Genomic Analysis of Cadmium-Resistant and Plant Growth-Promoting Burkholderia alba Isolated from Plant Rhizosphere
Abstract
1. Introduction
2. Materials and Methods
2.1. DNA Extraction, Identification, and Sequencing of Strain YIM B08401
2.2. Genome Assembly and Annotation of Strain YIM B08401
2.3. Experiment on Root Colonization Ability of Strain YIM B08401
2.4. Qualitative Assays for Enzyme Production by Strain YIM B08401
2.5. Cd Adsorption Assays of Strain YIM B08401
2.5.1. Cd2+ Adsorption by Strain YIM B08401
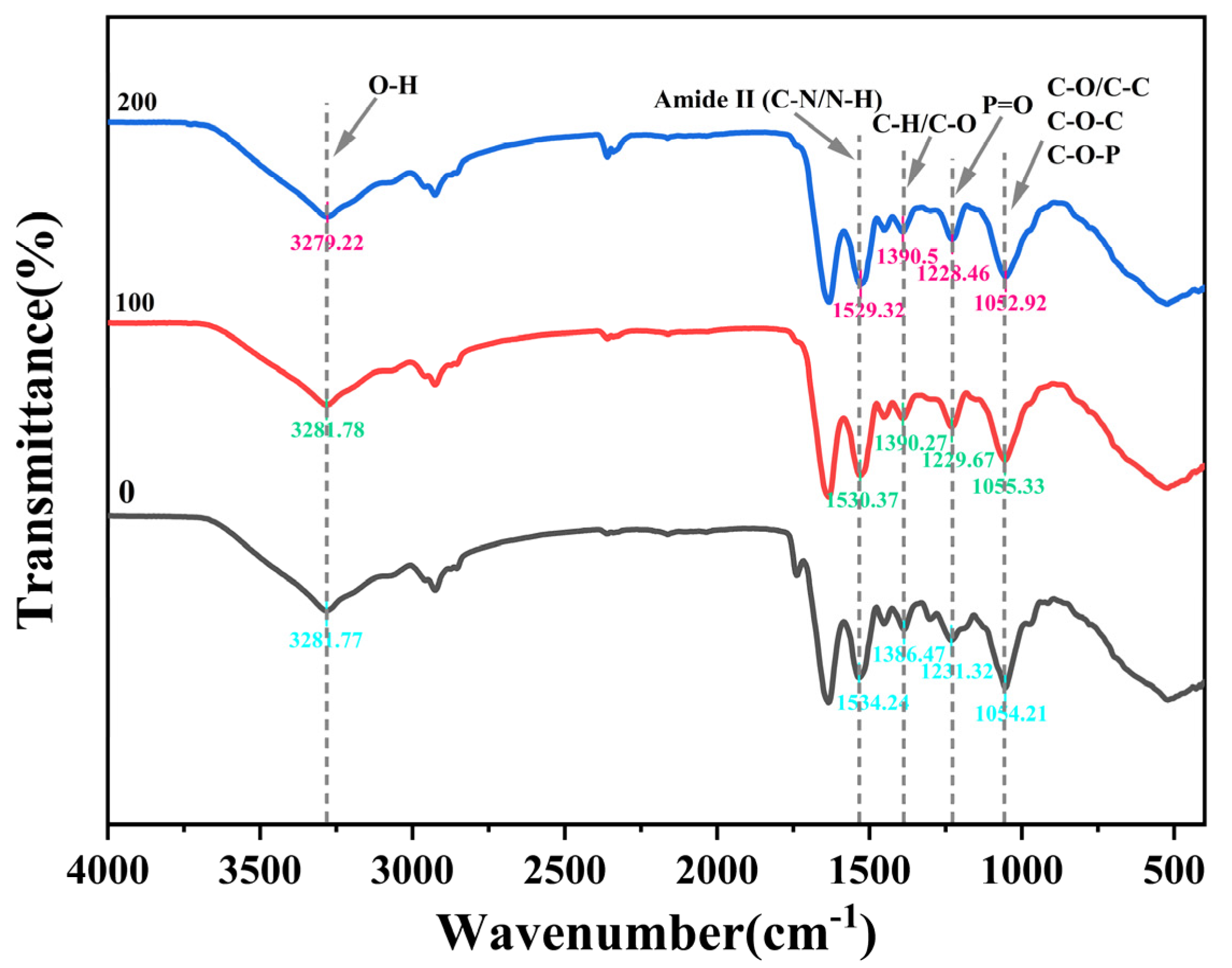
2.5.2. Fourier Transform Infrared Spectroscopy (FTIR) Analysis
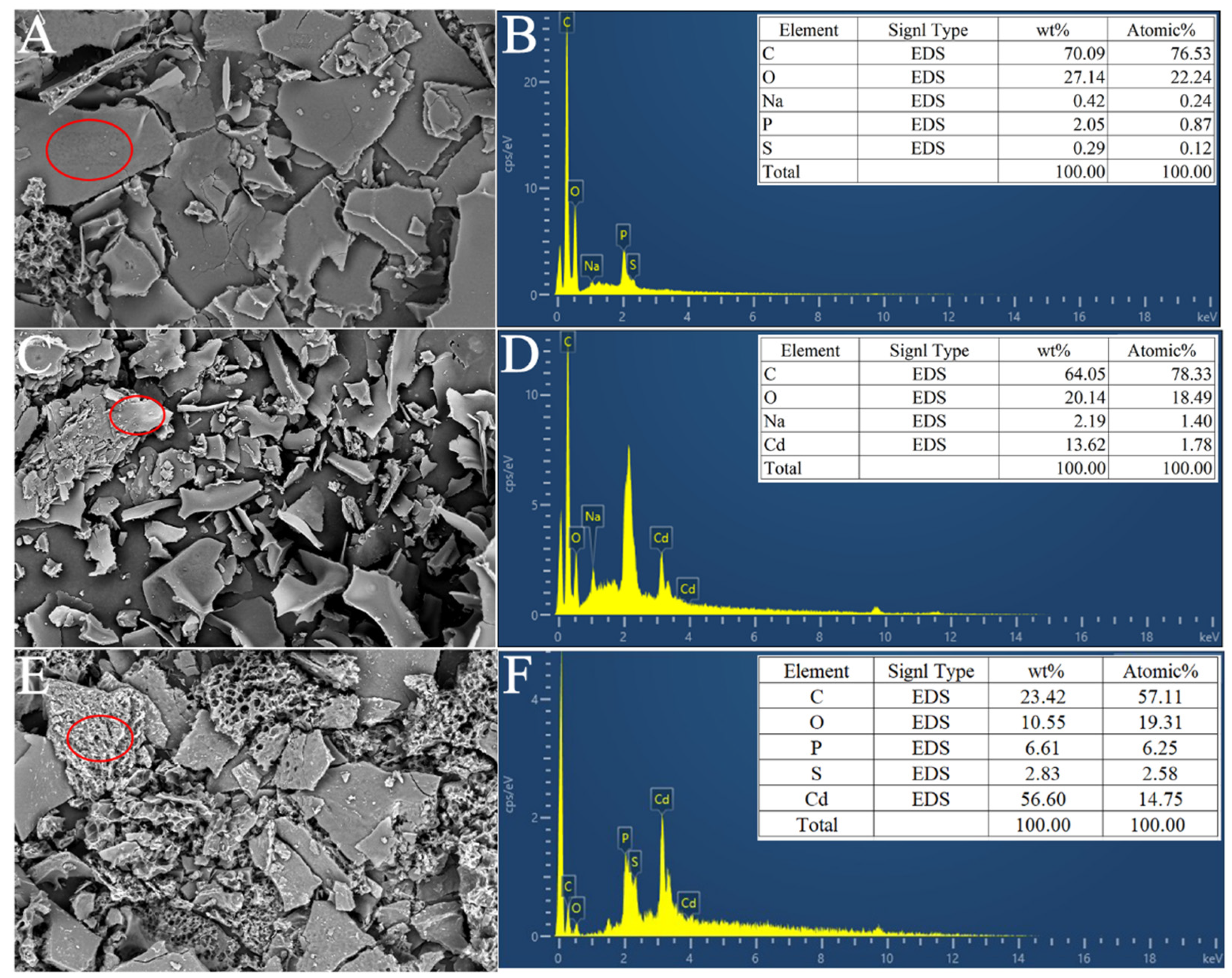
2.5.3. SEM-EDS Analysis
3. Results and Discussion
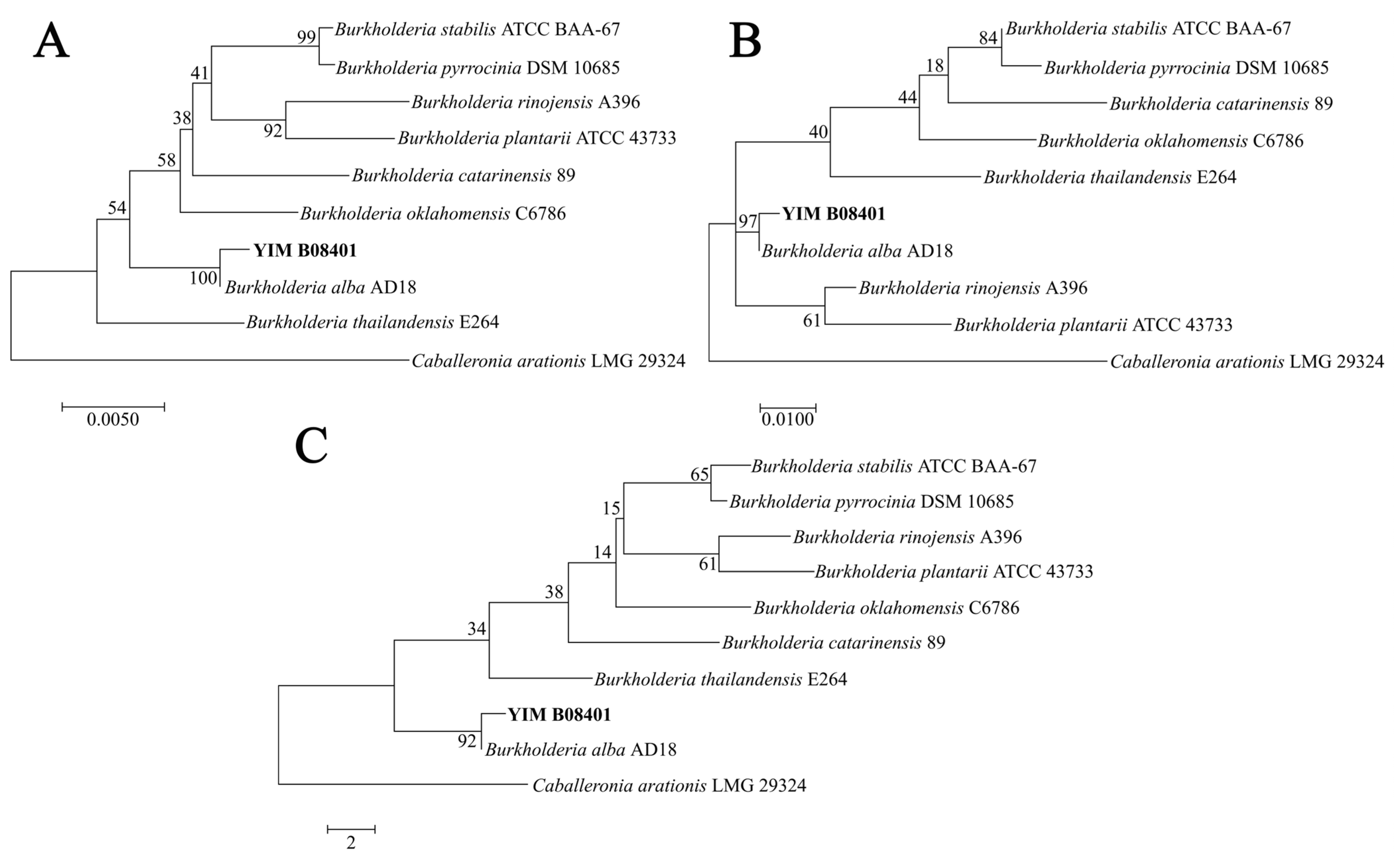
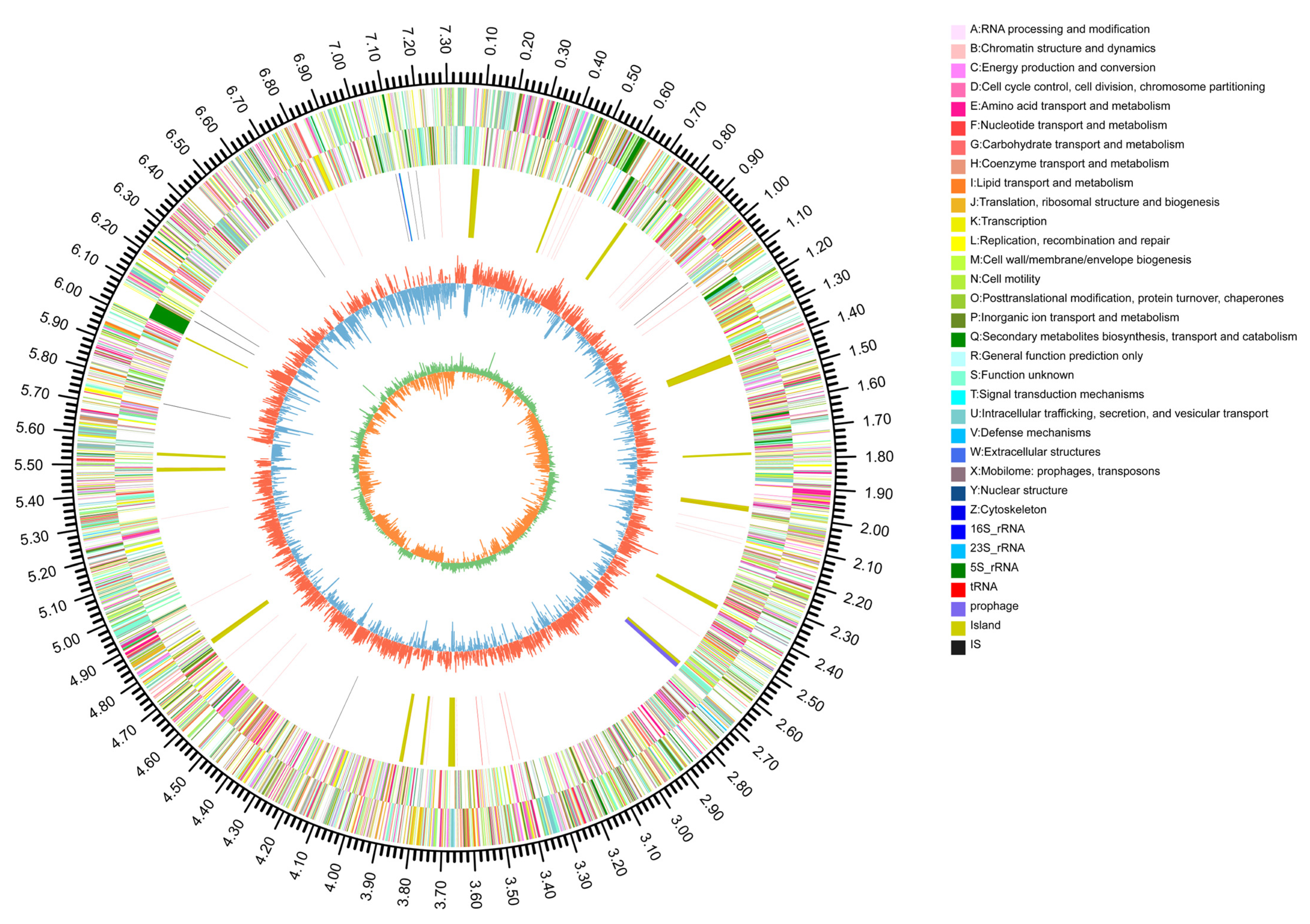
3.1. General Genome Characteristics and Strain Identification of YIM B08401
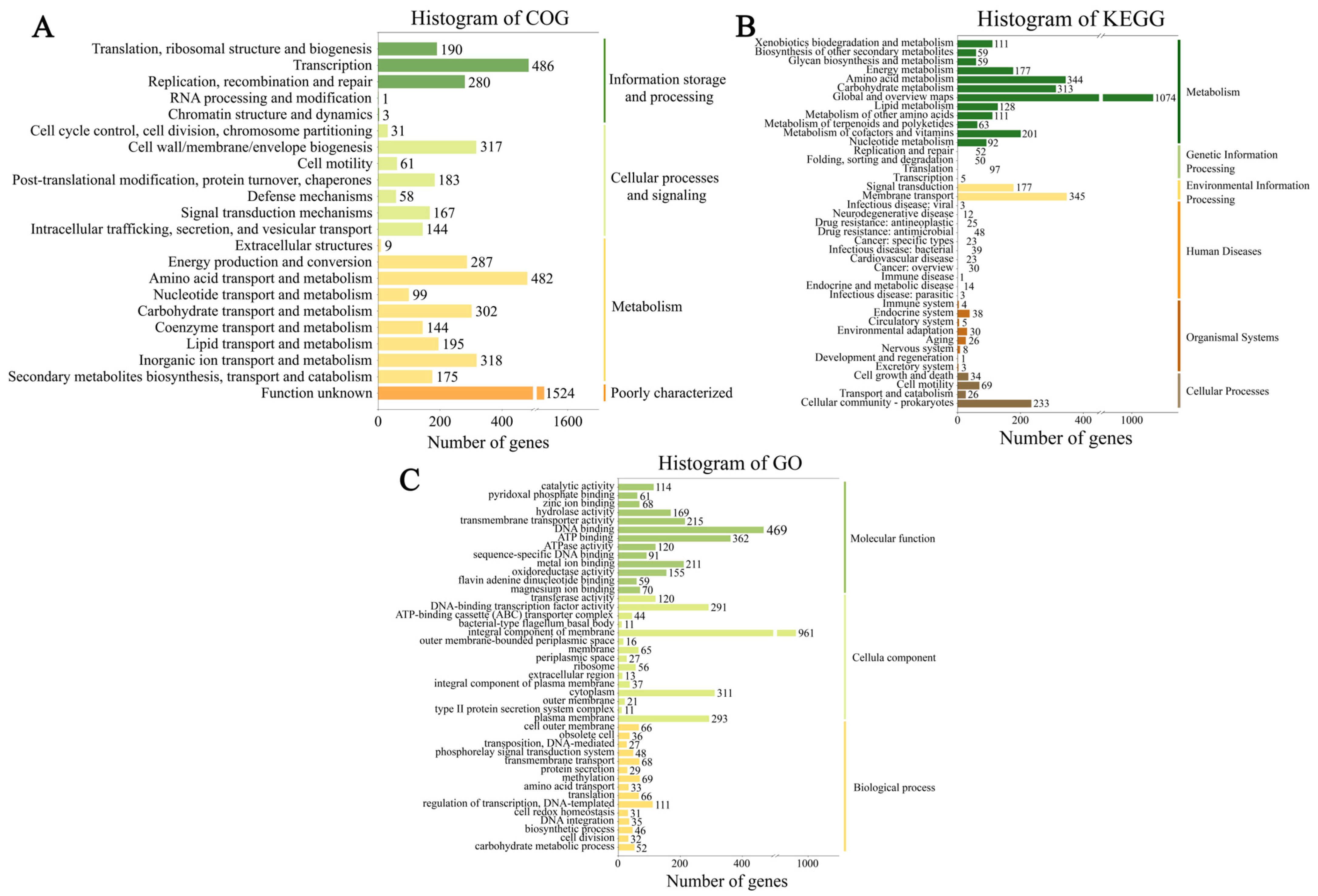
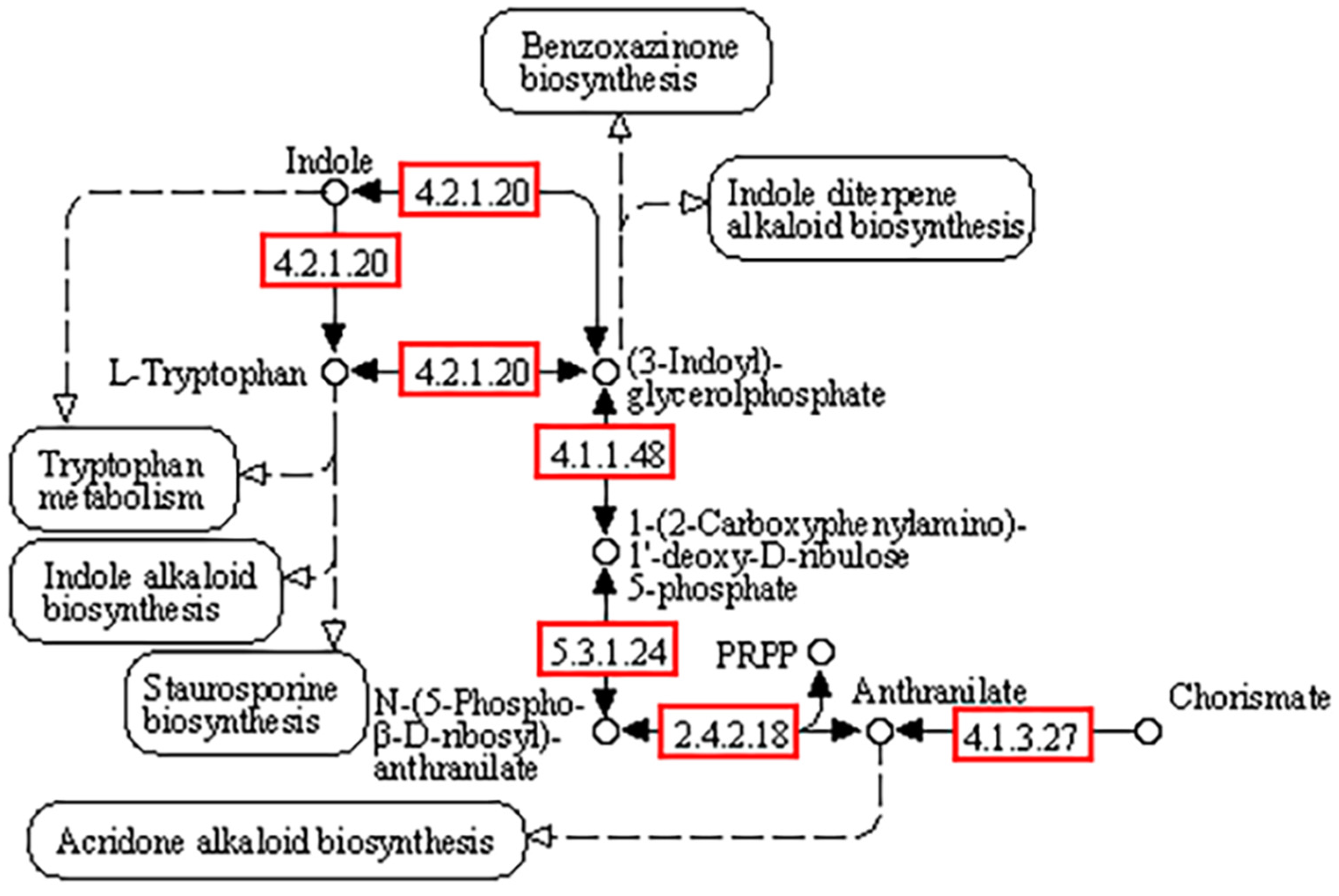
3.2. Genome Functional Annotation
3.3. CAZyme Database Analysis
3.4. antiSMASH Database Analysis
3.5. Plant Growth Promotion Potential
3.5.1. Phosphorus Transformation
3.5.2. Nitrogen Fixation
3.5.3. Sulfur Metabolism
3.5.4. Zinc Solubilization
3.5.5. Siderophore Production
3.5.6. Indole-3-Acetic Acid (IAA) Production
3.5.7. Ethylene Regulation
3.5.8. Degradation of Phenolic Acids
3.5.9. Root Colonization
3.5.10. Heavy Metal Adsorption
3.6. Cd2+ Adsorption Efficiency and Characteristics of Strain YIM B08401
3.7. FTIR Spectroscopy of Strain YIM B08401
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ning, C.; Gao, P.; Wang, B.; Wang, B.; Lin, W.; Jiang, N.; Cai, K. Impacts of chemical fertilizer reduction and organic amendments supplementation on soil nutrient, enzyme activity and heavy metal content. J. Integr. Agric. 2017, 16, 1819–1831. [Google Scholar] [CrossRef]
- Bao, L.; Liu, Y.; Ding, Y.; Shang, J.; Wei, Y.; Tan, Y.; Zi, F. Interactions Between Phenolic Acids and Microorganisms in Rhizospheric Soil from Continuous Cropping of Panax notoginseng. Front. Microbiol. 2022, 13, 791603. [Google Scholar] [CrossRef] [PubMed]
- Jia, M.; Wang, X.; Zhu, X.; Du, Y.; Zhou, P.; Wang, G.; Wang, N.; Bai, Y. Accumulation of coumaric acid is a key factor in tobacco continuous cropping obstacles. Front. Plant. Sci. 2024, 15, 1477324. [Google Scholar] [CrossRef]
- Li, X.; Yu, Y.; He, R.; Zhen, Q.; She, D. Synergistic effects of aged lignin-based biochar and selenium fertilization on heavy metal remediation in agricultural soils. Ind. Crops. Prod. 2025, 225, 120464. [Google Scholar] [CrossRef]
- Ouyang, X.; Ma, J.; Zhang, R.; Li, P.; Gao, M.; Sun, C.; Weng, L.; Chen, Y.; Yan, S.; Li, Y. Uptake of atmospherically deposited cadmium by leaves of vegetables: Subcellular localization by NanoSIMS and potential risks. J. Hazard. Mater. 2022, 431, 128624. [Google Scholar] [CrossRef]
- Roberts, T. Cadmium and phosphorous fertilizers: The issues and the science. Procedia. Eng. 2014, 83, 52–59. [Google Scholar] [CrossRef]
