Functional Characterization of a Synthetic Bacterial Community (SynCom) and Its Impact on Gene Expression and Growth Promotion in Tomato
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation and Construction of the Rhizosphere Consortium and Growth Conditions
2.2. Extraction of DNA, Shotgun Metagenome Sequencing, and Genomes Reconstruction
2.3. Bacterial Soil Community Analysis
2.4. Plant RNA Isolation and Transcriptomic Analysis
2.5. Statistical Analyses
3. Results
3.1. SynCom Design and Construction
3.2. Genomes Reconstruction (MAGs) and Functional Analysis
3.3. Effect of SynCom on Soil and Rhizosphere Bacterial Communities’ Composition and Diversity
3.4. Differential Gene Expression in Tomato Plants After Application of the SynCom
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
| PGP | Plant growth promoting |
| PGPR | Plant growth promoting rhizobacteria |
| SynCom | Synthetic bacterial communities |
| SA | Sucrose-Asparagine |
| PCA | Plate Count Agar |
| YMA | Yeast Mannitol Agar |
| MM | Minimal Salt Medium |
| PAS | Phosphate-buffered mineral medium salts |
| TYGS | Type-Strain Genome Server |
| DEGs | Differentially expressed genes |
| MAGs | Metagenome-assembled genomes |
| DAPG | 2,4-diacetylphloroglucinol |
| P | Phosphate |
| N | Nitrogen |
| IAA | Auxin indole-3-acetic acid |
| IAM | Indole-3-acetamide |
| PAA | Degradation of auxin phenylacetic acid |
| ACC | 1-aminocyclopropane-1-carboxylicacid |
| HCN | Hydrogen cyanide |
| PQQ | Pyrroloquinoline quinone |
References
- Jeyanthi, V.; Kanimozhi, S. Plant Growth Promoting Rhizobacteria (PGPR)-Prospective and Mechanisms: A Review. J. Pure Appl. Microbiol. 2018, 12, 733–749. [Google Scholar] [CrossRef]
- Zahid, M. Isolation and Identification of Indigenous Plant Growth Promoting Rhizobacteria from Himalayan Region of Kashmir and Their Effect on Improving Growth and Nutrient Contents of Maize (Zea mays L.). Front. Microbiol. 2015, 6, 207. [Google Scholar] [CrossRef] [PubMed]
- Vessey, J.K. Plant Growth Promoting Rhizobacteria as Biofertilizers. Plant Soil 2003, 255, 571–586. [Google Scholar] [CrossRef]
- Kumar, P.; Pandey, P.; Dubey, R.C.; Maheshwari, D.K. Bacteria Consortium Optimization Improves Nutrient Uptake, Nodulation, Disease Suppression and Growth of the Common Bean (Phaseolus vulgaris) in Both Pot and Field Studies. Rhizosphere 2016, 2, 13–23. [Google Scholar] [CrossRef]
- Lally, R.D.; Galbally, P.; Moreira, A.S.; Spink, J.; Ryan, D.; Germaine, K.J.; Dowling, D.N. Application of Endophytic Pseudomonas fluorescens and a Bacterial Consortium to Brassica napus Can Increase Plant Height and Biomass under Greenhouse and Field Conditions. Front. Plant Sci. 2017, 8, 2193. [Google Scholar] [CrossRef] [PubMed]
- Rosa, P.A.L.; Galindo, F.S.; Oliveira, C.E.d.S.; Jalal, A.; Mortinho, E.S.; Fernandes, G.C.; Marega, E.M.R.; Buzetti, S.; Teixeira Filho, M.C.M. Inoculation with Plant Growth-Promoting Bacteria to Reduce Phosphate Fertilization Requirement and Enhance Technological Quality and Yield of Sugarcane. Microorganisms 2022, 10, 192. [Google Scholar] [CrossRef]
- Ríos-Ruiz, W.F.; Torres-Chávez, E.E.; Torres-Delgado, J.; Rojas-García, J.C.; Bedmar, E.J.; Valdez-Nuñez, R.A. Inoculation of Bacterial Consortium Increases Rice Yield (Oryza sativa L.) Reducing Applications of Nitrogen Fertilizer in San Martin Region, Peru. Rhizosphere 2020, 14, 100200. [Google Scholar] [CrossRef]
- de Souza, L.G.M. Biotechnological Potential of Growth-Promoting Bacteria and Microbial Metabolites in Soybean Crop. UNESP. 2020. Available online: http://hdl.handle.net/11449/202661 (accessed on 9 June 2025).
- Timofeeva, A.M.; Galyamova, M.R.; Sedykh, S.E. Plant Growth-Promoting Bacteria of Soil: Designing of Consortia Beneficial for Crop Production. Microorganisms 2023, 11, 2864. [Google Scholar] [CrossRef]
- Hao, J.; Li, B.; Tan, J.; Zhang, Y.; Gu, X.; Wang, S.; Deng, Y.; Zhang, X.; Li, J. Double Advantages of Nutrients and Biostimulants Derived from Sewage Sludge by Alkaline Thermal Hydrolysis Process for Agricultural Use: Quality Promotion of Soil and Crop. Adv. Sci. 2024, 11, 2307793. [Google Scholar] [CrossRef]
- Nowrotek, M.; Jałowiecki, Ł.; Harnisz, M.; Płaza, G.A. Culturomics and Metagenomics: In Understanding of Environmental Resistome. Front. Environ. Sci. Eng. 2019, 13, 40. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Redondo-Nieto, M.; Guirado, M.; Pindado Jiménez, O.; Millán, R.; Martin, M.; Rivilla, R. Metagenomic Insights into the Bacterial Functions of a Diesel-Degrading Consortium for the Rhizoremediation of Diesel-Polluted Soil. Genes 2019, 10, 456. [Google Scholar] [CrossRef]
- Teeling, H.; Glöckner, F.O. Current Opportunities and Challenges in Microbial Metagenome Analysis—A Bioinformatic Perspective. Brief. Bioinform. 2012, 13, 728–742. [Google Scholar] [CrossRef]
- Brown, B.L.; Watson, M.; Minot, S.S.; Rivera, M.C.; Franklin, R.B. MinIONTM Nanopore Sequencing of Environmental Metagenomes: A Synthetic Approach. GigaScience 2017, 6, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Urich, T.; Lanzén, A.; Qi, J.; Huson, D.H.; Schleper, C.; Schuster, S.C. Simultaneous Assessment of Soil Microbial Community Structure and Function through Analysis of the Meta-Transcriptome. PLoS ONE 2008, 3, e2527. [Google Scholar] [CrossRef] [PubMed]
