High-Resolution 3D Reconstruction of Individual Rice Tillers for Genetic Studies
Abstract
1. Introduction
2. Materials and Methods
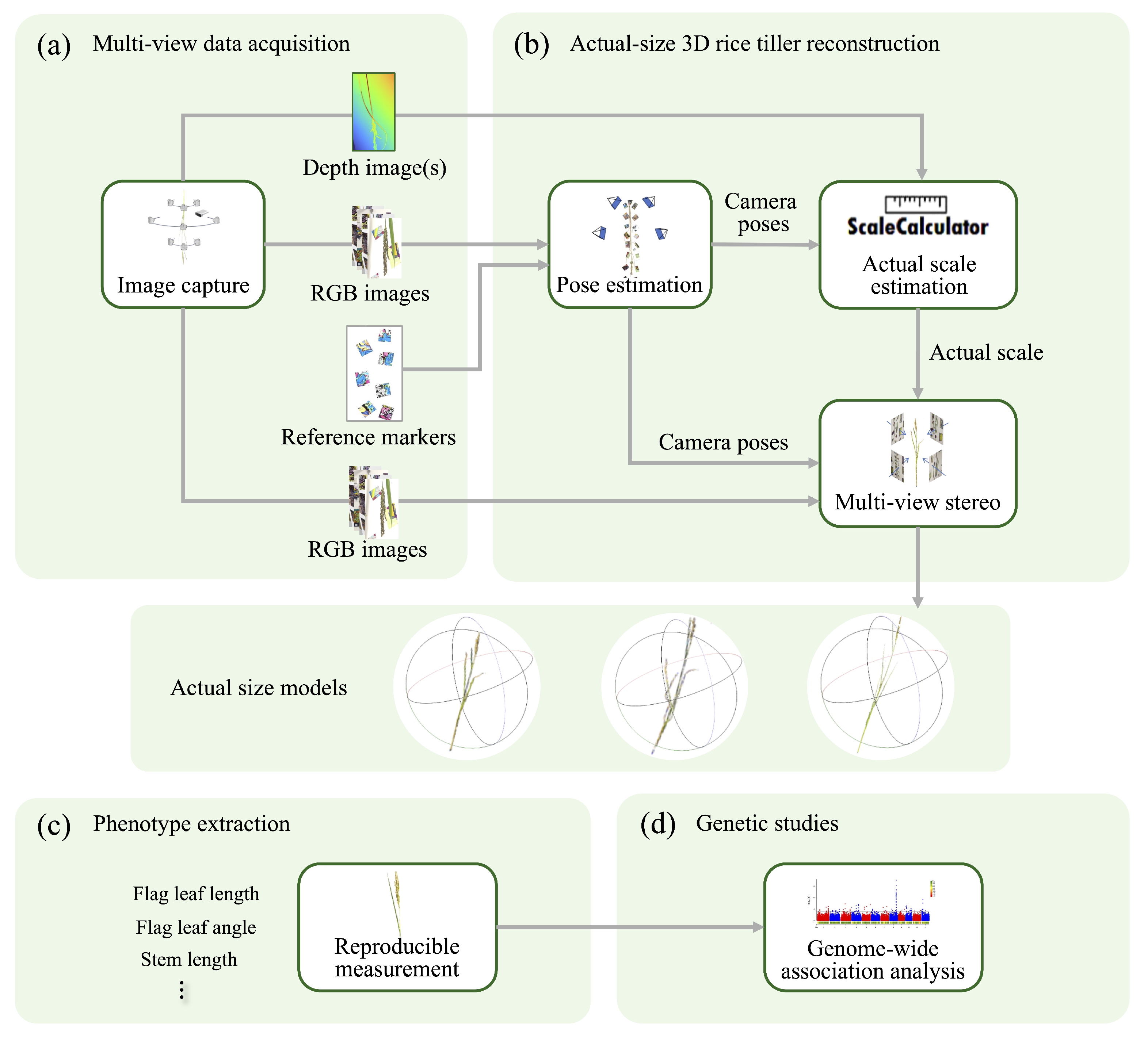
2.1. Overview of 3D Tiller Reconstruction for Genetic Study
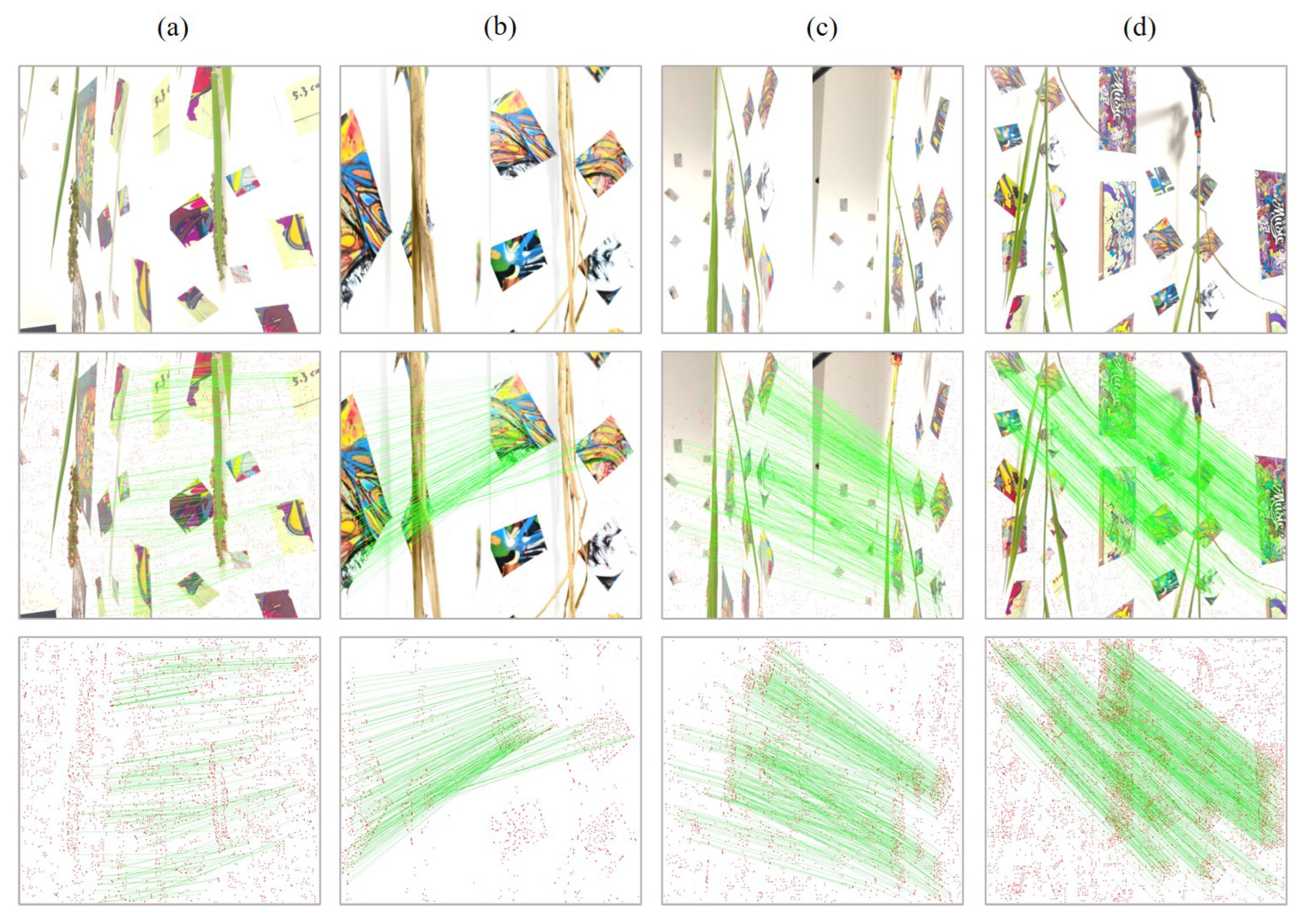
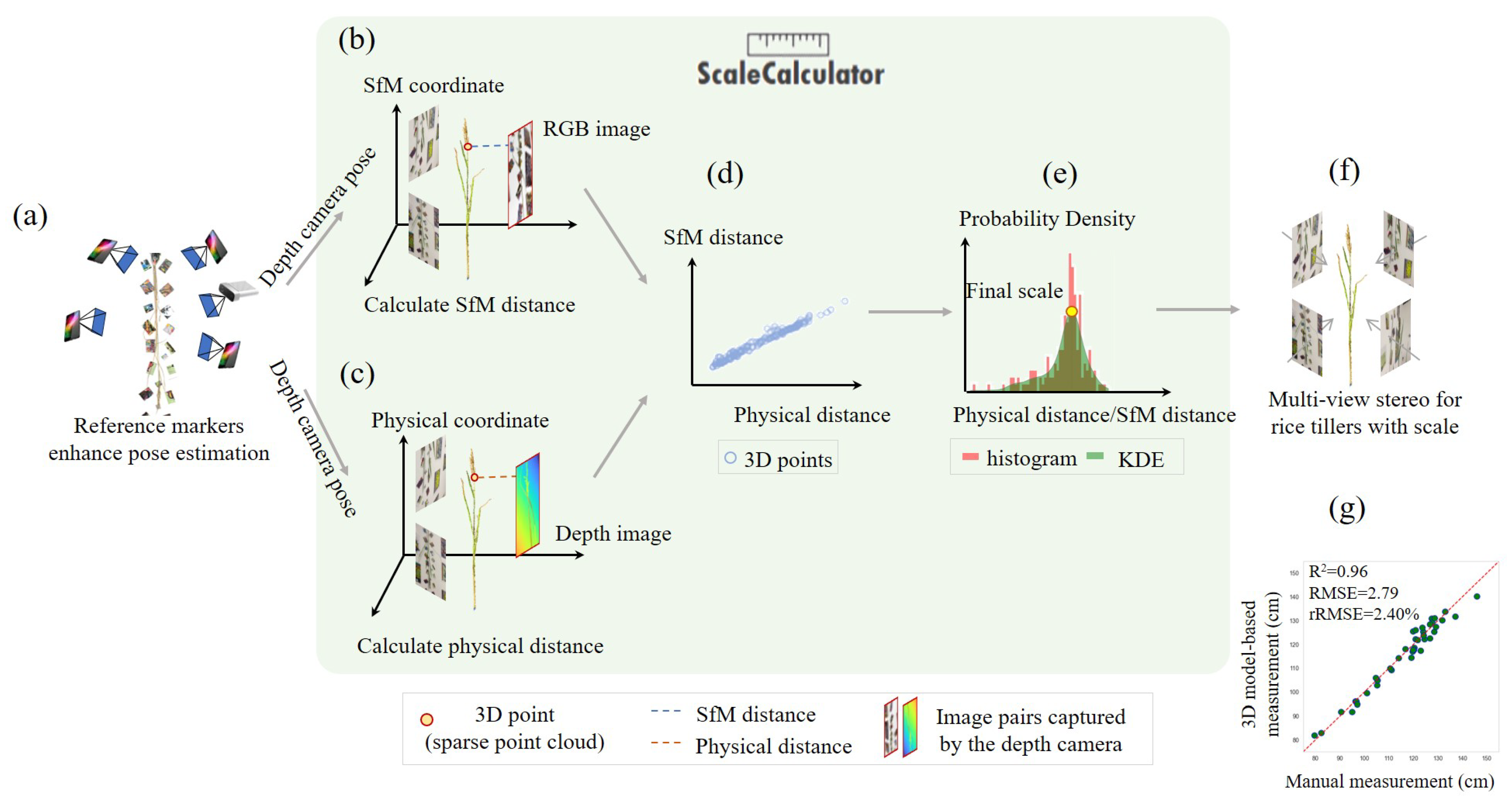
2.2. Reference Markers for Feature Points Matching Enhancement
2.3. Plant Materials
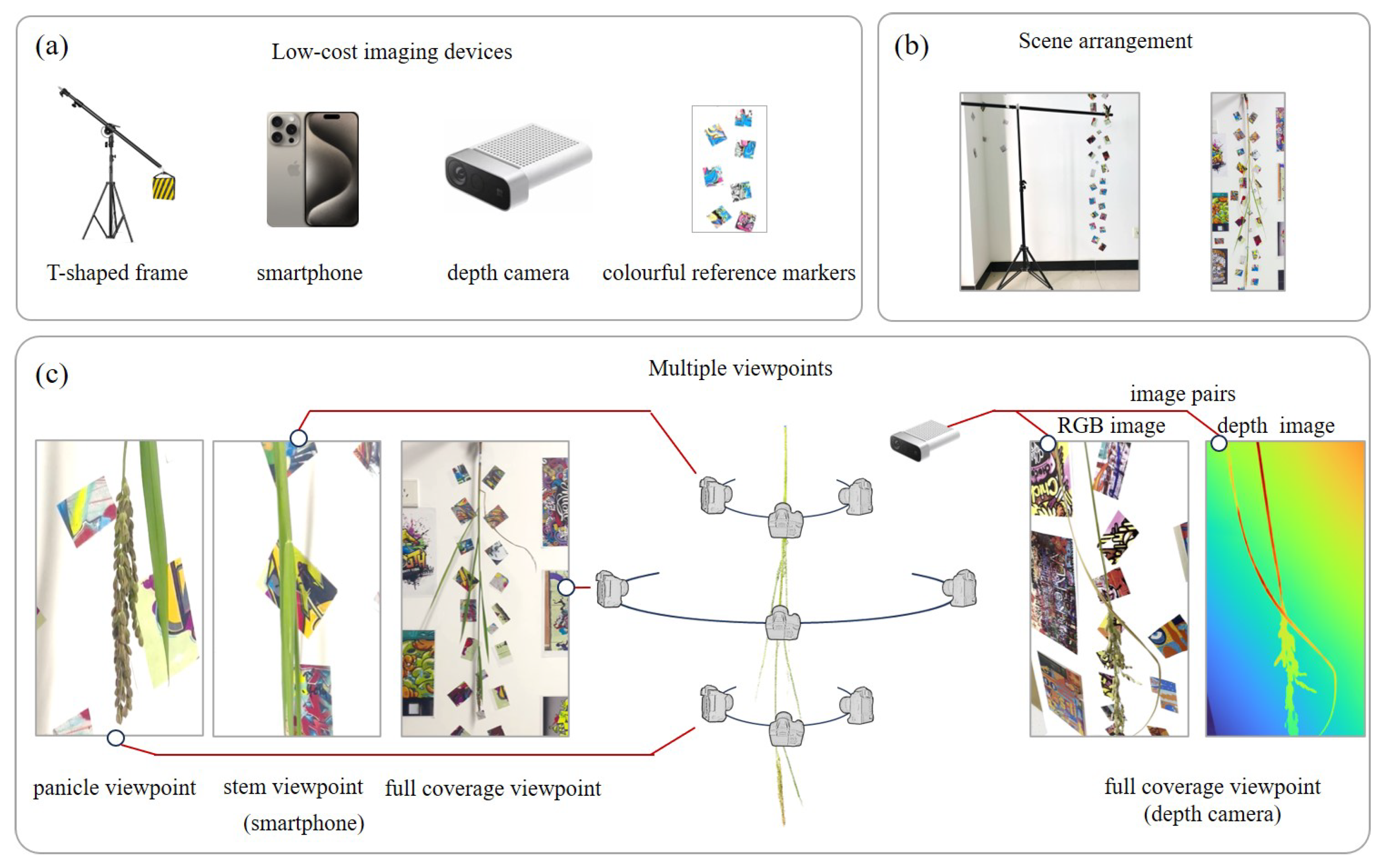
2.4. Multi-View Image Capturing
2.5. Recovering the Physical Scale for 3D Models
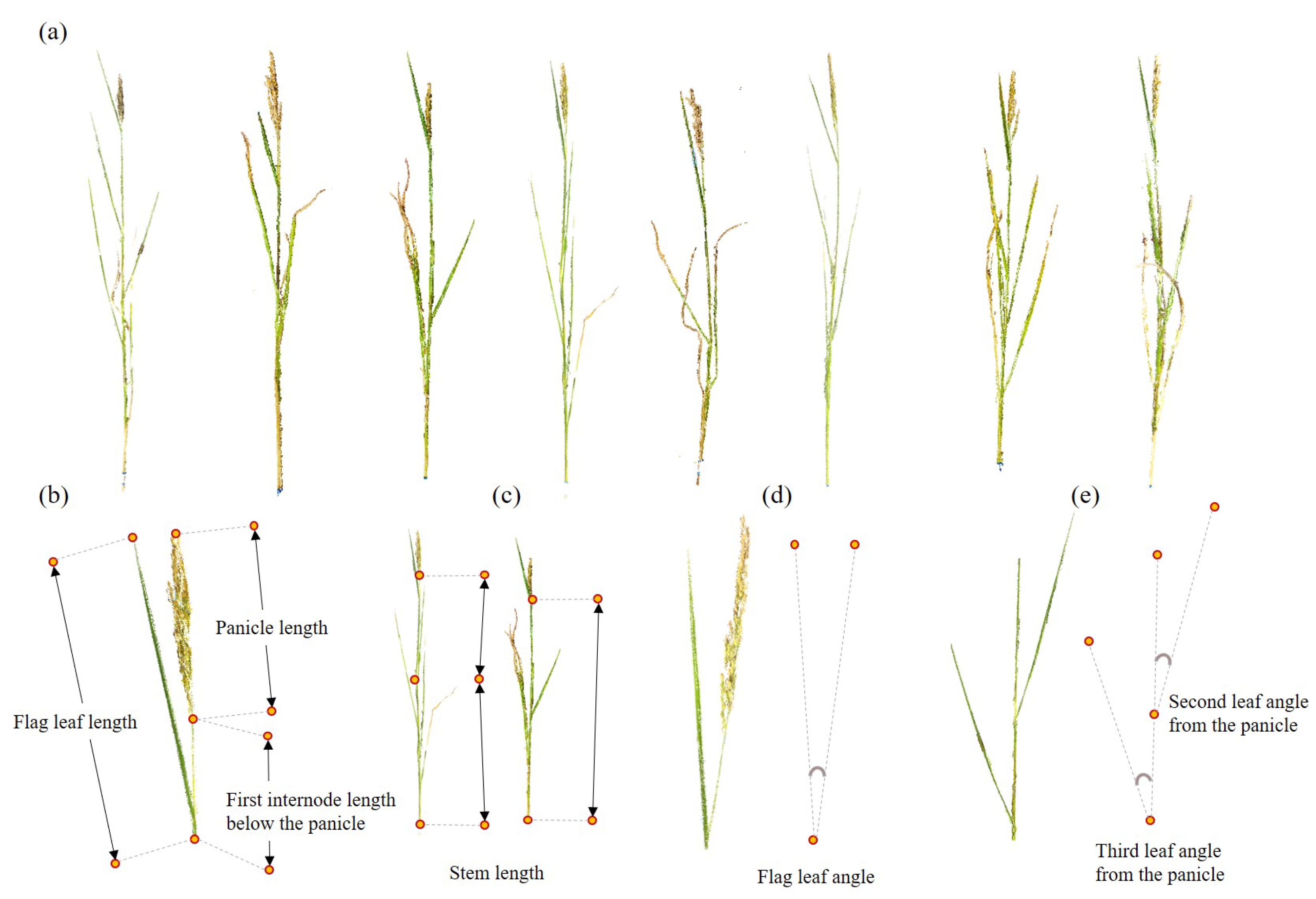
2.6. Three-Dimensional Model-Based Phenotypic Trait Extraction
2.7. Genome-Wide Association Study
3. Results
3.1. High-Resolution 3D Rice Tiller Models
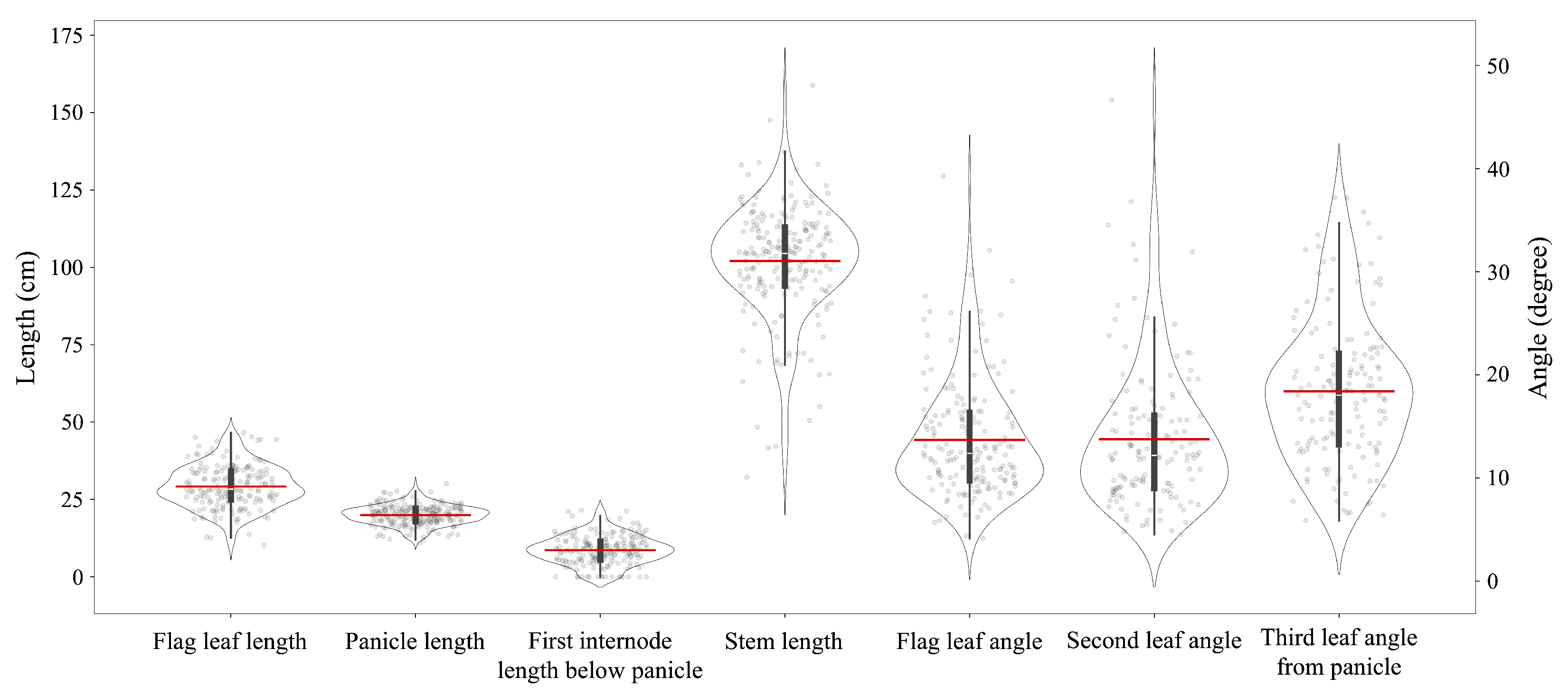
3.2. Phenotypic Traits by 3D Rice Tiller Models