- Mandal, R.; Bashir, Z.; Raj, D. Microbe-assisted phytoremediation for sustainable management of heavy metal in wastewater—A green approach to escalate the remediation of heavy metals. J. Environ. Manag. 2025, 375, 124199. [Google Scholar] [CrossRef]
- Sahu, O.; Chaudhari, P. Review on chemical treatment of industrial waste water. J. Appl. Sci. Environ. Manag. 2013, 17, 241–257. [Google Scholar] [CrossRef]
- Wang, Y.; Zhao, Q.; Sun, Z.; Li, Y.; He, H.; Zhang, Y.; Yang, X.; Wang, D.; Dong, B.; Zhou, H.; et al. Whole-genome analysis revealed the growth-promoting mechanism of endophytic bacterial strain Q2H1 in potato plants. Front. Microbiol. 2022, 13, 1035901. [Google Scholar] [CrossRef]
- Luo, Y.; Chen, L.; Lu, Z.; Zhang, W.; Liu, W.; Chen, Y.; Wang, X.; Du, W.; Luo, J.; Wu, H. Genome sequencing of biocontrol strain Bacillus amyloliquefaciens Bam1 and further analysis of its heavy metal resistance mechanism. Bioresour. Bioprocess. 2022, 9, 74. [Google Scholar] [CrossRef]
- Chlebek, D.; Płociniczak, T.; Gobetti, S.; Kumor, A.; Hupert-Kocurek, K.; Pacwa-Płociniczak, M. Analysis of the Genome of the Heavy Metal Resistant and Hydrocarbon-Degrading Rhizospheric Pseudomonas qingdaonensis ZCR6 Strain and Assessment of Its Plant-Growth-Promoting Traits. Int. J. Mol. Sci. 2021, 23, 214. [Google Scholar] [CrossRef] [PubMed]
- Janaki, M.; Kirupanantha-Rajan, P.; Senthil-Nathan, S.; Stanley-Raja, V.; Al Farraj, D.; Aljeidi, R.; Arokiyaraj, S. Beneficial role of Burkholderia cepacia in heavy metal bioremediation in metal-polluted soils and enhance the tomato plant growth. Biocatal. Agric. Biotechnol. 2024, 57, 103032. [Google Scholar] [CrossRef]
- Lan, Y.; Liu, M.; Song, Y.; Cao, Y.; Li, F.; Luo, D.; Qiao, D. Distribution, characterization, and evolution of heavy metal resistance genes and Tn7-like associated heavy metal resistance Gene Island of Burkholderia. Front. Microbiol. 2023, 14, 1252127. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Huang, Y.; Yang, X.; Xue, W.; Zhang, X.; Zhang, Y.; Pang, J.; Liu, Y.; Liu, Z. Burkholderia sp. Y4 inhibits cadmium accumulation in rice by increasing essential nutrient uptake and preferentially absorbing cadmium. Chemosphere 2020, 252, 126603. [Google Scholar] [CrossRef]
- Jiang, C.; Sheng, X.; Qian, M.; Wang, Q. Isolation and characterization of a heavy metal-resistant Burkholderia sp. from heavy metal-contaminated Paddy field soil and its potential in promoting plant growth and heavy metal accumulation in metal-polluted soil. Chemosphere 2008, 72, 157–164. [Google Scholar] [CrossRef]
- Huang, G.; Tian, H.; Liu, H.; Fan, X.; Liang, Y.; Li, Y. Characterization of plant-growth-promoting effects and concurrent promotion of heavy metal accumulation in the tissues of the plants grown in the polluted soil by Burkholderia strain LD-11. Int. J. Phytoremediation 2013, 15, 991–1009. [Google Scholar] [CrossRef]
- Chaudhry, V.; Runge, P.; Sengupta, P.; Doehlemann, G.; Parker, J.; Kemen, E. Shaping the leaf microbiota: Plant-microbe-microbe interactions. J. Exp. Bot. 2021, 72, 36–56. [Google Scholar] [CrossRef]
