Diversity and Functional Differences in Soil Bacterial Communities in Wind–Water Erosion Crisscross Region Driven by Microbial Agents
Abstract
1. Introduction
- The combined application of AMF, BM, and TH with Populus simonii Carr will synergistically improve soil microbial community structure and function in the wind–water erosion zone, leading to enhanced enzyme activity and microbial biomass.
- The inoculants will differentially modulate the relative abundance of key bacterial phyla (e.g., Firmicutes and Actinobacteria) and fungal taxa, thereby driving soil remediation.
- To quantify the effects of AMF, BM, and TH treatments on soil microbial diversity, enzyme activity, and biomass carbon/nitrogen in eroded soils.
- To identify the dominant microbial taxa and functional shifts induced by each inoculant combination using 16S sequencing.
- To elucidate the mechanistic links between inoculant-driven microbial changes and soil remediation potential.
2. Materials and Methods
2.1. Study Area
2.2. Experimental Materials and Design
2.3. Sample Collection
2.4. Index
2.4.1. Soil Physicochemical Properties
- Soil Organic Matter (SOM)Method: Modified Walkley–Black wet oxidation method [21].A quantity of 0.5 g air-dried soil was mixed with 5 mL 0.8 M K2Cr2O7 and 5 mL concentrated H2SO4.The sample was heated at 135 °C for 30 min, cooled, and diluted with 10 mL deionized water.
- Total Nitrogen (TN)Method: Automated Kjeldahl digestion and distillation (FOSS Kjeltec 8400) [22].Digestion with H2SO4-H2O2 at 420 °C for 2 h.Distillation with 40% NaOH, absorbed in 2% boric acid, and titrated with 0.01 M HCl.
- Phosphorus, Potassium, Calcium, and MagnesiumMethod: Inductively Coupled Plasma Optical Emission Spectrometry [23].Soil digested with HNO3-HF-HClO4 (3:1:1) at 180 °C for 6 h.Wavelengths (nm): P 213.618, K 766.490, Ca 317.933, Mg 285.213.
2.4.2. Soil Enzyme Activities
- UreaseMethod: Phenol–sodium hypochlorite colorimetry [24].Incubation with 10% urea at 37 °C for 24 h.Color development with phenol (1.5%) and NaOCl (0.9%), measured at 635 nm.
- SucraseMethod: 3,5-Dinitrosalicylic acid (DNS) assay [25].Substrate: 8% sucrose solution, reacted at 37 °C for 24 h.Absorbance measured at 540 nm after DNS reagent addition.
- DehydrogenaseMethod: Triphenyltetrazolium chloride (TTC) reduction [26].Soil incubated with 1% TTC (pH 7.4) at 37 °C for 6 h in darkness.Formazan extracted with methanol, measured at 485 nm. Activity expressed as μg Formazan·g−1·h−1.
- PhosphataseMethod: p-Nitrophenyl phosphate (pNPP) hydrolysis [27].Substrate: 5 mM pNPP in acetate buffer (pH 5.5), incubated at 37 °C for 1 h.Reaction stopped with 0.5 M NaOH, measured at 405 nm.