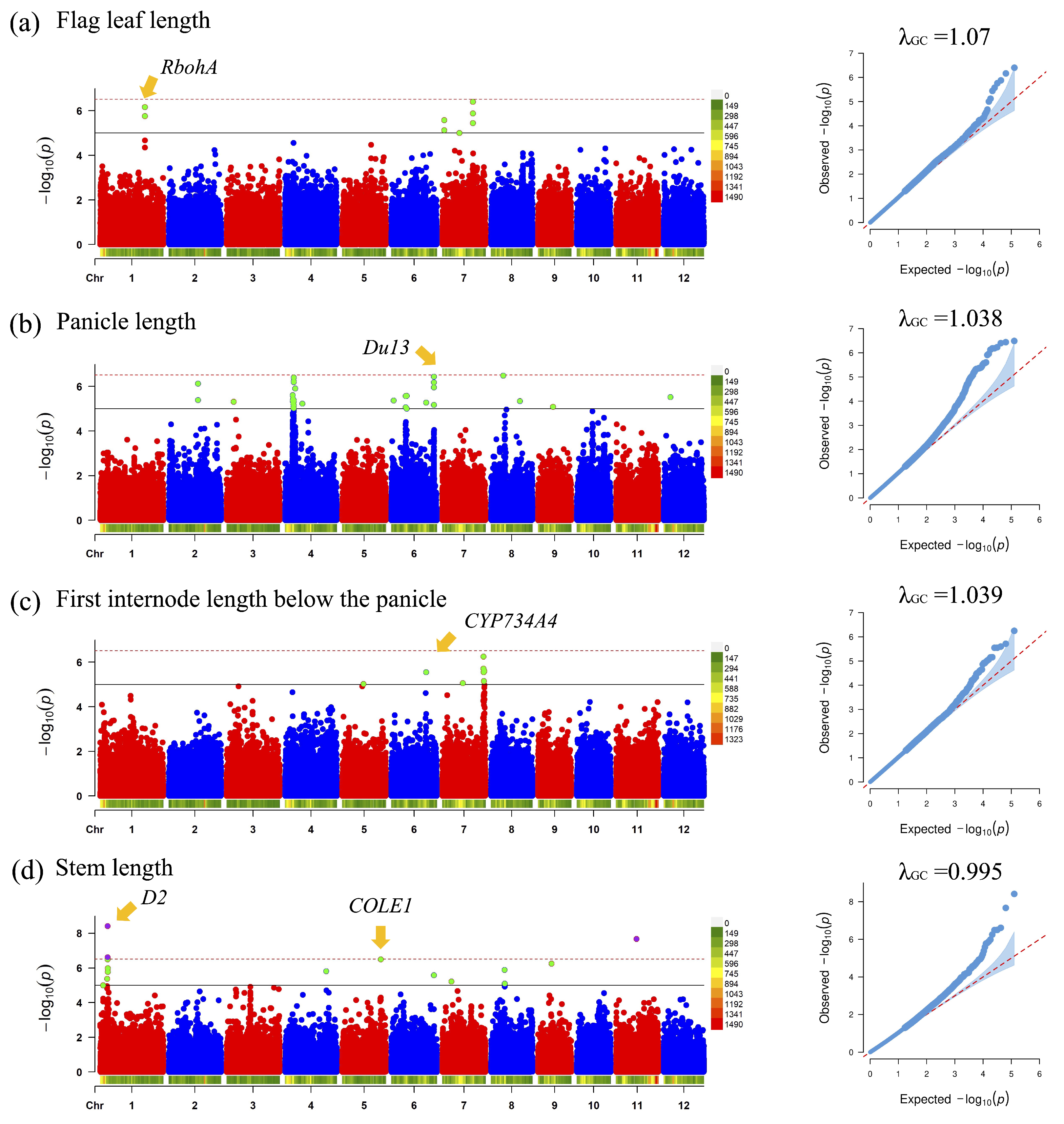
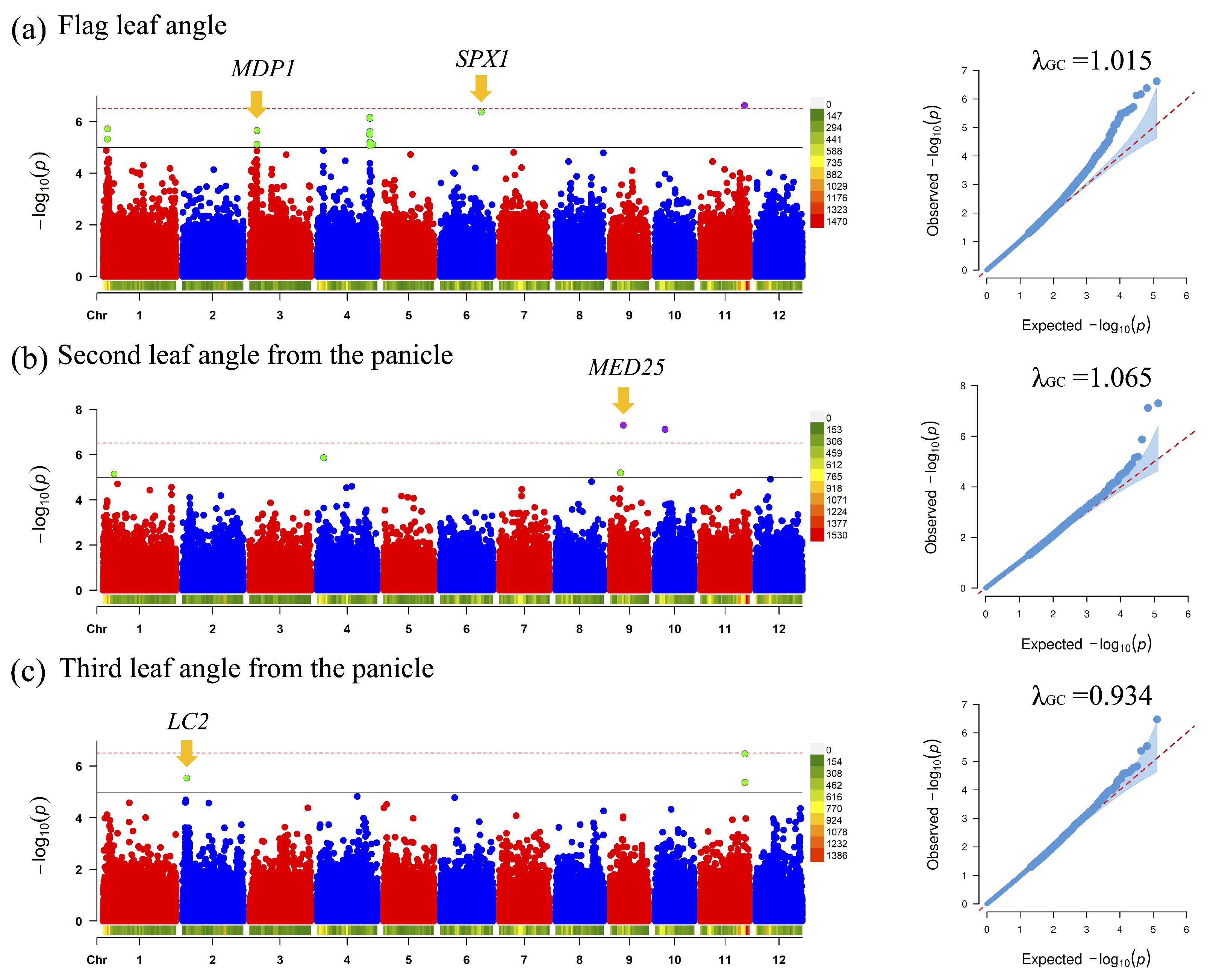
3.3. GWAS for 3D Rice Tiller Phenotypes
4. Discussion
4.1. Three-Dimensional Models for Rice Production
4.2. Applicability to Other Species
4.3. Challenges and Future Directions
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Food and Agriculture Organization of the United Nations (FAO). FAOSTAT: Production Quantity (Crops). Available online: https://www.fao.org/faostat/en/#data/QCL (accessed on 5 July 2025).
- Yuan, R.; Mao, Y.; Zhang, D.; Wang, S.; Zhang, H.; Wu, M.; Ye, M.; Zhang, Z. The formation of rice tillers and factors influencing it. Agronomy 2024, 14, 2904. [Google Scholar] [CrossRef]