- Prashar, D.; Sharma, M.; Sharma, D.; Mir, Z.; Sood, H. Exploring the complexity of plant stress response: Role of metabolomics and signalling pathways in metabolic diversification. J. Plant Biochem. Biotechnol. 2025, 1–14. [Google Scholar] [CrossRef]
- Pan, H.; Ren, Y.; Sun, P.; Jing, X. Toward better understanding of denitrifying phosphorus accumulating organisms (DPAOs) via Single-cell Raman spectroscopy: A review. Environ. Technol. Innov. 2025, 38, 104205. [Google Scholar] [CrossRef]
- Chavan, A.; Khardenavis, A. Annotating multiple prebiotic synthesizing capabilities through whole genome sequencing of Fusarium strain HFK-74. Appl. Biochem. Biotechnol. 2023, 196, 4993–5012. [Google Scholar] [CrossRef]
- Bhanse, P.; Singh, L.; Qureshi, A. Functional and Genomic Potential of Burkholderia contaminans PB_AQ24 Isolate for Boosting the Growth of Bamboo Seedlings in Heavy Metal Contaminated Soils. Appl. Biochem. Biotechnol. 2025, 197, 2437–2456. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Gupta, R.; Purohit, H.; Khardenavis, A. Genomic characterization of denitrifying methylotrophic Pseudomonas aeruginosa strain AAK/M5 isolated from municipal solid waste landfill soil. World J. Microbiol. Biotechnol. 2022, 38, 140. [Google Scholar] [CrossRef]
- Shen, W.; Zhang, J.; Geng, B.; Qiu, M.; Hu, M.; Yang, Q.; Bao, W.; Xiao, Y.; Zheng, Y.; Peng, W.; et al. Establishment and application of a CRISPR-Cas12a assisted genome-editing system in Zymomonas mobilis. Microb. Cell Fact. 2019, 18, 162. [Google Scholar] [CrossRef] [PubMed]
- Feng, L.; Zhao, J.; Shi, Z.; Mo, Y.; Yang, T.; Shen, Y.; He, F.; Li, M.; Yang, P. Isolation and Identification of Bacteria in Forest Rhizosphere Soil and Their Biological Activity Screening. Biotechnol. Bull. 2024, 40, 294–307. [Google Scholar]
- Qin, G.; Wang, D.; Luo, K.; Liu, Y.; Xie, Y.; Wang, M.; Li, C.; Fan, R.; Tian, X. Evaluation of probiotic properties and safety of a Bacillus strain for shrimp farming: Integrating in vitro testing, genomic analysis and in vivo validation. Microbiol. Res. 2025, 297, 128179. [Google Scholar] [CrossRef]
- Shen, Y.; Shi, Z.; Zhao, J.; Ling, M.; Tang, J.; Wang, N.; Mo, Y.; Yang, T.; Zhou, X.; Chen, Q.; et al. Whole genome sequencing provides evidence for Bacillus velezensis SH-1471 as a beneficial rhizosphere bacterium in plants. Sci. Rep. 2023, 13, 20929. [Google Scholar] [CrossRef]
- Ding, H.; Mo, W.; Yu, S.; Cheng, H.; Peng, L.; Liu, Z. Whole genome sequence of Bacillus velezensis strain GUMT319: A potential biocontrol agent against tobacco black shank disease. Front. Microbiol. 2021, 12, 658113. [Google Scholar] [CrossRef]
- Rishad, K.; Rebello, S.; Shabanamol, P.; Jisha, M. Biocontrol potential of Halotolerant bacterial chitinase from high yielding novel Bacillus pumilus MCB-7 autochthonous to mangrove ecosystem. Pestic. Biochem. Physiol. 2017, 137, 36–41. [Google Scholar] [CrossRef]