- Scher, F.M.; Baker, R. Effect of Pseudomonas Putida and a Synthetic Iron Chelator on Induction of Soil Suppressiveness to Fusarium Wilt Pathogens. Phytopathology 1982, 72, 1567–1573. [Google Scholar] [CrossRef]
- Corry, J.E.L.; Curtis, G.D.W.; Baird, R.M. Handbook of Culture Media for Food and Water Microbiology; Royal Society of Chemistry: London, UK, 2012; ISBN 978-1-84755-916-6. [Google Scholar]
- Schwartz, W.J.M. Vincent, A Manual for the Practical Study of the Root-Nodule Bacteria (IBP Handbuch No. 15 Des International Biology Program, London). XI u. 164 S., 10 Abb., 17 Tab., 7 Taf. Oxford-Edinburgh 1970: Blackwell Scientific Publ., 45 s. Z. Allg. Mikrobiol. 1972, 12, 440. [Google Scholar] [CrossRef]
- Brazil, G.M.; Kenefick, L.; Callanan, M.; Haro, A.; de Lorenzo, V.; Dowling, D.N.; O’Gara, F. Construction of a Rhizosphere Pseudomonad with Potential to Degrade Polychlorinated Biphenyls and Detection of Bph Gene Expression in the Rhizosphere. Appl. Environ. Microbiol. 1995, 61, 1946–1952. [Google Scholar] [CrossRef]
- Bedard, D.L.; Unterman, R.; Bopp, L.H.; Brennan, M.J.; Haberl, M.L.; Johnson, C. Rapid Assay for Screening and Characterizing Microorganisms for the Ability to Degrade Polychlorinated Biphenyls. Appl. Environ. Microbiol. 1986, 51, 761–768. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Farrell, C.M.; Feldgarden, M.; Fine, A.M.; Funk, K.; et al. Database Resources of the National Center for Biotechnology Information in 2023. Nucleic Acids Res. 2023, 51, D29–D38. Available online: https://academic.oup.com/nar/article/51/D1/D29/6825348?login=false (accessed on 12 February 2024). [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega. Curr. Protoc. Bioinform. 2014, 48, 3.13.1–3.13.16. [Google Scholar] [CrossRef]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural Sustainability and Intensive Production Practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef]
- Pradhan, N.; Sukla, L.B. Solubilization of Inorganic Phosphates by Fungi Isolated from Agriculture Soil. Afr. J. Biotechnol. 2006, 5, 850–854. [Google Scholar]
- Prigigallo, M.I.; Gómez-Lama Cabanás, C.; Mercado-Blanco, J.; Bubici, G. Designing a Synthetic Microbial Community Devoted to Biological Control: The Case Study of Fusarium Wilt of Banana. Front. Microbiol. 2022, 13, 967885. [Google Scholar] [CrossRef] [PubMed]
- Lozano-González, J.M.; Valverde, S.; Montoya, M.; Martín, M.; Rivilla, R.; Lucena, J.J.; López-Rayo, S. Evaluation of Siderophores Generated by Pseudomonas Bacteria and Their Possible Application as Fe Biofertilizers. Plants 2023, 12, 4054. [Google Scholar] [CrossRef] [PubMed]
- Meier-Kolthoff, J.P.; Carbasse, J.S.; Peinado-Olarte, R.L.; Göker, M. TYGS and LPSN: A Database Tandem for Fast and Reliable Genome-Based Classification and Nomenclature of Prokaryotes. Nucleic Acids Res. 2022, 50, D801–D807. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Augustijn, H.E.; Reitz, Z.L.; Biermann, F.; Alanjary, M.; Fetter, A.; Terlouw, B.R.; Metcalf, W.W.; Helfrich, E.J.N.; et al. antiSMASH 7.0: New and Improved Predictions for Detection, Regulation, Chemical Structures and Visualisation. Nucleic Acids Res. 2023, 51, W46–W50. [Google Scholar] [CrossRef]
- Razum, I.; Miko, S.; Rubinić, V.; Hasan, O.; Ilijanić, N.; Brunović, D.; Durn, G. Modelling End-Member Contributions in Terra Rossa Soils of the Eastern Adriatic Coast Using Heavy Mineral Assemblages Unravels the Role of Aeolian Processes in Their Formation. CATENA 2025, 250, 108736. [Google Scholar] [CrossRef]
- Fåhraeus, G. The Infection of Clover Root Hairs by Nodule Bacteria Studied by a Simple Glass Slide Technique. Microbiology 1957, 16, 374–381. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2—Approximately Maximum-Likelihood Trees for Large Alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E. qiime2R: Importing QIIME2 Artifacts and Associated Data into R Sessions. 2018. Available online: https://github.com/jbisanz/qiime2R/blob/master/inst/CITATION (accessed on 22 July 2025).
- McMurdie, P.J.; Holmes, S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef]
- Logemann, J.; Schell, J.; Willmitzer, L. Improved Method for the Isolation of RNA from Plant Tissues. Anal. Biochem. 1987, 163, 16–20. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG Tools for Functional Characterization of Genome and Metagenome Sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef]
- Puente-Sánchez, F.; García-García, N.; Tamames, J. SQMtools: Automated Processing and Visual Analysis of ’omics Data with R and Anvi’o. BMC Bioinform. 2020, 21, 358. [Google Scholar] [CrossRef]
- Oksanen, J. Vegan: Community Ecology Package. Ordination Methods, Diversity Analysis and Other Functions for Community and Vegetation Ecologists. R Package Version 2015. Available online: https://cir.nii.ac.jp/crid/1370564064030507943 (accessed on 19 December 2024).