- Huang, M.; Shan, S.; Cao, J.; Fang, S.; Tian, A.; Liu, Y.; Cao, F.; Yin, X.; Zou, Y. Primary-tiller panicle number is critical to achieving high grain yields in machine-transplanted hybrid rice. Sci. Rep. 2020, 10, 2811. [Google Scholar] [CrossRef]
- Tsugawa, S.; Shima, H.; Ishimoto, Y.; Ishikawa, K. Thickness-stiffness trade-off improves lodging resistance in rice. Sci. Rep. 2023, 13, 10828. [Google Scholar] [CrossRef]
- Wu, D.; Guo, Z.; Ye, J.; Feng, H.; Liu, J.; Chen, G.; Zheng, J.; Yan, D.; Yang, X.; Xiong, X.; et al. Combining high-throughput micro-CT-RGB phenotyping and genome-wide association study to dissect the genetic architecture of tiller growth in rice. J. Exp. Bot. 2019, 70, 545–561. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xia, P.; Gong, L.; Chen, B.; Li, Y.; Liu, C. Designing an interactively cognitive humanoid field-phenotyping robot for in-field rice tiller counting. Agriculture 2022, 12, 1966. [Google Scholar] [CrossRef]
- Van Crombrugge, I.; Sels, S.; Ribbens, B.; Steenackers, G.; Penne, R.; Vanlanduit, S. Accuracy assessment of joint angles estimated from 2D and 3D camera measurements. Sensors 2022, 22, 1729. [Google Scholar] [CrossRef] [PubMed]
- Ayaz, I.; Shaheen, E.; Aly, M.; Shujaat, S.; Gallo, G.; Coucke, W.; Politis, C.; Jacobs, R. Accuracy and reliability of 2-dimensional photography versus 3-dimensional soft tissue imaging. Imaging Sci. Dent. 2020, 50, 15–22. [Google Scholar] [CrossRef]
- Kenchanmane Raju, S.K.; Adkins, M.; Enersen, A.; Santana de Carvalho, D.; Studer, A.J.; Ganapathysubramanian, B.; Schnable, P.S.; Schnable, J.C. Leaf Angle eXtractor: A high-throughput image processing framework for leaf angle measurements in maize and sorghum. Appl. Plant Sci. 2020, 8, e11385. [Google Scholar] [CrossRef]