- Ninawe, S.; Lal, R.; Kuhad, R. Isolation of three xylanase-producing strains of actinomycetes and their identification using molecular methods. Curr. Microbiol. 2006, 53, 178–182. [Google Scholar] [CrossRef]
- Iqbal, M.; Saeed, A.; Zafar, S. FTIR spectrophotometry, kinetics and adsorption isotherms modeling, ion exchange, and EDX analysis for understanding the mechanism of Cd2+ and Pb2+ removal by mango peel waste. J. Hazard. Mater. 2009, 164, 161–171. [Google Scholar] [CrossRef]
- Liu, S.; Huang, Y.; Zheng, Q.; Zhan, M.; Hu, Z.; Ji, H.; Zhu, D.; Zhao, X. Cd-Resistant Plant Growth-Promoting Rhizobacteria Bacillus siamensis R27 Absorbed Cd and Reduced Cd Accumulation in Lettuce (Lactuca sativa L.). Microorganisms 2024, 12, 2321. [Google Scholar] [CrossRef]
- Tatusov, R.; Koonin, E.; Lipman, D. A genomic perspective on protein families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Szczerba, H.; Komoń-Janczara, E.; Krawczyk, M.; Dudziak, K.; Nowak, A.; Kuzdraliński, A.; Waśko, A.; Targoński, Z. Genome analysis of a wild rumen bacterium Enterobacter aerogenes LU2-a novel bio-based succinic acid producer. Sci. Rep. 2020, 10, 1986. [Google Scholar] [CrossRef]
- Eida, A.; Bougouffa, S.; L’Haridon, F.; Alam, I.; Weisskopf, L.; Bajic, V.; Saad, M.; Hirt, H. Genome Insights of the Plant-Growth Promoting Bacterium Cronobacter muytjensii JZ38 With Volatile-Mediated Antagonistic Activity Against Phytophthora infestans. Front. Microbiol. 2020, 11, 369. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.; Blake, J.; Botstein, D.; Butler, H.; Cherry, J.; Davis, A.; Dolinski, K.; Dwight, S.; Eppig, J.; et al. Gene Ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Yu, D.; Li, N.; Liu, S.; Xu, H.; Li, J.; He, F.; Zou, L.; Cao, Z.; Wen, J. Biological Characterization and Whole-Genome Analysis of Bacillus subtilis MG-1 Isolated from Mink Fecal Samples. Microorganisms 2023, 11, 2821. [Google Scholar] [CrossRef]
- Mannaa, M.; Han, G.; Jeong, T.; Kang, M.; Lee, D.; Jung, H.; Seo, Y. Taxonomy-guided selection of Paraburkholderia busanensis sp. nov.: A versatile biocontrol agent with mycophagy against Colletotrichum scovillei causing pepper anthracnose. Microbiol. Spectr. 2023, 11, e0242623. [Google Scholar] [CrossRef]
- Meena, B.; Rajan, L.; Vinithkumar, N.; Kirubagaran, R. Novel marine actinobacteria from emerald Andaman & Nicobar Islands: A prospective source for industrial and pharmaceutical byproducts. BMC Microbiol. 2013, 13, 145. [Google Scholar] [CrossRef]
- Bach, E.; Passaglia, L.; Jiao, J.; Gross, H. Burkholderia in the genomic era: From taxonomy to the discovery of new antimicrobial secondary metabolites. Crit. Rev. Microbiol. 2022, 48, 121–160. [Google Scholar] [CrossRef]
- Blin, K.; Shaw, S.; Kloosterman, A.; Charlop-Powers, Z.; van Wezel, G.; Medema, M.; Weber, T. antiSMASH 6.0: Improving cluster detection and comparison capabilities. Nucleic Acids Res. 2021, 49, W29–W35. [Google Scholar] [CrossRef]
- Naughton, L.; Romano, S.; O’Gara, F.; Dobson, A. Identification of Secondary Metabolite Gene Clusters in the Pseudovibrio Genus Reveals Encouraging Biosynthetic Potential toward the Production of Novel Bioactive Compounds. Front. Microbiol. 2017, 8, 1494. [Google Scholar] [CrossRef]