- Wickham, H. Ggplot2. WIREs Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Jeffries, P.; Gianinazzi, S.; Perotto, S.; Turnau, K.; Barea, J.-M. The Contribution of Arbuscular Mycorrhizal Fungi in Sustainable Maintenance of Plant Health and Soil Fertility. Biol. Fertil. Soils 2003, 37, 1–16. [Google Scholar] [CrossRef]
- Kloepper, J.W.; Schroth, M.N. Relationship of in Vitro Antibiosis of Plant Growth-Promoting Rhizobacteria to Plant Growth and the Displacement of Root Microflora. Phytopathology 1981, 71, 1020–1024. [Google Scholar] [CrossRef]
- García de Salamone, I.E.; Esquivel-Cote, R.; Hernández-Melchor, D.J.; Alarcón, A. Manufacturing and Quality Control of Inoculants from the Paradigm of Circular Agriculture. In Microbial Interventions in Agriculture and Environment: Volume 2: Rhizosphere, Microbiome and Agro-Ecology; Singh, D.P., Gupta, V.K., Prabha, R., Eds.; Springer: Singapore, 2019; pp. 37–74. ISBN 9789811383830. [Google Scholar]
- Verma, J.P.; Jaiswal, D.K.; Krishna, R.; Prakash, S.; Yadav, J.; Singh, V. Characterization and Screening of Thermophilic Bacillus Strains for Developing Plant Growth Promoting Consortium from Hot Spring of Leh and Ladakh Region of India. Front. Microbiol. 2018, 9, 1293. [Google Scholar] [CrossRef] [PubMed]
- Bal, H.B.; Das, S.; Dangar, T.K.; Adhya, T.K. ACC Deaminase and IAA Producing Growth Promoting Bacteria from the Rhizosphere Soil of Tropical Rice Plants. J. Basic Microbiol. 2013, 53, 972–984. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharyya, P.N.; Jha, D.K. Plant Growth-Promoting Rhizobacteria (PGPR): Emergence in Agriculture. World J. Microbiol. Biotechnol. 2012, 28, 1327–1350. [Google Scholar] [CrossRef] [PubMed]
- Allison, S.D.; Martiny, J.B.H. Resistance, Resilience, and Redundancy in Microbial Communities. Proc. Natl. Acad. Sci. USA 2008, 105, 11512–11519. [Google Scholar] [CrossRef]
- Yin, B.; Crowley, D.; Sparovek, G.; De Melo, W.J.; Borneman, J. Bacterial Functional Redundancy along a Soil Reclamation Gradient. Appl. Environ. Microbiol. 2000, 66, 4361–4365. [Google Scholar] [CrossRef]
- Wang, X.Q.; Zhao, D.L.; Shen, L.L.; Jing, C.L.; Zhang, C.S. Application and Mechanisms of Bacillus Subtilis in Biological Control of Plant Disease. In Role of Rhizospheric Microbes in Soil: Volume 1: Stress Management and Agricultural Sustainability; Meena, V.S., Ed.; Springer: Singapore, 2018; pp. 225–250. ISBN 978-981-10-8402-7. [Google Scholar]
- Singh, J.S.; Singh, D.P. Plant Growth Promoting Rhizobacteria (PGPR): Microbes in Sustainable Agriculture. In Management of Microbial Resources in the Environment; Malik, A., Grohmann, E., Alves, M., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2013; pp. 361–385. ISBN 978-94-007-5931-2. [Google Scholar]
- Haas, D.; Défago, G. Biological Control of Soil-Borne Pathogens by Fluorescent Pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef]
- de Souza, J.T.; Arnould, C.; Deulvot, C.; Lemanceau, P.; Gianinazzi-Pearson, V.; Raaijmakers, J.M. Effect of 2,4-Diacetylphloroglucinol on Pythium: Cellular Responses and Variation in Sensitivity Among Propagules and Species. Phytopathology 2003, 93, 966–975. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Vesga, P.; Heiman, C.M.; Altenried, A.; Keel, C.; Vacheron, J. Relation of Pest Insect-Killing and Soilborne Pathogen-Inhibition Abilities to Species Diversification in Environmental Pseudomonas protegens. ISME J. 2023, 17, 1369–1381. [Google Scholar] [CrossRef]
- Almario, J.; Bruto, M.; Vacheron, J.; Prigent-Combaret, C.; Moënne-Loccoz, Y.; Muller, D. Distribution of 2,4-Diacetylphloroglucinol Biosynthetic Genes among the Pseudomonas spp. Reveals Unexpected Polyphyletism. Front. Microbiol. 2017, 8, 1218. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Moënne-Loccoz, Y.; Dubost, A.; Gonçalves-Martins, M.; Muller, D.; Prigent-Combaret, C. Fluorescent Pseudomonas Strains with Only Few Plant-Beneficial Properties Are Favored in the Maize Rhizosphere. Front. Plant Sci. 2016, 7, 1212. [Google Scholar] [CrossRef]
- Calderón, C.E.; de Vicente, A.; Cazorla, F.M. Role of 2-Hexyl, 5-Propyl Resorcinol Production by Pseudomonas chlororaphis PCL1606 in the Multitrophic Interactions in the Avocado Rhizosphere during the Biocontrol Process. FEMS Microbiol. Ecol. 2014, 89, 20–31. [Google Scholar] [CrossRef] [PubMed]