- Tross, M.C.; Gaillard, M.; Zwiener, M.; Miao, C.; Grove, R.J.; Li, B.; Benes, B.; Schnable, J.C. 3D reconstruction identifies loci linked to variation in angle of individual sorghum leaves. PeerJ 2021, 9, e12628. [Google Scholar] [CrossRef]
- Xu, J. GrainShape: A landmark-annotated image dataset of japonica rice grains for geometric morphometric analysis. Data Brief 2025, 61, 111781. [Google Scholar] [CrossRef]
- Paturkar, A.; Sen Gupta, G.; Bailey, D. Making use of 3D models for plant physiognomic analysis: A review. Remote Sens. 2021, 13, 2232. [Google Scholar] [CrossRef]
- Wang, K.; Pu, X.; Li, B. Automated phenotypic trait extraction for rice plant using terrestrial laser scanning data. Sensors 2024, 24, 4322. [Google Scholar] [CrossRef]
- Pound, M.P.; French, A.P.; Fozard, J.A.; Murchie, E.H.; Pridmore, T.P. A patch-based approach to 3D plant shoot phenotyping. Mach. Vis. Appl. 2016, 27, 767–779. [Google Scholar] [CrossRef]
- Schonberger, J.L.; Frahm, J.M. Structure-from-motion revisited. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 27–30 June 2016; pp. 4104–4113. [Google Scholar]
- Schönberger, J.L.; Zheng, E.; Frahm, J.M.; Pollefeys, M. Pixelwise view selection for unstructured multi-view stereo. In Proceedings of the Computer Vision-ECCV 2016: 14th European Conference, Amsterdam, The Netherlands, 11–14 October 2016; pp. 501–518. [Google Scholar]
- Cernea, D. OpenMVS: Multi-View Stereo Reconstruction Library. Available online: https://github.com/cdcseacave/openMVS (accessed on 28 December 2024).
