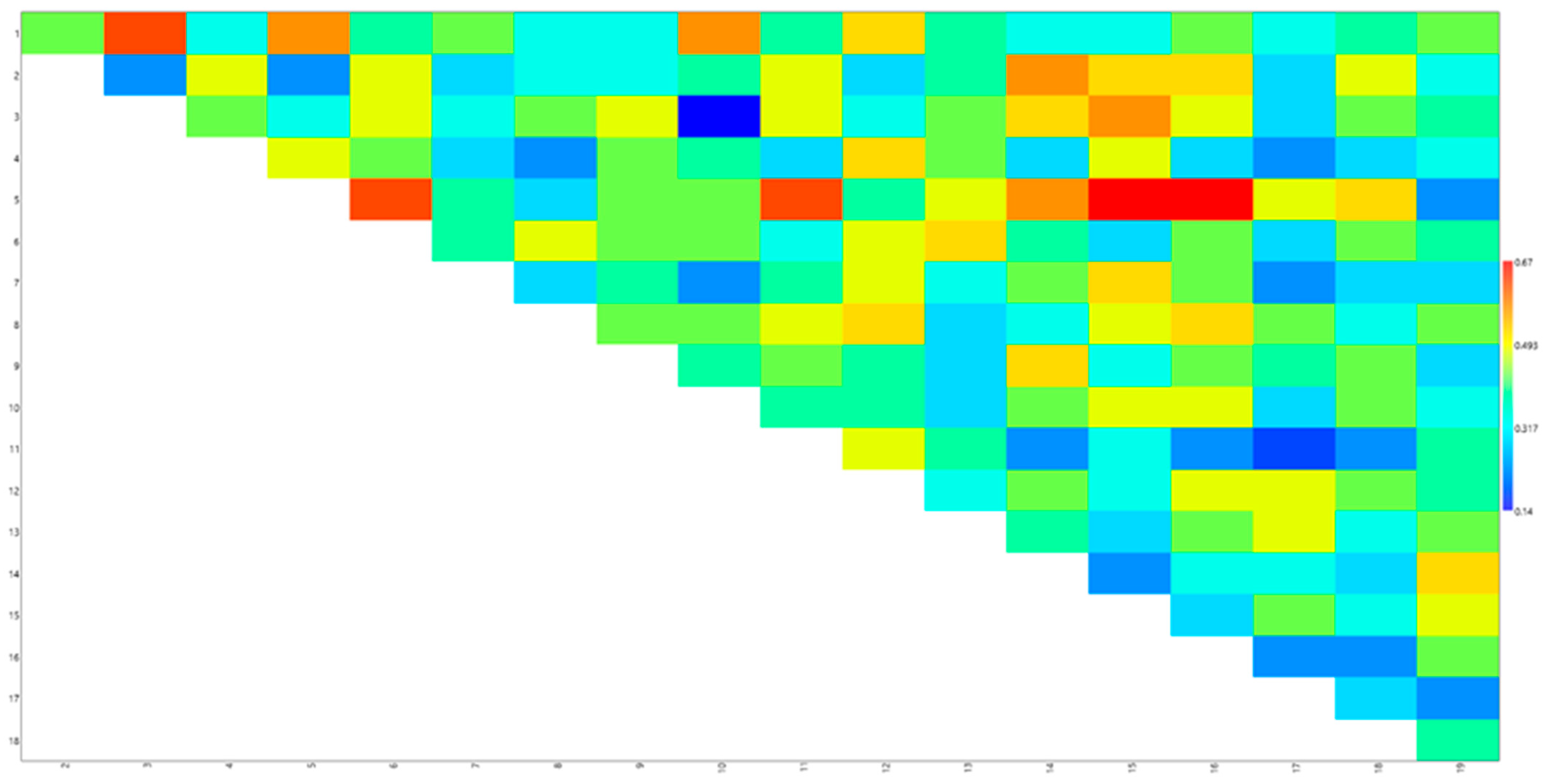
3.2. Genetic Distance Matrix
3.3. UPGMA Method
3.4. Dispersion Analysis
4. Discussion
4.1. Features of Similarity Groups
4.1.1. Group I
4.1.2. Group II
4.1.3. Group III
4.1.4. Group IV
4.1.5. Common Traits Across Groups
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wu, X.; Yang, P.; Dai, S.; Wen, Z. Effect of Dietary Stevioside Supplementation on Growth Performance, Nutrient Digestibility, Serum Parameters, and Intestinal Microflora in Broilers. Food Funct. 2019, 10, 2340–2346. [Google Scholar] [CrossRef]
- Gunasena, M.; Senarath, R.; Senarath, W. A Review on Chemical Composition, Biosynthesis of Steviol Glycosides, Application, Cultivation, and Phytochemical Screening of Stevia rebaudiana (Bert.) Bertoni. J. Pharm. Res. Int. 2021, 33, 85–104. [Google Scholar] [CrossRef]
- GVR Grand View Reserach. Stevia Market Size, Share & Trends Analysis Report By Application (Beverages, Food, Pharmaceutical, Tabletop Sweeteners), By Region (North America, Asia Pacific, Europe, Latin America, Middle East and Africa), and Segment Forecasts, 2024–2030. 2025. Available online: https://www.grandviewresearch.com/industry-analysis/stevia-market (accessed on 9 May 2025).