2.4.3. Microbial Biomass Carbon and Nitrogen
2.4.4. Microbial Community Diversity
2.5. Statistics and Analysis
3. Results
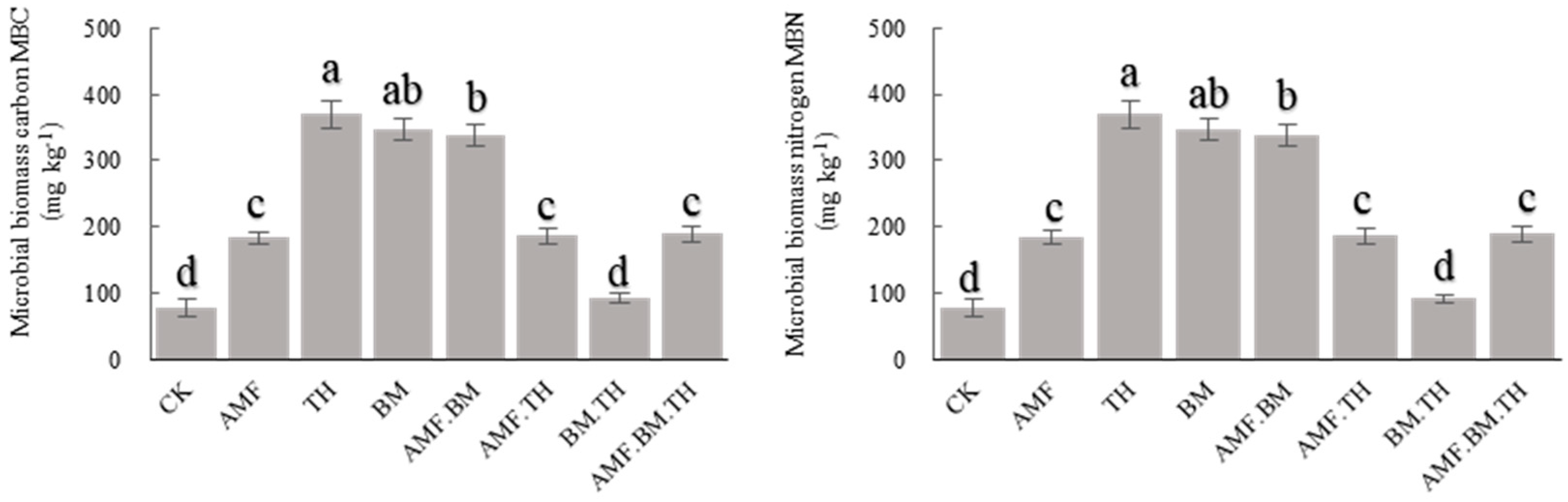