- Jenul, C.; Sieber, S.; Daeppen, C.; Mathew, A.; Lardi, M.; Pessi, G.; Hoepfner, D.; Neuburger, M.; Linden, A.; Gademann, K.; et al. Biosynthesis of fragin is controlled by a novel quorum sensing signal. Nat. Commun. 2018, 9, 1297. [Google Scholar] [CrossRef]
- Deng, P.; Foxfire, A.; Xu, J.; Baird, S.; Jia, J.; Delgado, K.; Shin, R.; Smith, L.; Lu, S. The Siderophore Product Ornibactin Is Required for the Bactericidal Activity of Burkholderia contaminans MS14. Appl. Environ. Microbiol. 2017, 83, e00051-17. [Google Scholar] [CrossRef] [PubMed]
- Dose, B.; Niehs, S.; Scherlach, K.; Flórez, L.; Kaltenpoth, M.; Hertweck, C. Unexpected Bacterial Origin of the Antibiotic Icosalide: Two-Tailed Depsipeptide Assembly in Multifarious Burkholderia Symbionts. ACS Chem. Biol. 2018, 13, 2414–2420. [Google Scholar] [CrossRef] [PubMed]
- Adeniji, A.; Ayangbenro, A.; Loots, D. Genome Sequence Resource of Pseudomonas fulva HARBPS9.1-Candidate Biocontrol Agent. Phytopathology 2021, 111, 896–898. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhou, H.; Chen, H.; Jing, X.; Zheng, W.; Li, R.; Sun, T.; Liu, J.; Fu, J.; Huo, L.; et al. Discovery of recombinases enables genome mining of cryptic biosynthetic gene clusters in Burkholderiales species. Proc. Natl. Acad. Sci. USA 2018, 115, E4255–E4263. [Google Scholar] [CrossRef]
- Suarez-Moreno, Z.; Caballero-Mellado, J.; Coutinho, B.; Mendonca-Previato, L.; James, E.; Venturi, V. Common Features of Environmental and Potentially Beneficial Plant-Associated Burkholderia. Microb. Ecol. 2012, 63, 249–266. [Google Scholar] [CrossRef]
- Rawat, P.; Das, S.; Shankhdhar, D.; Shankhdhar, S. Phosphate-Solubilizing Microorganisms: Mechanism and Their Role in Phosphate Solubilization and Uptake. J. Soil Sci. Plant Nutr. 2021, 21, 49–68. [Google Scholar] [CrossRef]
- Nacoon, S.; Jogloy, S.; Riddech, N.; Mongkolthanaruk, W.; Kuyper, T.; Boonlue, S. Interaction between Phosphate Solubilizing Bacteria and Arbuscular Mycorrhizal Fungi on Growth Promotion and Tuber Inulin Content of Helianthus tuberosus L. Sci. Rep. 2020, 10, 4916. [Google Scholar] [CrossRef]
- Goldstein, A. Recent progress in understanding the molecular genetics and biochemistry of calcium phosphate solubilization by gram negative bacteria. Biol. Agric. Horti. 1995, 12, 185–193. [Google Scholar] [CrossRef]
- Stosiek, N.; Talma, M.; Klimek-Ochab, M. Carbon-Phosphorus Lyase-the State of the Art. Appl. Biochem. Biotechnol. 2020, 190, 1525–1552. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, P.; Johnson, D.; Ragle, B.; Unciuleac, M.; Dean, D. Controlled expression of nif and isc iron-sulfur protein maturation components reveals target specificity and limited functional replacement between the two systems. J. Bacteriol. 2007, 189, 2854–2862. [Google Scholar] [CrossRef]