- Cosson, P.; Hastoy, C.; Errazzu, L.E.; Budeger, C.J.; Boutié, P.; Rolin, D.; Schurd-Levraud, V. Genetic diversity and population structure of the sweet leaf herb, Stevia rebaudiana B., cultivated and landraces germplasm assessed by EST-SSRs genotyping and steviol glycosides phenotyping. BMC Plant Biol. 2019, 19, 436. [Google Scholar] [CrossRef]
- Tavarini, S.; Passera, B.; Angelini, L.G. Chapter 1: Crop and Steviol Glycoside Improvement in Stevia by Breeding. In Food Chemistry, Function and Analysis; Royal Society of Chemistry: London, UK, 2018; pp. 1–31. [Google Scholar]
- Wölwer-Rieck, U. Steviol Glycosides. In Food Chemistry, Function and Analysis; Royal Society of Chemistry: London, UK, 2018; pp. 1–32. [Google Scholar]
- Santos, M.G.A. Adaptação da Stevia rebaudiana Bertoni em Modo de Produção Biológico em Ponte de Lima. Master’s Thesis, Instituto Politécnico de Viana do Castelo, Viana do Castelo, Portugal, 2016. [Google Scholar]
- Burle, M.L.; Oliveira, M. Manual de Curadores de Germoplasma-Vegetal: Caracterização Morfológica; Embrapa Recursos Genéticos e Biotecnologia Documentos (INFOTECA-E): Brasília, Brazil, 2010. [Google Scholar]
- Faleiro, F.G.; da Silva Oliveira, J.; de Jesus, O.N.; da Fonseca, K.G.; Junqueira, N.T.V. Importância da Caracterização dos Recursos Genéticos do Maracujazeiro. In Descritores Morfoagronômicos Ilustrados para Passiflora spp.; Jesus, O.N., Oliveira, E.J., Faleiro, F.G., Soares, T.L., Girardi, E.A., Eds.; Embrapa: Brasília, Brazil, 2016; pp. 8–17. [Google Scholar]
- Karimi, M.; Ahmadi, A.; Hashemi, J.; Abbasi, A.; Tavarini, S.; Pompeiano, A.; Angelini, L.G. The Positive Role of Steviol Glycosides in Stevia (Stevia rebaudiana Bertoni) under Drought Stress Condition. Plant Biosyst. 2015, 150, 1323–1331. [Google Scholar] [CrossRef]
- MAPA (Ministério da Agricultura, Pecuária e Abastecimento). Proteção de Cultivares, 2019. Available online: http://www.agricultura.gov.br (accessed on 24 March 2019).
- Brasil Ministério da Agricultura, Pecuária e Abastecimento. Ato n. 1, de 13 de janeiro de 2009. Diário Oficial da União, Brasília, 16 January 2009. Available online: http://www.agricultura.gov.br/assuntos/insumos-agropecuarios/insumos-agricolas/protecao-de-cultivar/arquivos-medicinais-e-aromaticas/estevia_18dez2008p.doc (accessed on 19 October 2024).