- Sandhu, J.; Zhu, F.; Paul, P.; Gao, T.; Dhatt, B.K.; Ge, Y.; Staswick, P.; Yu, H.; Walia, H. PI-Plat: A high-resolution image-based 3D reconstruction method to estimate growth dynamics of rice inflorescence traits. Plant Methods 2019, 15, 162. [Google Scholar] [CrossRef]
- Dharni, J.S.; Dhatt, B.K.; Paul, P.; Gao, T.; Awada, T.; Bacher, H.; Peleg, Z.; Staswick, P.; Hupp, J.; Yu, H.; et al. A non-destructive approach for measuring rice panicle-level photosynthetic responses using 3D-image reconstruction. Plant Methods 2022, 18, 126. [Google Scholar] [CrossRef] [PubMed]
- Mildenhall, B.; Srinivasan, P.P.; Tancik, M.; Barron, J.T.; Ramamoorthi, R.; Ng, R. NeRF: Representing scenes as neural radiance fields for view synthesis. In Lecture Notes in Computer Science; Springer International Publishing: Cham, Switzerland, 2020; pp. 405–421. [Google Scholar]
- Yang, X.; Lu, X.; Xie, P.; Guo, Z.; Fang, H.; Fu, H.; Hu, X.; Sun, Z.; Cen, H. PanicleNeRF: Low-cost, high-precision in-field phenotyping of rice panicles with smartphone. Plant Phenomics 2024, 6, 0279. [Google Scholar] [CrossRef] [PubMed]
- Gong, L.; Du, X.; Zhu, K.; Lin, K.; Lou, Q.; Yuan, Z.; Huang, G.; Liu, C. Panicle-3D: Efficient phenotyping tool for precise semantic segmentation of rice Panicle point cloud. Plant Phenomics 2021, 2021, 9838929. [Google Scholar] [CrossRef]
- Cui, D.; Liu, P.; Liu, Y.; Zhao, Z.; Feng, J. Automated Phenotypic Analysis of Mature Soybean Using Multi-View Stereo 3D Reconstruction and Point Cloud Segmentation. Agriculture 2025, 15, 175. [Google Scholar] [CrossRef]
- Yang, D.; Yang, H.; Liu, D.; Wang, X. Research on Automatic 3D Reconstruction of Plant Phenotype Based on Multi-View Images. Comput. Electron. Agric. 2024, 220, 108866. [Google Scholar] [CrossRef]
- He, W.; Ye, Z.; Li, M.; Yan, Y.; Lu, W.; Xing, G. Extraction of soybean plant trait parameters based on SfM–MVS algorithm combined with GRNN. Front. Plant Sci. 2023, 14, 1181322. [Google Scholar] [CrossRef]
- Wu, S.; Wen, W.; Gou, W.; Lu, X.; Zhang, W.; Zheng, C.; Xiang, Z.; Chen, L.; Guo, X. A miniaturized phenotyping platform for individual plants using multi-view stereo 3D reconstruction. Front. Plant Sci. 2022, 13, 897746. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; Yang, T.; A, X.; Liu, J. Incremental Structure from Motion for Small-Scale Scenes Based on Auxiliary Calibration. Sensors 2025, 25, 415. [Google Scholar] [CrossRef]
- Xu, J.; Zhao, W.; Tang, Z.; Gan, X. A One Stop 3D Target Reconstruction and multilevel Segmentation Method. arXiv 2023, arXiv:2308.06974. [Google Scholar] [CrossRef]
- Laina, I.; Rupprecht, C.; Belagiannis, V.; Tombari, F.; Navab, N. Deeper depth prediction with fully convolutional residual networks. In Proceedings of the 2016 4th International Conference on 3D Vision (3DV), Stanford, CA, USA, 25–28 October 2016; pp. 239–248. [Google Scholar]
- Tölgyessy, M.; Dekan, M.; Chovanec, Ľ.; Hubinský, P. Evaluation of the Azure Kinect and its comparison to Kinect V1 and Kinect V2. Sensors 2021, 21, 413. [Google Scholar] [CrossRef] [PubMed]
- CloudCompare (Version 2.14.alpha): 3D Point Cloud and Mesh Processing Software. Available online: https://www.danielgm.net/cc/ (accessed on 6 July 2025).