- Omoboye, O.O.; Oni, F.E.; Batool, H.; Yimer, H.Z.; De Mot, R.; Höfte, M. Pseudomonas Cyclic Lipopeptides Suppress the Rice Blast Fungus Magnaporthe oryzae by Induced Resistance and Direct Antagonism. Front. Plant Sci. 2019, 10, 901. [Google Scholar] [CrossRef]
- Lin, R.; Zhang, Q.; Yin, L.; Zhang, Y.; Yang, Q.; Liu, K.; Wang, Y.; Han, S.; Zhao, H.; Zhao, H. Isolation and Characterization of a Mycosubtilin Homologue Antagonizing Verticillium dahliae Produced by Bacillus subtilis Strain Z15. PLoS ONE 2022, 17, e0269861. [Google Scholar] [CrossRef]
- Grossi, C.E.M.; Fantino, E.; Serral, F.; Zawoznik, M.S.; Fernandez Do Porto, D.A.; Ulloa, R.M. Methylobacterium sp. 2A Is a Plant Growth-Promoting Rhizobacteria That Has the Potential to Improve Potato Crop Yield Under Adverse Conditions. Front. Plant Sci. 2020, 11, 71. [Google Scholar] [CrossRef]
- Khan, M.S.; Zaidi, A.; Ahemad, M.; Oves, M.; Wani, P.A. Plant Growth Promotion by Phosphate Solubilizing Fungi—Current Perspective. Arch. Agron. Soil Sci. 2010, 56, 73–98. [Google Scholar] [CrossRef]
- Chen, Y.P.; Rekha, P.D.; Arun, A.B.; Shen, F.T.; Lai, W.-A.; Young, C.C. Phosphate Solubilizing Bacteria from Subtropical Soil and Their Tricalcium Phosphate Solubilizing Abilities. Appl. Soil Ecol. 2006, 34, 33–41. [Google Scholar] [CrossRef]
- Corretto, E.; Antonielli, L.; Sessitsch, A.; Höfer, C.; Puschenreiter, M.; Widhalm, S.; Swarnalakshmi, K.; Brader, G. Comparative Genomics of Microbacterium Species to Reveal Diversity, Potential for Secondary Metabolites and Heavy Metal Resistance. Front. Microbiol. 2020, 11, 1869. [Google Scholar] [CrossRef]
- Yasmeen, S.; and Bano, A. Combined Effect of Phosphate-Solubilizing Microorganisms, Rhizobium and Enterobacter on Root Nodulation and Physiology of Soybean (Glycine max L.). Commun. Soil Sci. Plant Anal. 2014, 45, 2373–2384. [Google Scholar] [CrossRef]
- Yadav, A.; Yadav, K.; Vashistha, A. Phosphate Solubilizing Activity of Pseudomonas fluorescens PSM1 Isolated from Wheat Rhizosphere. J. Appl. Nat. Sci. 2016, 8, 93–96. [Google Scholar] [CrossRef]
- Stajkovic, O.; Delic, D.; Josic, D.; Kuzmanovic, D.; Rasulic, N.; Knezevic-Vukcevic, J. Improvement of Common Bean Growth by Co-Inoculation with Rhizobium and Plant Growth-Promoting Bacteria. Rom. Biotechnol. Lett. 2011, 16, 5919–5926. [Google Scholar]
- Olanrewaju, O.S.; Babalola, O.O. Bacterial Consortium for Improved Maize (Zea mays L.) Production. Microorganisms 2019, 7, 519. [Google Scholar] [CrossRef] [PubMed]
- Hider, R.C.; Kong, X. Iron: Effect of Overload and Deficiency. In Interrelations between Essential Metal Ions and Human Diseases; Metal Ions in Life Sciences; Sigel, A., Sigel, H., Sigel, R.K.O., Eds.; Springer Netherlands: Dordrecht, The Netherlands, 2013; pp. 229–294. ISBN 978-94-007-7500-8. [Google Scholar]
- Rajendran, G.; Patel, M.H.; Joshi, S.J. Isolation and Characterization of Nodule-Associated Exiguobacterium sp. from the Root Nodules of Fenugreek (Trigonella Foenum-Graecum) and Their Possible Role in Plant Growth Promotion. Int. J. Microbiol. 2012, 2012, e693982. [Google Scholar] [CrossRef]
- Fu, S.-F.; Wei, J.-Y.; Chen, H.-W.; Liu, Y.-Y.; Lu, H.-Y.; Chou, J.-Y. Indole-3-Acetic Acid: A Widespread Physiological Code in Interactions of Fungi with Other Organisms. Plant Signal Behav. 2015, 10, e1048052. [Google Scholar] [CrossRef] [PubMed]
- Mohite, B. Isolation and Characterization of Indole Acetic Acid (IAA) Producing Bacteria from Rhizospheric Soil and Its Effect on Plant Growth. J. Soil Sci. Plant Nutr. 2013, 13, 638–649. [Google Scholar] [CrossRef]
- Egamberdieva, D. Indole-Acetic Acid Production by Root Associated Bacteria and Its Role in Plant Growth and Development. In Auxins: Structure, Biosynthesis and Functions; Soil Nutrients, Nova Science Publishers: Hauppauge, NY, USA, 2011. [Google Scholar]
- Pacurar, D.I.; Perrone, I.; Bellini, C. Auxin Is a Central Player in the Hormone Cross-Talks That Control Adventitious Rooting. Physiol. Plant. 2014, 151, 83–96. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Ruan, Y.-L. Regulation of Cell Division and Expansion by Sugar and Auxin Signaling. Front. Plant Sci. 2013, 4, 163. [Google Scholar] [CrossRef]