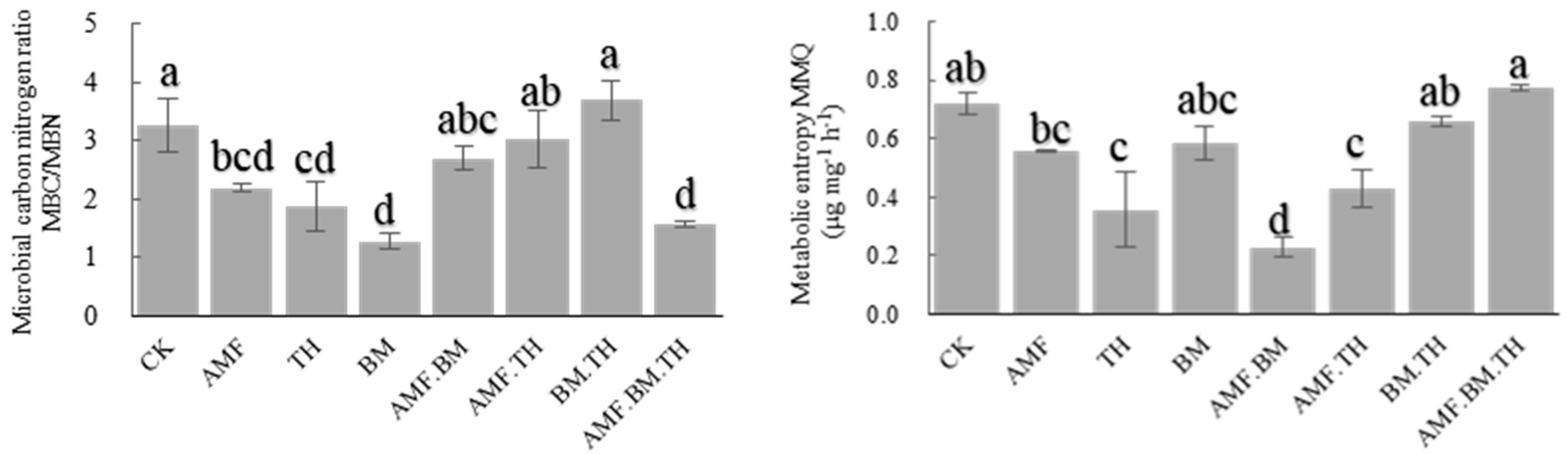
3.1. Effects of Different Bacterial Combinations on Soil Microbial Carbon and Nitrogen and Metabolic Entropy
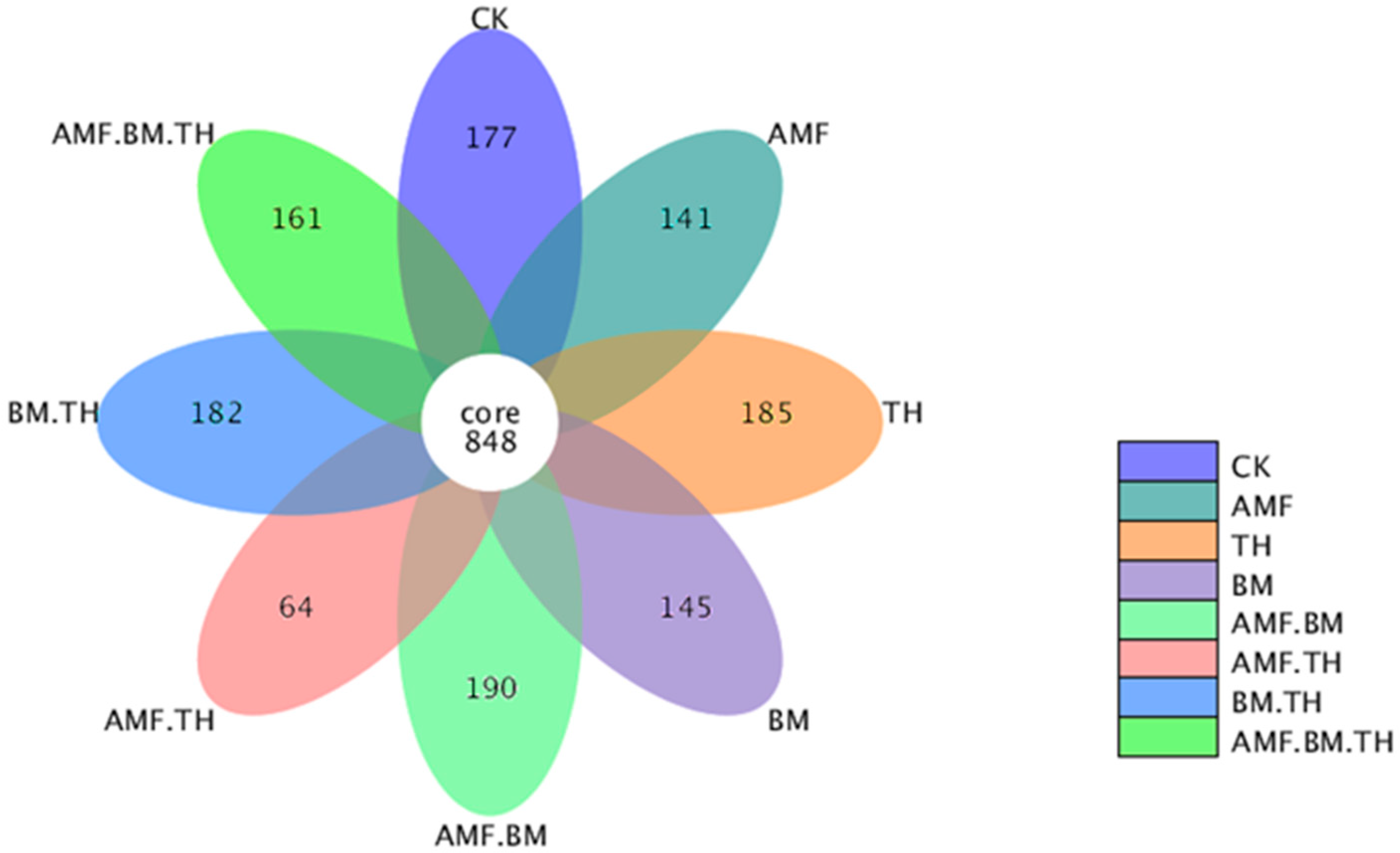