- Tokumoto, U.; Kitamura, S.; Fukuyama, K.; Takahashi, Y. Interchangeability and distinct properties of bacterial Fe-S cluster assembly systems: Functional replacement of the isc and suf operons in Escherichia coli with the nifSU-like operon from Helicobacter pylori. J. Biochem. 2004, 136, 199–209. [Google Scholar] [CrossRef]
- Yusuf, M.; Saeed, T.; Almarri, H.; Khan, T.; Faizan, M.; Elsayed, N. Hydrogen sulfide counteract copper induced inhibition of photosynthetic performance through altered proline metabolism and enhanced antioxidants in Cucumis sativus. Plant Stress 2023, 10, 100222. [Google Scholar] [CrossRef]
- Pourebrahimi, M.; Eshghi, S.; Ramezanian, A.; Faghih, S. Effect of combined application of selenium and hydrogen sulfide under salinity stress on yield, physiological traits and biofortification of strawberries in hydroponic cultivation. Sci. Hortic. 2023, 315, 111982. [Google Scholar] [CrossRef]
- Wang, L.; Huang, X.; Liu, C.; Zhang, C.; Shi, K.; Wang, M.; Wang, Y.; Song, Q.; Chen, X.; Jin, P.; et al. Hydrogen sulfide alleviates chilling injury by modulating respiration and energy metabolisms in cold-stored peach fruit. Postharvest Biol. Technol. 2023, 199, 112291. [Google Scholar] [CrossRef]
- Wang, L.; Chen, S.; Shao, J.; Zhang, C.; Mei, L.; Wang, K.; Jin, P.; Zheng, Y. Hydrogen sulfide alleviates chilling injury in peach fruit by maintaining cell structure integrity via regulating endogenous H2S, antioxidant and cell wall metabolisms. Food Chem. 2022, 391, 133283. [Google Scholar] [CrossRef]
- Hu, L.; Wang, Y.; Ci, M.; Long, Y. Unravelling microbial drivers of the sulfate-reduction process inside landfill using metagenomics. Chemosphere 2023, 313, 137537. [Google Scholar] [CrossRef]
- Roberts, C.; Ni, F.; Mitra, B. The Zinc and Iron Binuclear Transport Center of ZupT, a ZIP Transporter from Escherichia coli. Biochem. 2021, 60, 3738–3752. [Google Scholar] [CrossRef]
- Dubois, M.; Van den Broeck, L.; Inzé, D. The Pivotal Role of Ethylene in Plant Growth. Trends Plant Sci. 2018, 23, 311–323. [Google Scholar] [CrossRef]
- Yang, S. Biosynthesis and action of ethylene. HortScience 1985, 20, 41–45. [Google Scholar] [CrossRef]
- Karmakar, R. State of the art of bacterial chemotaxis. J. Basic Microbiol. 2021, 61, 366–379. [Google Scholar] [CrossRef] [PubMed]
- Eisenbach, M.; Caplan, S. Bacterial chemotaxis: Unsolved mystery of the flagellar switch. Curr. Biol. 1998, 8, R444–R446. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Jia, Q.; Jing, W.; Dahms, H.; Wang, L. Screening strains for microbial biosorption technology of cadmium. Chemosphere 2020, 251, 126428. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.; Zheng, X.; Zhang, D.; Iqbal, W.; Liu, C.; Yang, B.; Zhao, X.; Lu, X.; Mao, Y. Microbial characterization of heavy metal resistant bacterial strains isolated from an electroplating wastewater treatment plant. Ecotoxicol. Environ. Saf. 2019, 181, 472–480. [Google Scholar] [CrossRef]
- Khanna, K.; Kohli, S.; Kaur, R.; Handa, N.; Bakshi, P.; Sharma, P.; Ohri, P.; Bhardwaj, R. Reconnoitering the efficacy of plant growth promoting rhizobacteria in expediting phytoremediation potential of heavy metals. J. Plant Growth. Regul. 2023, 42, 6474–6502. [Google Scholar] [CrossRef]