- Peyvandi, M.; Farahani, F.; Mazinani, M.H.; Noormohamadi, Z.; Ataii, S.; Asgharzade, A. Pseudomonas fluorescent and Its Ability to Promote Root Formation of Olive Microshoots. Int. J. Plant Prod. 2012, 4, 63–66. [Google Scholar] [CrossRef]
- Ghosh, S.; Sengupta, C.; Maiti, T.K.; Basu, P.S. Production of 3-Indolylacetic Acid in Root Nodules and Culture by a Rhizobium Species Isolated from Root Nodules of the Leguminous Pulse Phaseolus mungo. Folia Microbiol. 2008, 53, 351–355. [Google Scholar] [CrossRef]
- Cook, S.D.; Nichols, D.S.; Smith, J.; Chourey, P.S.; McAdam, E.L.; Quittenden, L.; Ross, J.J. Auxin Biosynthesis: Are the Indole-3-Acetic Acid and Phenylacetic Acid Biosynthesis Pathways Mirror Images? Plant Physiol. 2016, 171, 1230–1241. [Google Scholar] [PubMed]
- Sugawara, S.; Mashiguchi, K.; Tanaka, K.; Hishiyama, S.; Sakai, T.; Hanada, K.; Kinoshita-Tsujimura, K.; Yu, H.; Dai, X.; Takebayashi, Y.; et al. Distinct Characteristics of Indole-3-Acetic Acid and Phenylacetic Acid, Two Common Auxins in Plants. Plant Cell Physiol. 2015, 56, 1641–1654. [Google Scholar] [CrossRef] [PubMed]
- Hooppaw, A.J.; McGuffey, J.C.; Di Venanzio, G.; Ortiz-Marquez, J.C.; Weber, B.S.; Lightly, T.J.; van Opijnen, T.; Scott, N.E.; Cardona, S.T.; Feldman, M.F. The Phenylacetic Acid Catabolic Pathway Regulates Antibiotic and Oxidative Stress Responses in Acinetobacter. mBio 2022, 13, e01863-21. [Google Scholar] [CrossRef] [PubMed]
- Cafiero, J.H.; Salvetti Casasco, M.; Lozano, M.J.; Vacca, C.; López García, S.L.; Draghi, W.O.; Lagares, A.; Del Papa, M.F. Genomic Analysis of Sinorhizobium meliloti LPU63, an Acid-Tolerant and Symbiotically Efficient Alfalfa-Nodulating Rhizobia. Front. Agron. 2023, 5, 1175524. [Google Scholar] [CrossRef]
- Mohamed, M.; Ismail, W.; Heider, J.; Fuchs, G. Aerobic Metabolism of Phenylacetic Acids in Azoarcus evansii. Arch. Microbiol. 2002, 178, 180–192. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xiao, Y.; Gao, D.; Long, Y.; Xie, Z. The Gene paaZ of the Phenylacetic Acid (PAA) Catabolic Pathway Branching Point and Ech Outside the PAA Catabolon Gene Cluster Are Synergistically Involved in the Biosynthesis of the Iron Scavenger 7-Hydroxytropolone in Pseudomonas Donghuensis HYS. Int. J. Mol. Sci. 2023, 24, 12632. [Google Scholar] [CrossRef]
- Magnucka, E.G.; Pietr, S.J. Various Effects of Fluorescent Bacteria of the Genus Pseudomonas Containing ACC Deaminase on Wheat Seedling Growth. Microbiol. Res. 2015, 181, 112–119. [Google Scholar] [CrossRef]
- Saravanakumar, D.; Samiyappan, R. ACC Deaminase from Pseudomonas Fluorescens Mediated Saline Resistance in Groundnut (Arachis Hypogea) Plants. J. Appl. Microbiol. 2007, 102, 1283–1292. [Google Scholar] [CrossRef]
- Choi, O.; Kim, J.; Kim, J.-G.; Jeong, Y.; Moon, J.S.; Park, C.S.; Hwang, I. Pyrroloquinoline Quinone Is a Plant Growth Promotion Factor Produced by Pseudomonas fluorescens B16. Plant Physiol. 2008, 146, 657–668. [Google Scholar] [CrossRef]
- Carreño-López, R.; Alatorre-Cruz, J.M.; Marín-Cevada, V. Pyrroloquinoline Quinone (PQQ): Role in Plant-Microbe Interactions. In Secondary Metabolites of Plant Growth Promoting Rhizomicroorganisms: Discovery and Applications; Singh, H.B., Keswani, C., Reddy, M.S., Sansinenea, E., García-Estrada, C., Eds.; Springer: Singapore, 2019; pp. 169–184. ISBN 978-981-13-5862-3. [Google Scholar]
- Jonscher, K.R.; Chowanadisai, W.; Rucker, R.B. Pyrroloquinoline-Quinone Is More Than an Antioxidant: A Vitamin-like Accessory Factor Important in Health and Disease Prevention. Biomolecules 2021, 11, 1441. [Google Scholar] [CrossRef]
- Lo, S.-C.; Tsai, S.-Y.; Chang, W.-H.; Wu, I.-C.; Sou, N.-L.; Hung, S.-H.W.; Chiang, E.-P.I.; Huang, C.-C. Characterization of the Pyrroloquinoline Quinone Producing Rhodopseudomonas palustris as a Plant Growth-Promoting Bacterium under Photoautotrophic and Photoheterotrophic Culture Conditions. Int. J. Mol. Sci. 2023, 24, 14080. [Google Scholar] [CrossRef]
- Kumazawa, T.; Sato, K.; Seno, H.; Ishii, A.; Suzuki, O. Levels of Pyrroloquinoline Quinone in Various Foods. Biochem. J. 1995, 307, 331–333. [Google Scholar] [CrossRef] [PubMed]