- Machado, I.; Ramalho, W.P.; Batista, V.G.; Ferraz, D.; Filho, J.A.F.D. Desvendando as Lacunas de Conhecimento sobre a Anurofauna do Cerrado: Diagnósticos, Diretrizes e Perspectivas. CBH 2019, 2, 105504. [Google Scholar]
- Dutra, R.; Souza, M. Cerrado, Revolução Verde e Evolução do Consumo de Agrotóxicos. Soc. Nat. 2022, 29, 473–488. [Google Scholar]
- Ferreira, F.; Rodrigues, L.; Silva, F. Performance Evaluation of Climate Models in the Simulation of Precipitation and Average Temperature in the Brazilian Cerrado. Theor. Appl. Climatol. 2024, 155, 845–857. [Google Scholar] [CrossRef]
- Castro, F.C.d. Variabilidade Genética, Morfológica e Agronômica e Seleção de Genótipos Elite de Stevia Rebaudiana Visando ao Desenvolvimento de Cultivares Adaptadas para o Cerrado. Dissertação Mestrado em Agronomia, Universidade de Brasília, Brasília-DF, Brazil, 14 February 2020. Available online: http://repositorio.unb.br/handle/10482/38576 (accessed on 14 January 2022).
- Renyi, A. On Measures of Entropy and Information. In Fourth Berkeley Symposium; University of California Press: Berkeley, CA, USA, 1960; pp. 547–561. [Google Scholar]
- Pillar, V.P. Multivariate Exploratory Analysis and Randomization Testing Using Multiv. Coenoses 1997, 12, 145–148. [Google Scholar]
- Cruz, C.D. GENES—A Software Package for Analysis in Experimental Statistics and Quantitative Genetics. Acta Sci. Agron. 2013, 35, 271–276. [Google Scholar] [CrossRef]
- Hammer, Ø.; Harper, D. Paleontological Data Analysis, 2nd ed.; Wiley: Hoboken, NJ, USA, 2024. [Google Scholar]
- SAS Institute. SAS/STAT User’s Guide, 4th ed.; SAS Institute: Cary, NC, USA, 1989; Volume 6, p. 846. [Google Scholar]
- Dziak, J.J.; Li, R.; Wagner, A.T. Weighted TVEM SAS Macro Users’ Guide (Version 2.6); The Methodology Center, Penn State: University Park, PA, USA, 2017. [Google Scholar]
- StatSoft Inc. Statistica for Windows [Computer Program Manual]; StatSoft Inc.: Tulsa, OK, USA, 1999. [Google Scholar]
- Morgan, G.A.; Barrett, K.C.; Leech, N.L.; Gloeckner, G.W. IBM SPSS for Introductory Statistics: Use and Interpretation; Routledge: New York, NY, USA, 2019. [Google Scholar]
- Othman, H.S.; Osman, M.; Zainuddin, Z. Genetic Variabilities of Stevia rebaudiana Bertoni Cultivated in Malaysia as Revealed by Morphological, Chemical and Molecular Characterisations. AGRIVITA J. Agric. Sci. 2018, 40, 267–283. [Google Scholar] [CrossRef]
- Yadav, A.K.; Singh, S.; Dhyani, D.; Ahuja, P.S. A Review on the Improvement of Stevia [Stevia rebaudiana (Bertoni)]. Can. J. Plant Sci. 2011, 91, 1–27. [Google Scholar] [CrossRef]
- Haida, Z.; Asikin, A.; Hakiman, M. Health Benefits of Stevia rebaudiana Bertoni as Zero Calorie Natural Sweetener: A Review. Int. Food Res. J. 2020, 27, 783–789. [Google Scholar]
- Monteiro, R. Taxonomia e Biologia da Reprodução de Stevia Rebaudiana (Bert.) Bertoni; Dissertação (Mestrado), Universidade Estadual de Campinas, Campinas, 1980. Available online: https://repositorio.unicamp.br/Acervo/Detalhe/48636 (accessed on 2 January 2025).