- Margaryan, A.; Panosyan, H.; Birkeland, N. Heavy metal resistance in prokaryotes: Mechanism and application. In Microbial Communities and Their Interactions in the Extreme Environment; Springer: London, UK, 2021; pp. 273–313. [Google Scholar]
- Rauser, W. Phytochelatins and related peptides. Structure, biosynthesis, and function. Plant Physiol. 1995, 109, 1141. [Google Scholar] [CrossRef]
- Lu, S. Glutathione synthesis. Biochim. Biophys. Acta. 2013, 1830, 3143–3153. [Google Scholar] [CrossRef]
- Li, M.; Stragliati, L.; Bellini, E.; Ricci, A.; Saba, A.; Sanità di Toppi, L.; Varotto, C. Evolution and functional differentiation of recently diverged phytochelatin synthase genes from Arundo donax L. J. Exp. Bot. 2019, 70, 5391–5405. [Google Scholar] [CrossRef]
- Nies, D. Efflux-mediated heavy metal resistance in prokaryotes. FEMS Microbio. Rev. 2003, 27, 313–339. [Google Scholar] [CrossRef]
- Gaballa, A.; Helmann, J. Bacillus subtilis CPx-type ATPases: Characterization of Cd, Zn, Co and Cu efflux systems. Biometals 2003, 16, 497–505. [Google Scholar] [CrossRef]
- Nies, D. Resistance to cadmium, cobalt, zinc, and nickel in microbes. Plasmid 1992, 27, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Nies, D. The cobalt, zinc, and cadmium efflux system CzcABC from Alcaligenes eutrophus functions as a cation-proton antiporter in Escherichia coli. J. Bacteriol. 1995, 177, 2707–2712. [Google Scholar] [CrossRef] [PubMed]
- Decho, A.; Gutierrez, T. Microbial Extracellular Polymeric Substances (EPSs) in Ocean Systems. Front. Microbiol. 2017, 8, 922. [Google Scholar] [CrossRef]
- Sreedevi, P.; Suresh, K.; Jiang, G. Bacterial bioremediation of heavy metals in wastewater: A review of processes and applications. J. Water Pro. Engin. 2022, 48, 102884. [Google Scholar] [CrossRef]
- Abbas, S.; Ismail, I.; Mostafa, T.; Sulaymon, A. Biosorption of heavy metals: A review. J. Chem. Sci. Technol. 2014, 3, 74–102. [Google Scholar]
- Zeng, Q.; Zhu, T.; Wen, Y.; Li, F.; Cheng, Y.; Chen, S.; Lan, T.; Yang, Y.; Liao, J.; Sun, Q.; et al. The dynamic behavior and mechanism of uranium (VI) biomineralization in Enterobacter sp. X57. Chemosphere 2022, 298, 134196. [Google Scholar] [CrossRef]
- Alidoust, L.; Zahiri, H.; Maleki, H.; Soltani, N.; Vali, H.; Noghabi, K. Nostoc entophytum cell response to cadmium exposure: A possible role of chaperon proteins GroEl and HtpG in cadmium-induced stress. Ecotoxicol. Environ. Saf. 2019, 169, 40–49. [Google Scholar] [CrossRef]
- Xie, Y.; He, N.; Wei, M.; Wen, T.; Wang, X.; Liu, H.; Zhong, S.; Xu, H. Cadmium biosorption and mechanism investigation using a novel Bacillus subtilis KC6 isolated from pyrite mine. J. Clean. Prod. 2021, 312, 127749. [Google Scholar] [CrossRef]
- Qiao, W.; Zhang, Y.; Xia, H.; Luo, Y.; Liu, S.; Wang, S.; Wang, W. Bioimmobilization of lead by Bacillus subtilis X3 biomass isolated from lead mine soil under promotion of multiple adsorption mechanisms. R. Soc. Open Sci. 2019, 6, 181701. [Google Scholar] [CrossRef]