- Zheng, H.; Tang, W.; Yang, T.; Zhou, M.; Guo, C.; Cheng, T.; Cao, W.; Zhu, Y.; Zhang, Y.; Yao, X. Grain protein content phenotyping in rice via hyperspectral imaging technology and a genome-wide association study. Plant Phenomics 2024, 6, 0200. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Stephens, M. Genome-wide efficient mixed-model analysis for association studies. Nat. Genet. 2012, 44, 821–824. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; De Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Alamin, M.; Sultana, M.H.; Lou, X.; Jin, W.; Xu, H. Dissecting Complex Traits Using Omics Data: A Review on the Linear Mixed Models and Their Application in GWAS. Plants 2022, 11, 3277. [Google Scholar] [CrossRef]
- Castel, S.E.; Tluway, F.D.; Emde, A.K.; Smyth, N.; Karim, M.; Sengupta, D.; Gray, O.A.; Hendershott, M.; LeBaron von Baeyer, S.; Burke, E.E.; et al. A map of blood regulatory variation in South Africans enables GWAS interpretation. Nat. Genet. 2025, 57, 1628–1637. [Google Scholar] [CrossRef]
- Xu, X.; Ye, J.; Yang, Y.; Zhang, M.; Xu, Q.; Feng, Y.; Yuan, X.; Yu, H.; Wang, Y.; Yang, Y.; et al. Genome-wide association study of rice rooting ability at the seedling stage. Rice 2020, 13, 59. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, M.M.; Wang, Y.J.; Gao, Y.T.; Li, R.; Wang, G.F.; Li, W.Q.; Liu, W.T.; Chen, K.M. The plasma membrane NADPH oxidase OsRbohA plays a crucial role in developmental regulation and drought-stress response in rice. Physiol. Plant. 2016, 156, 421–443. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, W.; Fu, Y.; Shan, Z.; Xu, J.; Wang, P.; Kong, F.; Jin, J.; Yan, H.; Ge, X.; et al. Du13 encodes a C2 H2 zinc-finger protein that regulates Wxb pre-mRNA splicing and microRNA biogenesis in rice endosperm. Plant Biotechnol. J. 2022, 20, 1387–1401. [Google Scholar] [CrossRef]
- Zhan, C.; Hu, J.; Pang, Q.; Yang, B.; Cheng, Y.; Xu, E.; Zhu, P.; Li, Y.; Zhang, H.; Cheng, J. Genome-wide association analysis of panicle exsertion and uppermost internode in rice (Oryza sativa L.). Rice 2019, 12, 72. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Umemura, K.; Uozu, S.; Fujioka, S.; Takatsuto, S.; Yoshida, S.; Ashikari, M.; Kitano, H.; Matsuoka, M. A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 2003, 15, 2900–2910. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Zhang, L.; Luo, Y.; Xu, M.; Fan, Y.; Wang, L. Interactions of Oryza sativa Os CONTINUOUS VASCULAR RING-LIKE 1 (Os COLE1) and Os COLE1-INTERACTING PROTEIN reveal a novel intracellular auxin transport mechanism. New Phytol. 2016, 212, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Duan, K.; Li, L.; Hu, P.; Xu, S.P.; Xu, Z.H.; Xue, H.W. A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 2006, 47, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Ruan, W.; Guo, M.; Xu, L.; Wang, X.; Zhao, H.; Wang, J.; Yi, K. An SPX-RLI1 module regulates leaf inclination in response to phosphate availability in rice. Plant Cell 2018, 30, 853–870. [Google Scholar] [CrossRef]
- Karkute, S.G.; Kumar, V.; Tasleem, M.; Mishra, D.C.; Chaturvedi, K.K.; Rai, A.; Sevanthi, A.M.; Gaikwad, K.; Sharma, T.R.; Solanke, A.U. Genome-wide analysis of von Willebrand factor A gene family in rice for its role in imparting biotic stress resistance with emphasis on rice blast disease. Rice Sci. 2022, 29, 375–384. [Google Scholar] [CrossRef]
- Zhao, S.Q.; Hu, J.; Guo, L.B.; Qian, Q.; Xue, H.W. Rice leaf inclination2, a VIN3-like protein, regulates leaf angle through modulating cell division of the collar. Cell Res. 2010, 20, 935–947. [Google Scholar] [CrossRef]
- Organisation for Economic Co-operation and Development; Food and Agriculture Organization of the United Nations (OECD/FAO). OECD Agriculture Statistics (Database): OECD-FAO Agricultural Outlook 2022–2031. Available online: https://www.oecd.org/en/publications/oecd-fao-agricultural-outlook-2022-2031_f1b0b29c-en.html (accessed on 5 July 2025).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Lee, J.; Jiang, G.; Gan, X. High-Resolution 3D Reconstruction of Individual Rice Tillers for Genetic Studies. Agronomy 2025, 15, 1803. https://doi.org/10.3390/agronomy15081803
Xu J, Lee J, Jiang G, Gan X. High-Resolution 3D Reconstruction of Individual Rice Tillers for Genetic Studies. Agronomy. 2025; 15(8):1803. https://doi.org/10.3390/agronomy15081803
Chicago/Turabian StyleXu, Jiexiong, Jiyoung Lee, Gang Jiang, and Xiangchao Gan. 2025. "High-Resolution 3D Reconstruction of Individual Rice Tillers for Genetic Studies" Agronomy 15, no. 8: 1803. https://doi.org/10.3390/agronomy15081803
APA StyleXu, J., Lee, J., Jiang, G., & Gan, X. (2025). High-Resolution 3D Reconstruction of Individual Rice Tillers for Genetic Studies. Agronomy, 15(8), 1803. https://doi.org/10.3390/agronomy15081803