- Xie, S.-S.; Wu, H.-J.; Zang, H.-Y.; Wu, L.-M.; Zhu, Q.-Q.; Gao, X.-W. Plant Growth Promotion by Spermidine-Producing Bacillus subtilis OKB105. MPMI 2014, 27, 655–663. [Google Scholar] [CrossRef] [PubMed]
- Fujihara, S.; Harada, Y. Fast-Growing Root Nodule Bacteria Produce a Novel Polyamine, Aminobutylhomospermidine. Biochem. Biophys. Res. Commun. 1989, 165, 659–666. [Google Scholar] [CrossRef]
- Liu, Z.; Hossain, S.S.; Morales Moreira, Z.; Haney, C.H. Putrescine and Its Metabolic Precursor Arginine Promote Biofilm and C-Di-GMP Synthesis in Pseudomonas aeruginosa. J. Bacteriol. 2022, 204, e00297-21. [Google Scholar] [CrossRef] [PubMed]
- Altenburger, P.; KaMPFER, P.; Akimov, V.N.; Lubit, W.; Busse, H.-J. Polyamine Distribution in Actinomycetes with Group B Peptidoglycan and Species of the Genera Brevibacterium, Corynebacterium, and Tsukamurella. Int. J. Syst. Bacteriol. 1997, 47, 270–277. [Google Scholar] [CrossRef]
- Bueno, E.; Mesa, S.; Bedmar, E.J.; Richardson, D.J.; Delgado, M.J. Bacterial Adaptation of Respiration from Oxic to Microoxic and Anoxic Conditions: Redox Control. Antioxid. Redox Signal. 2012, 16, 819–852. [Google Scholar] [CrossRef]
- Butterbach-Bahl, K.; Baggs, E.M.; Dannenmann, M.; Kiese, R.; Zechmeister-Boltenstern, S. Nitrous Oxide Emissions from Soils: How Well Do We Understand the Processes and Their Controls? Philos. Trans. R. Soc. B Biol. Sci. 2013, 368, 20130122. [Google Scholar] [CrossRef]
- Hallin, S.; Philippot, L.; Löffler, F.E.; Sanford, R.A.; Jones, C.M. Genomics and Ecology of Novel N2O-Reducing Microorganisms. Trends Microbiol. 2018, 26, 43–55. [Google Scholar] [CrossRef]
- Garrido-Sanz, D.; Meier-Kolthoff, J.P.; Göker, M.; Martín, M.; Rivilla, R.; Redondo-Nieto, M. Correction: Genomic and Genetic Diversity within the Pseudomonas fluorescens Complex. PLoS ONE 2016, 11, e0153733. [Google Scholar] [CrossRef]
- Delgado, M.J.; Casella, S.; Bedmar, E.J. Chapter 6—Denitrification in Rhizobia-Legume Symbiosis. In Biology of the Nitrogen Cycle; Bothe, H., Ferguson, S.J., Newton, W.E., Eds.; Elsevier: Amsterdam, The Netherlands, 2007; pp. 83–91. ISBN 978-0-444-52857-5. [Google Scholar]
- Gasol, J.M.; Kirchman, D.L. Microbial Ecology of the Oceans; John Wiley & Sons: Hoboken, NJ, USA, 2018; ISBN 978-1-119-10719-4. [Google Scholar]
- Manriquez, B. Study of the Evolutionary Dynamics of Plant Beneficial Bacteria. Ph.D. Thesis, Université de Lyon, Lyon, France, 2021. [Google Scholar]
- Chukwuneme, C.F.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic Analyses of Plant Growth-Promoting and Carbon-Cycling Genes in Maize Rhizosphere Soils with Distinct Land-Use and Management Histories. Genes 2021, 12, 1431. [Google Scholar] [CrossRef] [PubMed]
- Nisrina, L.; Effendi, Y.; Pancoro, A. Revealing the Role of Plant Growth Promoting Rhizobacteria in Suppressive Soils against Fusarium oxysporum f. sp. cubense Based on Metagenomic Analysis. Heliyon 2021, 7, e07636. [Google Scholar] [CrossRef]
- Shen, X.; Hu, H.; Peng, H.; Wang, W.; Zhang, X. Comparative Genomic Analysis of Four Representative Plant Growth-Promoting Rhizobacteria in Pseudomonas. BMC Genom. 2013, 14, 271. [Google Scholar] [CrossRef] [PubMed]
- Jorrin, B.; Haskett, T.L.; Knights, H.E.; Martyn, A.; Underwood, T.J.; Dolliver, J.; Ledermann, R.; Poole, P.S. Stable, Fluorescent Markers for Tracking Synthetic Communities and Assembly Dynamics. Microbiome 2024, 12, 81. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus Amyloliquefaciens FZB42 on Lettuce Growth and Health under Pathogen Pressure and Its Impact on the Rhizosphere Bacterial Community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef] [PubMed]
- Amor, D.R.; Ratzke, C.; Gore, J. Transient Invaders Can Induce Shifts between Alternative Stable States of Microbial Communities. Sci. Adv. 2020, 6, eaay8676. [Google Scholar] [CrossRef]
- Fu, L.; Penton, C.; Ruan, Y.; Shen, Z.; Xue, C.; Li, R.; Shen, Q. Inducing the Rhizosphere Microbiome by Biofertilizer Application to Suppress Banana Fusarium wilt Disease. Soil Biol. Biochem. 2017, 104, 39–48. [Google Scholar] [CrossRef]