- Meza, B.E.; Peralta, J.M. Moisture Sorption Isotherms of Reduced-Sugar Confectionery Coatings Elaborated with Stevia: Mathematical Modeling and Thermodynamic Analysis. Food Process. Eng. 2023, 46, e14402. [Google Scholar] [CrossRef]
- De Souza, L.M.; Barbosa, M.R.; Nascimento, K.; Fonseca, P.; Houllou, L. Cultivo Fotoautotrófico e Fotomixotrófico de Plantas In Vitro: Uma Revisão Sistemática sobre o Uso da Técnica e Suas Implicações na Produção de Mudas de Espécies Arbóreas. Multidiscip. Sci. Rep. 2023, 3, 1–20. [Google Scholar] [CrossRef]
- Oliveira, V.M.; Forni-Martins, E.R.; Magalhães, P.M.; Alves, M.N. Chromosomal and morphological studies of diploid and polyploid cytotypes of Stevia rebaudiana (Bertoni) Bertoni (Eupatorieae, Asteraceae). Genet. Mol. Biol. 2004, 27, 215–222. [Google Scholar] [CrossRef]
- Vieira, E.A.; de Freitas Fialho, J.; Silva, M.S.; Fukuda, W.M.G.; Faleiro, F.G. Variabilidade Genética do Banco de Germoplasma de Mandioca da Embrapa Cerrados Acessada por Meio de Descritores Morfológicos. Científica 2009, 36, 56–67. [Google Scholar]
- Ogbonna, A.C.; de Andrade, L.R.B.; Mueller, L.A.; de Oliveira, E.J.; Bauchet, G.J. Comprehensive Genotyping of a Brazilian Cassava (Manihot esculenta Crantz) Germplasm Bank: Insights into Diversification and Domestication. Theor. Appl. Genet. 2021, 134, 1343–1362. [Google Scholar] [CrossRef]
- Mahdi, S.A.; Meena, C.M.; Tholakabavi, A. Induction of Genetic Variability by Colchicine Treatment in Stevia rebaudiana Bertoni. Al-Qadisiyah J. Pure Sci. 2018, 23, 161. [Google Scholar]
- Miyagawa, H.; Fujioka, N.; Kohda, H.; Yamasaki, K.; Taniguchi, K.; Tanaka, R. Studies on the Tissue Culture of Stevia rebaudiana and Its Components; (II) 1. Induction of Shoot Primordia. Planta Med. 1986, 52, 321–323. [Google Scholar] [CrossRef]
- Benelli, G.; Canale, A.; Romano, D.; Flamini, G. Flower Scent Bouquet Variation and Bee Pollinator Visits in Stevia rebaudiana Bertoni (Asteraceae), a Source of Natural Sweeteners. Arthropod-Plant Interact. 2017, 11, 381–388. [Google Scholar] [CrossRef]
- Masand, M.; Sharma, S.; Kumari, S.; Pal, P.; Majeed, A.; Singh, G.; Sharma, R.M. High-Quality Haplotype-Resolved Chromosome Assembly Provides Evolutionary Insights and Targeted Steviol Glycosides (SGs) Biosynthesis in Stevia rebaudiana Bertoni. Plant Biotechnol. J. 2024, 22, 3262–3277. [Google Scholar] [CrossRef] [PubMed]


| Local | Genotypes | Code |
|---|---|---|
| LA1 | 1 | 11 |
| LA2 | 2 | 13 |
| LA3 | 3 | 3016 |
| LA4 | 4 | 1102 |
| LA5 | 5 | 7 |
| LA6 | 6 | 3002 |
| LA7 | 7 | 3015 |
| LA8 | 8 | CPAC1 |
| LA9 | 9 | CPAC2 |
| LA10 | 10 | CPAC3 |
| LB1 | 11 | 3004 |
| LB3 | 12 | 3024 |
| LB4 | 13 | 3 _ 12 |
| LB5 | 14 | 3 _ 25 |
| LB6 | 15 | 12 |
| LB7 | 16 | 1 |
| LB8 | 17 | 3 = 6 |
| LB9 | 18 | CPAC4 |
| LB10 | 19 | CPAC5 |
| Features | Feature Description | Description Code | Frequency (%) | Entropy (H) |
|---|---|---|---|---|
| 1. Floor plan: type (TP) VS (+) | type I | 1 | - | 0.20 |
| type II | 2 | - | ||
| type III | 3 | 94.74% | ||
| type IV | 4 | 5.26% | ||