3.2. Bacterial Sequencing Statistics and Rationality Analysis
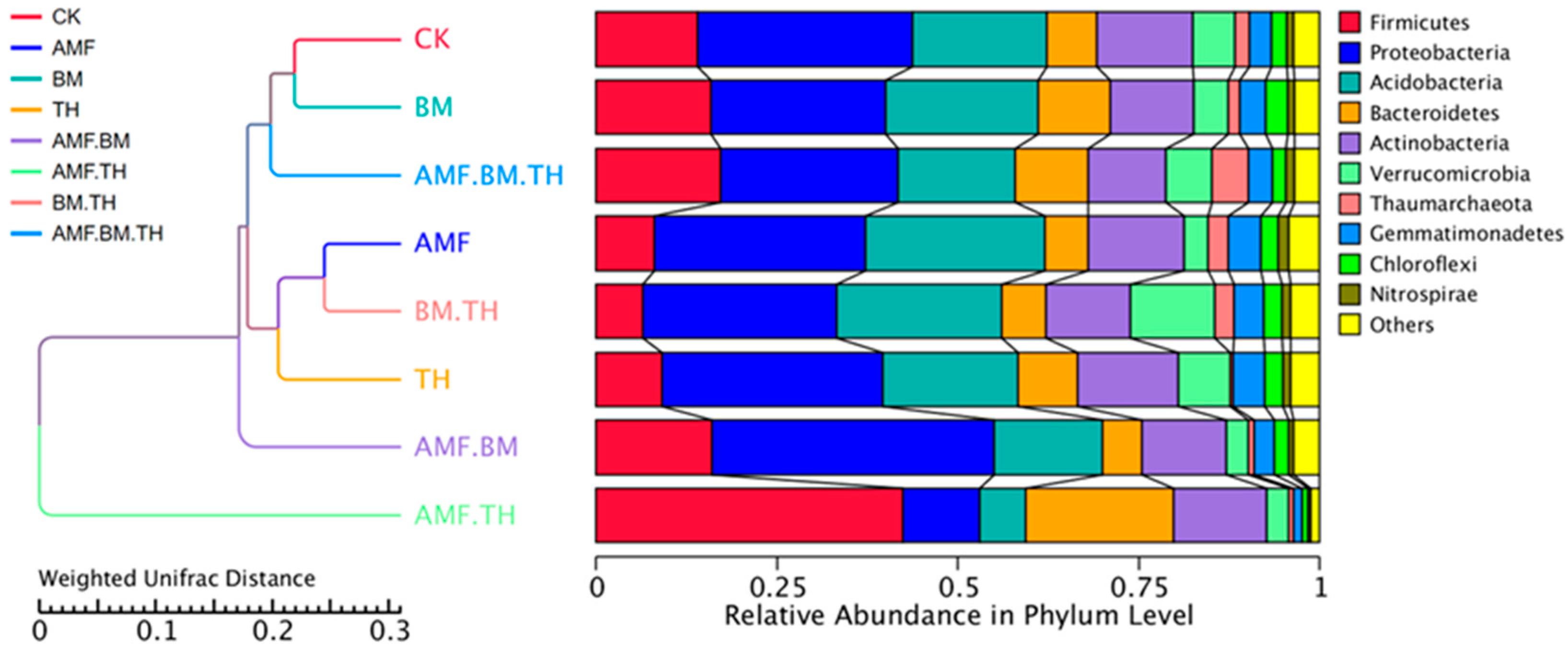
3.3. Effects of Different Microbial Agents on Soil Microbial Community Structure