- Wang, J.; Li, X.; Li, X.; Wang, H.; Su, Z.; Wang, X.; Zhang, H. Dynamic Changes in Microbial Communities during the Bioremediation of Herbicide (Chlorimuron-Ethyl and Atrazine) Contaminated Soils by Combined Degrading Bacteria. PLoS ONE 2018, 13, e0194753. [Google Scholar] [CrossRef]
- Yin, D.; Wang, N.; Xia, F.; Li, Q.; Wang, W. Impact of Biocontrol Agents Pseudomonas fluorescens 2P24 and CPF10 on the Bacterial Community in the Cucumber Rhizosphere. Eur. J. Soil Biol. 2013, 59, 36–42. [Google Scholar] [CrossRef]
- Mawarda, P.C.; Lakke, S.L.; Dirk van Elsas, J.; Salles, J.F. Temporal Dynamics of the Soil Bacterial Community Following Bacillus Invasion. iScience 2022, 25, 104185. [Google Scholar] [CrossRef]
- Zehra, A.; Raytekar, N.A.; Meena, M.; Swapnil, P. Efficiency of Microbial Bio-Agents as Elicitors in Plant Defense Mechanism under Biotic Stress: A Review. Curr. Res. Microb. Sci. 2021, 2, 100054. [Google Scholar] [CrossRef] [PubMed]
- Oleńska, E.; Małek, W.; Wójcik, M.; Swiecicka, I.; Thijs, S.; Vangronsveld, J. Beneficial Features of Plant Growth-Promoting Rhizobacteria for Improving Plant Growth and Health in Challenging Conditions: A Methodical Review. Sci. Total Environ. 2020, 743, 140682. [Google Scholar] [CrossRef]
- Yan, Z.; Reddy, M.; Ryu, C.-M.; McInroy, J.; Wilson, M.; Kloepper, J. Induced Systemic Protection Against Tomato Late Blight Elicited by Plant Growth-Promoting Rhizobacteria. Phytopathology 2003, 92, 1329–1333. [Google Scholar] [CrossRef]
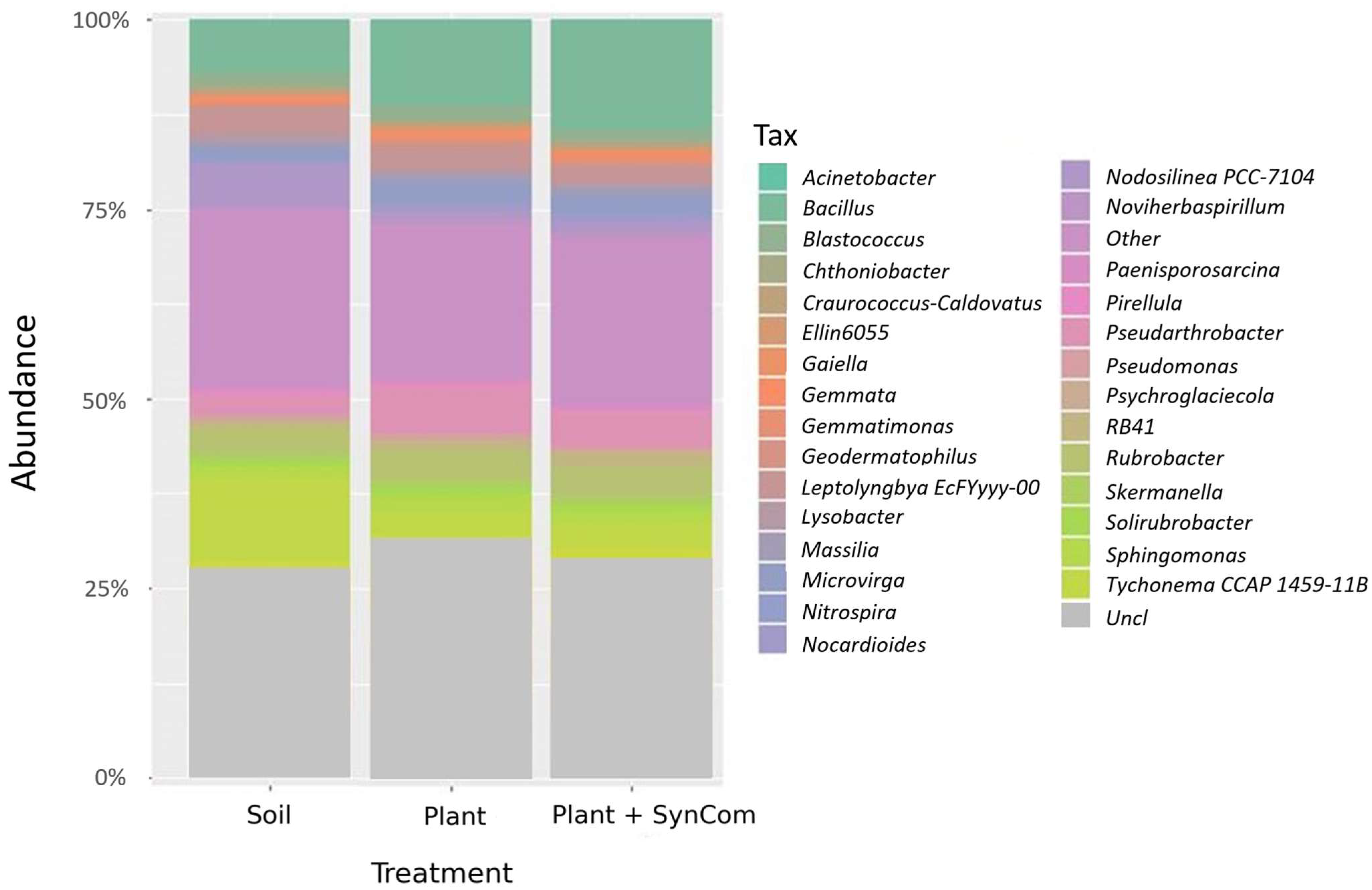
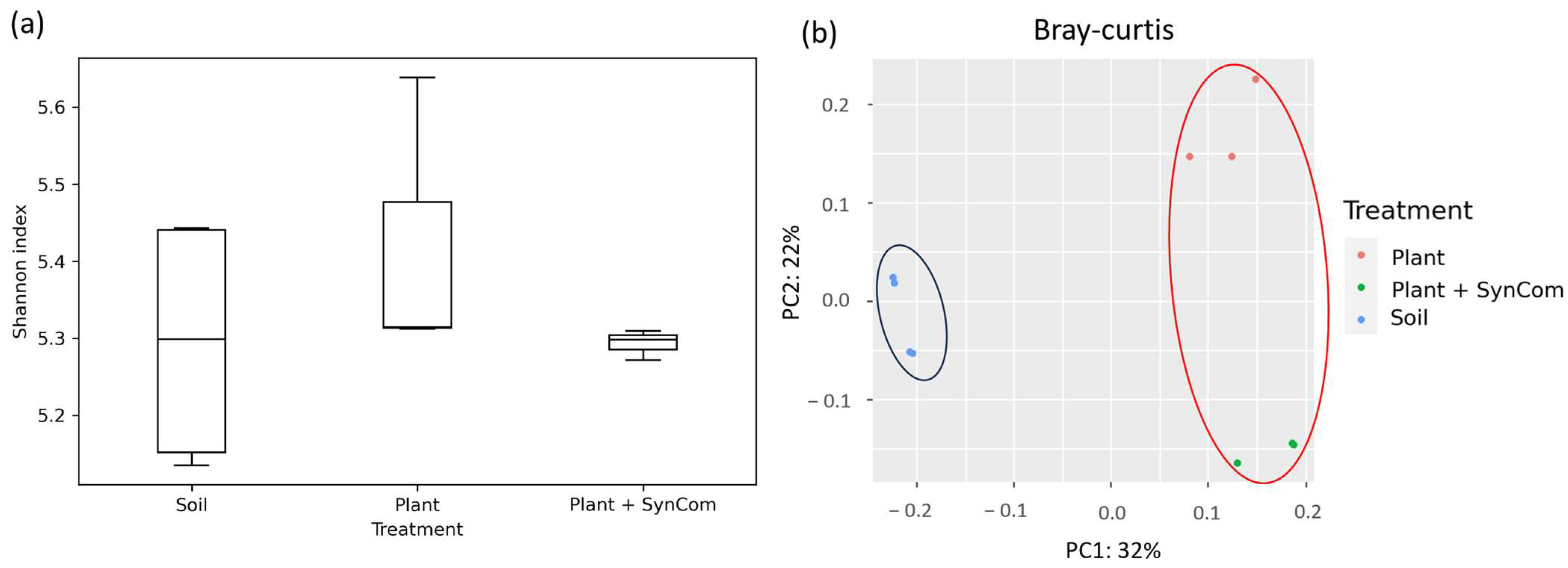
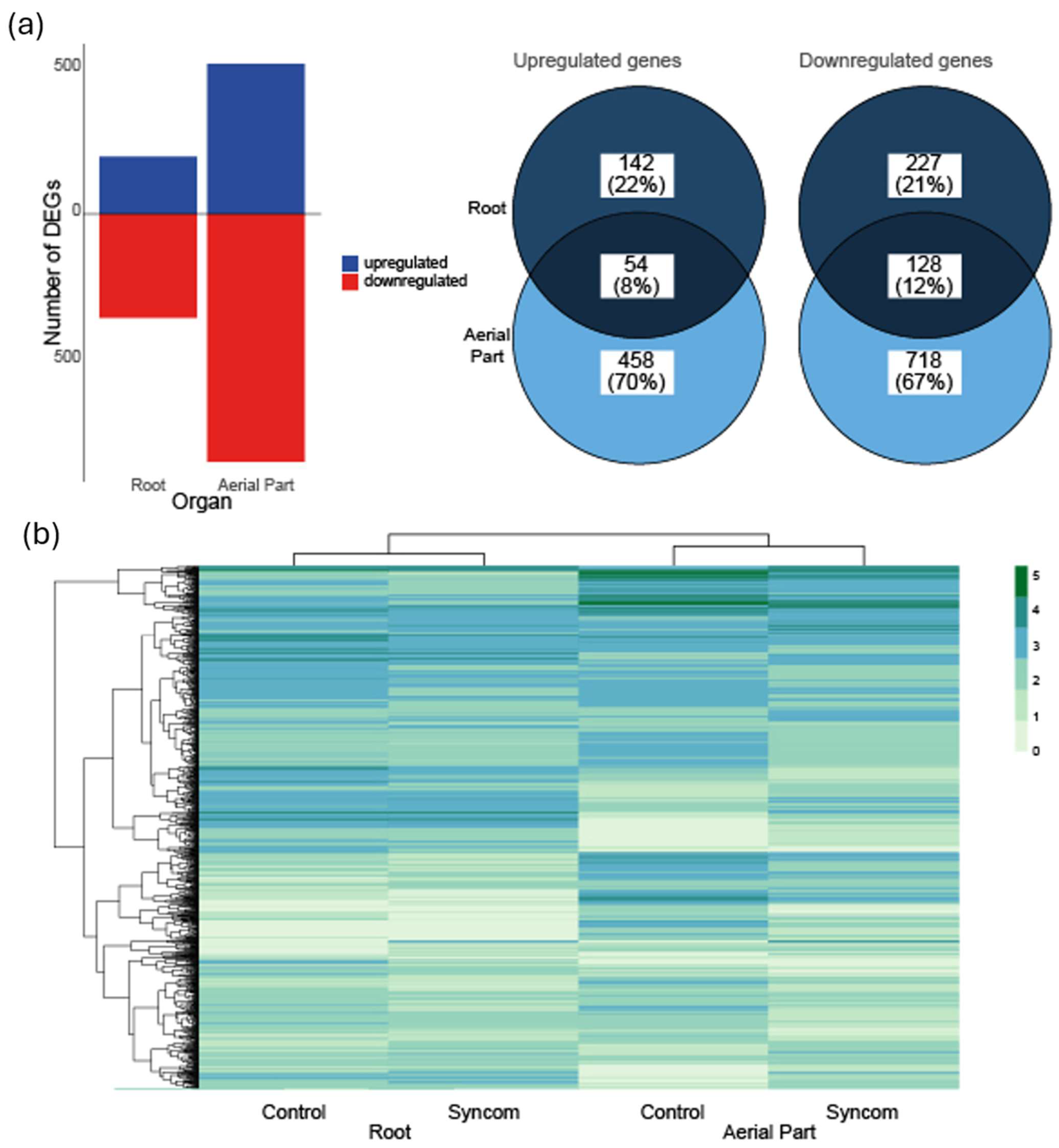
| Assembly | Closest Relative Genome | % dDDH | Length (bp) | Contigs | Completeness |
|---|---|---|---|---|---|
| Rhizobium | Rhizobium metallidurans | 31.9 | 5,818,479 | 5 | 99.00 |
| Pseudomonas | Pseudomonas monsensis | 94.8 | 6,443,679 | 16 | 98.40 |
| Agromyces | Agromyces mediolanus | 72.8 | 4,695,114 | 26 | 97.11 |
| Pseudomonas | Pseudomonas ogarae | 87.3 | 7,066,282 | 29 | 95.30 |
| Chryseobacterium | Chryseobacterium taeanense | 74.4 | 4,333,431 | 18 | 79.94 |
| Ensifer | Ensifer morelensis | 21.0 | 8,190,650 | 460 | 72.33 |
| Microbacterium | Microbacteium oxydans | 91.9 | 4,028,011 | 3 | 99.50 |
| Possible Specie | Genes/Clusters | Function | PGP Category |
|---|---|---|---|
| Pseudomonas monsensis | fecAR | Transport of iron dicitrate (III) | Siderophore |
| fitD | Insect toxin | Toxin | |
| hasDEF | Hemophore biosynthesis | Siderophore | |
| hcnABC | Hydrocyanic acid biosynthesis | Biocontrol | |
| hcp (T6SS) | Type VI secretion system | Biocontrol | |
| iaaHM | Auxin biosynthesis | Phytohormone modulation | |
| paaFIKY | Phenylacetic acid degradation | Phytohormone modulation | |
| phnBCDENWXZ | Phosphate transport | Nutrient mobilization (P) | |
| phoBDHH2LPQRU | Phosphate transport | Nutrient mobilization (P) | |
| pqqABCDE | Pyrroloquinoline quinone | Plant–bacteria interaction, antioxidant | |
| pstABCS | Phosphate transport | Nutrient mobilization (P) | |
| pvdAELMNOPRSY | Pyoverdine | Nutrient mobilization (Fe) | |
| ubiA | Production of 4-hydroxibenzoate | Antibiotic | |
| Cluster 1 | Type NRPS/lokisin | Antifungal | |
| Chryseobacterium taeanense | paaABCDEGHIKYZ | Phenylacetic acid degradation | Phytohormone modulation |
| phnABP | Phosphate transport | Nutrient mobilization (P) | |
| phoABB1DHLPR | Phosphate transport | Nutrient mobilization (P) | |
| ubiA | Production of 4-hydroxibenzoate | Antibiotic | |
| Cluster 1 | Type lantipeptide class I | Antimicrobial | |
| Cluster 2 | Type Aryl polyene, resorcinol | Biocontrol | |
| Rhizobium spp. | acsA | Achromobactin biosynthesis | Siderophore |
| napABCD | Nitrate reductase | Denitrification/nutrient mobilization(N) | |
| nirBCDFS | Nitrite reductase | Denitrification/nutrient mobilization(N) | |
| norBCDEGQR | Nitric oxide reductase | Denitrification | |
| nosDFLYZ | Nitrous oxide reductase | Denitrification/nutrient mobilization(N) | |
| paaABCDEFGHIKXYZ | Phenylacetic acid degradation | Phytohormone modulation | |
| pchR | Enantio-pyochelin biosynthesis | Siderophore | |
| phnABCDEFGHIJKLMNOPW | Phosphate transport | Nutrient mobilization (P) | |