| type V | 5 | - | ||
| type VI | 6 | - | ||
| 2. Plant: height (AP) MS | low (<40 cm) | 3 | 31.58% | 0.99 |
| medium (40–50 cm) | 5 | 52.63% | ||
| high (> 50 cm) | 7 | 15.79% | ||
| 3. Stem: number of primary branches (QRP) MS (+) | very low (<3) | 3 | 5.26% | 1.44 |
| low (3) | 4 | 31.58% | ||
| average (4 to 7) | 5 | 10.53% | ||
| high (8) | 6 | 31.58 | ||
| very high (>8) | 8 | 21.05 | ||
| 4. Stem: thickness (diameter at average height of main stem) (ESPC) DM (+) | thin (<0.20 cm) | 3 | 15.79% | 0.63 |
| medium (0.20 to 0.35 cm) | 5 | 78.95% | ||
| thick (>0.35 cm) | 7 | 5.26% | ||
| 5. Stem: anthocyanin pigmentation (PAC) VS/VG | absent | 1 | 63.16% | 0.65 |
| present | 2 | 36.84% | ||
| 6. Stem: pubescence (in the middle third) (PUBC) VS | absent | 1 | - | 0.00 |
| present | 2 | 100% | ||
| 7. Stem: intensity of pubescence (IPC) (in the middle third) VS | low | 3 | 68.42% | 0.62 |
| average | 5 | 31.58% | ||
| high | 7 | - | ||
| 8. Stem: number of nodes on the main stem (NNCP) MS | few (<6) | 3 | 5.26% | 0.70 |
| medium (6 to 9) | 5 | 73.68% | ||
| many (>9) | 7 | 21.05% | ||
| 9. Leaf: shape (FFOL) VS (+) | elliptical | 1 | - | 0.33 |
| rhomboid | 2 | - | ||
| obovada | 3 | 10.53% | ||
| oval | 4 | - | ||
| lanceolate | 5 | 89.47% | ||
| 10. Leaf: length (COMPF) MS | short (<4.0 cm) | 3 | 42.11% | 1.07 |
| medium (4.0 to 5.0 cm) | 5 | 31.58% | ||
| long (>5.0 cm) | 7 | 26.32% | ||
| 11. Leaf: width (LFOL) MS | narrow (<1.0 cm) | 3 | - | 0.65 |
| medium (1.0 cm to 1.5 cm) | 5 | 63.16% | ||
| wide (>1.5 cm) | 7 | 36.84% | ||
| 12. Leaf: intensity of green coloration (adaxial part) (ICVF) VG (+) | clear | 3 | 42.11% | 1.01 |
| average | 5 | 42.11% | ||
| dark | 7 | 15.79% | ||
| 13. Leaf: incisions on the margin (IMF) VS | absent | 1 | - | 0.00 |
| gifts | 2 | 100% | ||
| 14. Leaf: depth of incisions at the margin (PIMF) VS | shallow | 3 | 10.53% | 0.33 |
| average | 5 | 89.47% | ||
| deep | 7 | - | ||
| 15. Leaf: pubescence (PUBFOL) VS | absent | 1 | 52.63% | 0.69 |
| present | 2 | 47.37% | ||
| 16. Flower: coloration (in the center of the corolla) (CFLR) VS | white | 1 | 100% | 0.00 |
| light reddish purple | 3 | - | ||
| dark reddish purple | 5 | |||
| 17. Aquenium: intensity of brown coloration (ICMA) VS/VG | clear | 1 | 47.37% | 0.69 |
| average | 2 | 52.63% | ||
| dark | 3 | - | ||
| 18. Aquenium: spindle length (CFA) VS (+) | very short | 2 | 5.26% | 1.11 |
| short | 3 | 36.84% | ||
| medium | 5 | 47.37% | ||
| long | 7 | 10.53% | ||
| 19. Flowering cycle (when at least 50% of the plants have at least one flower) (CF50) MG/MS | early | 3 | 47.37% | 1.04 |
| medium | 5 | 31.58% | ||
| late | 7 | 21.05% | ||
| 20. Seeds: formation (FORS) VS | absent | 1 | - | 0.00 |
| present | 2 | 100% | ||
| 21. Seeds: number of seeds per plant (QSPL) MS | low | 3 | - | 0.51 |
| average | 5 | 21.05% | ||
| high | 7 | 78.95% |
| Genotypes |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
 | 0.43 | 0.62 | 0.33 | 0.57 | 0.38 | 0.43 | 0.33 | 0.33 | 0.57 | 0.38 | 0.52 | 0.38 | 0.33 | 0.33 | 0.43 | 0.33 | 0.38 | 0.43 |