3.4. Analysis of Soil Bacterial Community Diversity
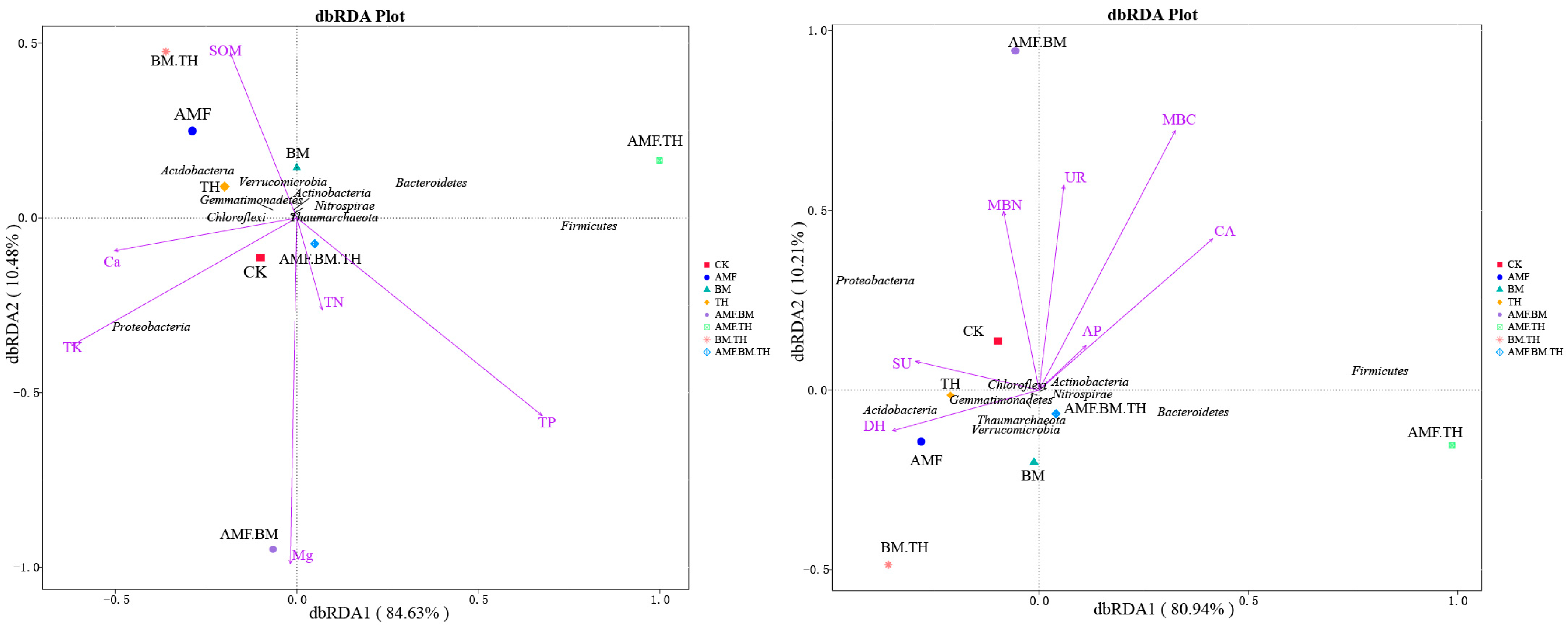
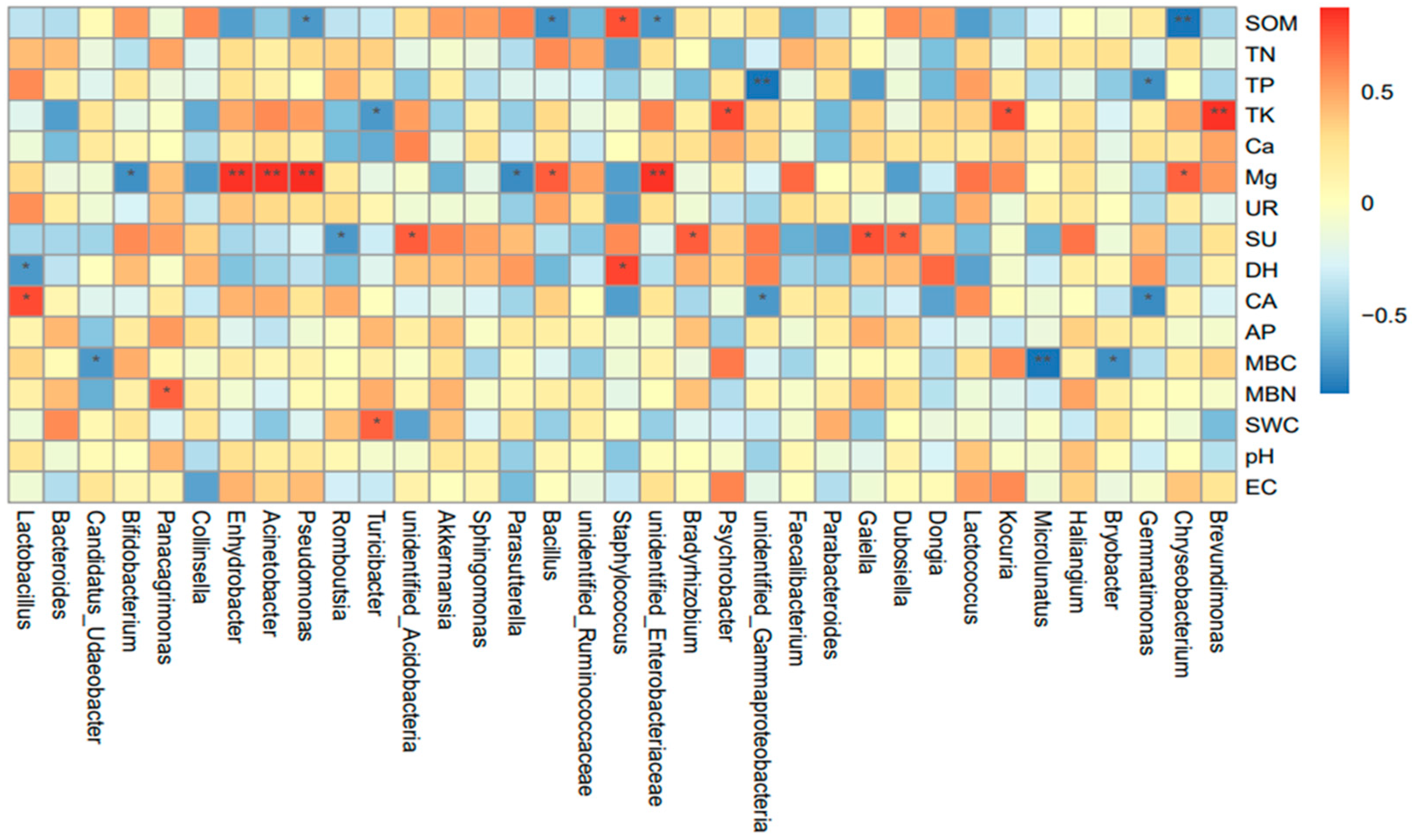
3.5. Correlation Analysis Between Community