- D’Souza, L.; Devi, P.; Shridhar, D.; Naik, C. Use of Fourier Transform Infrared (FTIR) spectroscopy to study cadmium-induced changes in Padina tetrastromatica (Hauck). Anal. Chem. Insights 2008, 3, 135–143. [Google Scholar] [CrossRef]
- Yee, N.; Benning, L.; Phoenix, V.; Ferris, F. Characterization of metal-cyanobacteria sorption reactions: A combined macroscopic and infrared spectroscopic investigation. Environ. Sci. Technol. 2004, 38, 775–782. [Google Scholar] [CrossRef] [PubMed]
- Naik, M.; Pandey, A.; Dubey, S. Biological characterization of lead-enhanced exopolysaccharide produced by a lead resistant Enterobacter cloacae strain P2B. Biodegradation 2012, 23, 775–783. [Google Scholar] [CrossRef] [PubMed]
- Kepenek, E.; Gozen, A.; Severcan, F. Molecular characterization of acutely and gradually heavy metal acclimated aquatic bacteria by FTIR spectroscopy. J. Biophot. 2019, 12, e201800301. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Fang, L.; Cai, P.; Huang, Q.; Chen, H.; Liang, W.; Rong, X. Influence of extracellular polymeric substances (EPS) on Cd adsorption by bacteria. Environ. Pollut. 2011, 159, 1369–1374. [Google Scholar] [CrossRef]

| The Characterization | Value |
|---|---|
| Assembly information | |
| The total length (bp) | 7,322,157 |
| Total scaffold No | 204 |
| Scaffold N50(bp) | 201,844 |
| Contigs | 193 |
| Conting N50 (bp) | 143,587 |
| G + C content (%) | 66.39 |
| Annotation information | |
| CDSs (total) | 6504 |
| rRNA | 3 |
| tRNA | 63 |
| Mobile Genetic Element (MGE) | |
| Gene island | 15 |
| Prophage regions | 2 |
| CRISPR-Cas | 22 |
| Insertion sequence | 9 |
| Tns | 62 |
| NCBI | |
| BioProject ID | PRJNA1027102 |
| BioSample ID | SAMN37776913 |
| GeneBank accession number | JAWJWL000000000 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, L.; Liu, X.; Wang, N.; Shi, Z.; Wang, Y.; Jia, J.; Shi, Z.; Pu, T.; Yang, P. Genomic Analysis of Cadmium-Resistant and Plant Growth-Promoting Burkholderia alba Isolated from Plant Rhizosphere. Agronomy 2025, 15, 1780. https://doi.org/10.3390/agronomy15081780
Feng L, Liu X, Wang N, Shi Z, Wang Y, Jia J, Shi Z, Pu T, Yang P. Genomic Analysis of Cadmium-Resistant and Plant Growth-Promoting Burkholderia alba Isolated from Plant Rhizosphere. Agronomy. 2025; 15(8):1780. https://doi.org/10.3390/agronomy15081780
Chicago/Turabian StyleFeng, Luyao, Xin Liu, Nan Wang, Zhuli Shi, Yu Wang, Jianpeng Jia, Zhufeng Shi, Te Pu, and Peiwen Yang. 2025. "Genomic Analysis of Cadmium-Resistant and Plant Growth-Promoting Burkholderia alba Isolated from Plant Rhizosphere" Agronomy 15, no. 8: 1780. https://doi.org/10.3390/agronomy15081780
APA StyleFeng, L., Liu, X., Wang, N., Shi, Z., Wang, Y., Jia, J., Shi, Z., Pu, T., & Yang, P. (2025). Genomic Analysis of Cadmium-Resistant and Plant Growth-Promoting Burkholderia alba Isolated from Plant Rhizosphere. Agronomy, 15(8), 1780. https://doi.org/10.3390/agronomy15081780