 | 0.24 | 0.48 | 0.24 | 0.48 | 0.29 | 0.33 | 0.33 | 0.38 | 0.48 | 0.29 | 0.38 | 0.57 | 0.52 | 0.52 | 0.29 | 0.48 | 0.33 | |
 | 0.43 | 0.33 | 0.48 | 0.33 | 0.43 | 0.48 | 0.14 | 0.48 | 0.33 | 0.43 | 0.52 | 0.57 | 0.48 | 0.29 | 0.43 | 0.38 | ||
 | 0.48 | 0.43 | 0.29 | 0.24 | 0.43 | 0.38 | 0.29 | 0.52 | 0.43 | 0.29 | 0.48 | 0.29 | 0.24 | 0.29 | 0.33 | |||
 | 0.62 | 0.38 | 0.29 | 0.43 | 0.43 | 0.62 | 0.38 | 0.48 | 0.57 | 0.67 | 0.67 | 0.48 | 0.52 | 0.24 | ||||
 | 0.38 | 0.48 | 0.43 | 0.43 | 0.33 | 0.48 | 0.52 | 0.38 | 0.29 | 0.43 | 0.29 | 0.43 | 0.38 | |||||
 | 0.29 | 0.38 | 0.24 | 0.38 | 0.48 | 0.33 | 0.43 | 0.52 | 0.43 | 0.24 | 0.29 | 0.29 | ||||||
 | 0.43 | 0.43 | 0.48 | 0.52 | 0.29 | 0.33 | 0.48 | 0.52 | 0.43 | 0.33 | 0.43 | |||||||
 | 0.38 | 0.43 | 0.38 | 0.29 | 0.52 | 0.33 | 0.43 | 0.38 | 0.43 | 0.29 | ||||||||
 | 0.38 | 0.38 | 0.29 | 0.43 | 0.48 | 0.48 | 0.29 | 0.43 | 0.33 | |||||||||
 | 0.48 | 0.38 | 0.24 | 0.33 | 0.24 | 0.19 | 0.24 | 0.38 | ||||||||||
 | 0.33 | 0.43 | 0.33 | 0.48 | 0.48 | 0.43 | 0.38 | |||||||||||
 | 0.38 | 0.29 | 0.43 | 0.48 | 0.33 | 0.43 | ||||||||||||
 | 0.24 | 0.33 | 0.33 | 0.29 | 0.52 | |||||||||||||
 | 0.29 | 0.43 | 0.33 | 0.48 | ||||||||||||||
 | 0.24 | 0.24 | 0.43 | |||||||||||||||
 | 0.29 | 0.24 | ||||||||||||||||
 | 0.38 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Castro, F.C.d.; Faleiro, F.G.; Amabile, R.F.; da Silva Oliveira, J.; Lopes da Luz, A.; Pinheiro Melo, J.V.; Fialho, A.R.; Soares, K.C.d.S.; Santos, G.B.C.; Bruno, L.P. Assessment of Genetic Diversity in Elite Stevia Genotypes Utilizing Distinguishability, Homogeneity and Stability (DHS) Through Morphological Descriptors. Agronomy 2025, 15, 1836. https://doi.org/10.3390/agronomy15081836
Castro FCd, Faleiro FG, Amabile RF, da Silva Oliveira J, Lopes da Luz A, Pinheiro Melo JV, Fialho AR, Soares KCdS, Santos GBC, Bruno LP. Assessment of Genetic Diversity in Elite Stevia Genotypes Utilizing Distinguishability, Homogeneity and Stability (DHS) Through Morphological Descriptors. Agronomy. 2025; 15(8):1836. https://doi.org/10.3390/agronomy15081836
Chicago/Turabian StyleCastro, Fellipe Celestino de, Fábio Gelape Faleiro, Renato Fernando Amabile, Jamile da Silva Oliveira, Adriana Lopes da Luz, João Victor Pinheiro Melo, Arlini Rodrigues Fialho, Kelly Cristina dos Santos Soares, Gustavo Barbosa Cobalchini Santos, and Lorena Portilho Bruno. 2025. "Assessment of Genetic Diversity in Elite Stevia Genotypes Utilizing Distinguishability, Homogeneity and Stability (DHS) Through Morphological Descriptors" Agronomy 15, no. 8: 1836. https://doi.org/10.3390/agronomy15081836
APA StyleCastro, F. C. d., Faleiro, F. G., Amabile, R. F., da Silva Oliveira, J., Lopes da Luz, A., Pinheiro Melo, J. V., Fialho, A. R., Soares, K. C. d. S., Santos, G. B. C., & Bruno, L. P. (2025). Assessment of Genetic Diversity in Elite Stevia Genotypes Utilizing Distinguishability, Homogeneity and Stability (DHS) Through Morphological Descriptors. Agronomy, 15(8), 1836. https://doi.org/10.3390/agronomy15081836