| phoABB1PDHLQRU | Phosphate transport | Nutrient mobilization (P) | |
| pstABCS | Phosphate transport | Nutrient mobilization (P) | |
| speE | Spermidine biosynthesis | Plant–bacteria interaction | |
| ubiA | Production of 4-hydroxibenzoate | Antibiotic | |
| Agromyces mediolanus | hcnABC | Hydrocyanic acid biosynthesis | Biocontrol |
| paaABCDEFGHIKYZ | Phenylacetic acid degradation | Phytohormone modulation | |
| phnBCDEF | Phosphate transport | Nutrient mobilization (P) | |
| phoABB1HPLH2UPR | Phosphate transport | Nutrient mobilization (P) | |
| pstABCS | Phosphate transport | Nutrient mobilization (P) | |
| speE | Spermidine biosynthesis | Plant–bacteria interaction | |
| Microbacterium oxydans | paaDFH | Phenylacetic acid degradation | Phytohormone modulation |
| phnBCDEO | Phosphate transport | Nutrient mobilization (P) | |
| phoBHLRU | Phosphate transport | Nutrient mobilization (P) | |
| Cluster 1 | Type NRP-metallophore/parabactin | Siderophore | |
| Pseudomonas ogarae | acdS | ACC deaminase | Phytohormone modulation |
| fecAR | Transport of iron dicitrate (III) | Siderophore | |
| fitD | Insect toxin | Toxin | |
| hasDEF | Hemophore biosynthesis | Siderophore | |
| hcnABC | Hydrocyanic acid biosynthesis | Biocontrol | |
| hcp (T6SS) | Type VI secretion system | Biocontrol | |
| narGHIJ | Nitrate reductase | Denitrification/nutrient mobilization (N) | |
| nirBCDFS | Nitrite reductase | Denitrification/nutrient mobilization (N) | |
| norBCDEGQR | Nitric oxide reductase | Denitrification | |
| nosDFLYZ | Nitrous oxide reductase | Denitrification/phytohormone modulation | |
| paaFGIKY | Phenylacetic acid degradation | Phytohormone modulation | |
| pchR | Enantio-pyochelin biosynthesis | Siderophore | |
| phnABCDEXZ | Phosphate transport | Nutrient mobilization (P) | |
| phoBDHH2LPQRU | Phosphate transport | Nutrient mobilization (Fe) | |
| pqqBCDE | Pyrroloquinoline quinone | Plant–bacteria interaction, antioxidant | |
| pstABCS | Phosphate transport | Nutrient mobilization (P) | |
| pvdAELMNOPRSY | Pyoverdine | Nutrient mobilization (Fe) | |
| speE | Spermidine biosynthesis | Plant–bacteria interaction | |
| ubiA | Production of 4-hydroxibenzoate | Antibiotic | |
| Cluster 1 | Type 2,4-diacetylphloroglucinol (DAPG) | Antifungal | |
| Cluster 2 | Type lantipeptide class II | Antimicrobial | |
| Ensifer spp. | paaFG | Phenylacetic acid degradation | Phytohormone modulation |
| phnAB | Phosphate transport | Nutrient mobilization (P) | |
| phoABQR | Phosphate transport | Nutrient mobilization (P) | |
| ubiA | Production of 4-hydroxibenzoate | Antibiotic | |
| Cluster 1 | Type β-lactone | Antimicrobial | |
| Cluster 2 | Type β-lactone/mycosubtilin | Antifungal | |
| Cluster 3 | Type Resorcinol | Biocontrol |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Montoya, M.; Durán-Wendt, D.; Garrido-Sanz, D.; Carrera-Ruiz, L.; Vázquez-Arias, D.; Redondo-Nieto, M.; Martín, M.; Rivilla, R. Functional Characterization of a Synthetic Bacterial Community (SynCom) and Its Impact on Gene Expression and Growth Promotion in Tomato. Agronomy 2025, 15, 1794. https://doi.org/10.3390/agronomy15081794
Montoya M, Durán-Wendt D, Garrido-Sanz D, Carrera-Ruiz L, Vázquez-Arias D, Redondo-Nieto M, Martín M, Rivilla R. Functional Characterization of a Synthetic Bacterial Community (SynCom) and Its Impact on Gene Expression and Growth Promotion in Tomato. Agronomy. 2025; 15(8):1794. https://doi.org/10.3390/agronomy15081794
Chicago/Turabian StyleMontoya, Mónica, David Durán-Wendt, Daniel Garrido-Sanz, Laura Carrera-Ruiz, David Vázquez-Arias, Miguel Redondo-Nieto, Marta Martín, and Rafael Rivilla. 2025. "Functional Characterization of a Synthetic Bacterial Community (SynCom) and Its Impact on Gene Expression and Growth Promotion in Tomato" Agronomy 15, no. 8: 1794. https://doi.org/10.3390/agronomy15081794
APA StyleMontoya, M., Durán-Wendt, D., Garrido-Sanz, D., Carrera-Ruiz, L., Vázquez-Arias, D., Redondo-Nieto, M., Martín, M., & Rivilla, R. (2025). Functional Characterization of a Synthetic Bacterial Community (SynCom) and Its Impact on Gene Expression and Growth Promotion in Tomato. Agronomy, 15(8), 1794. https://doi.org/10.3390/agronomy15081794