Structure of Soil Bacteria and Environmental Factors in Different Treatments
4. Discussion
4.1. Effects of Combined Application of Different Microbial Agents on Soil Microbial Carbon and Nitrogen
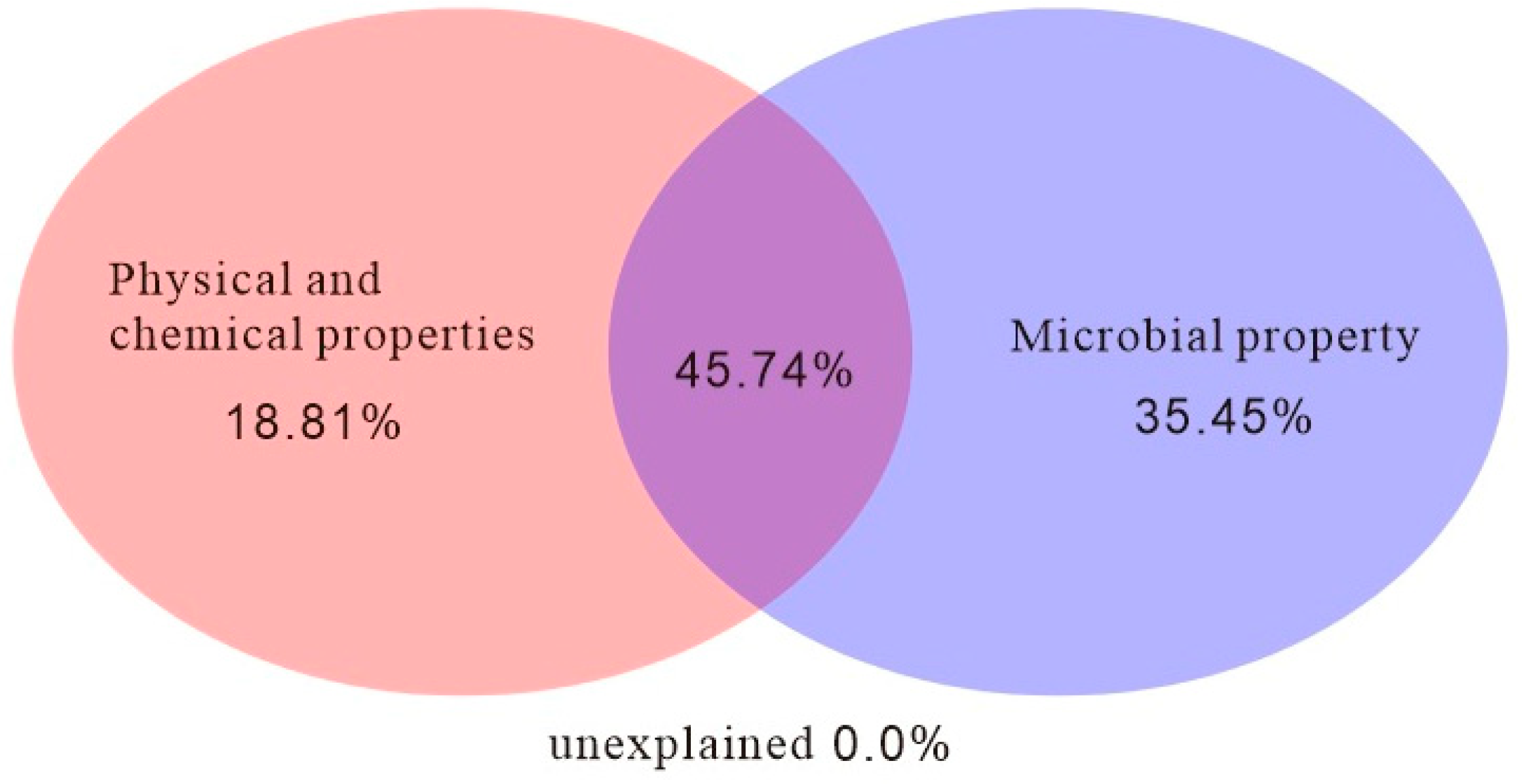
4.2. Relationship Between Soil Bacterial Community Structure and Soil Physicochemical Properties and Enzyme Activities
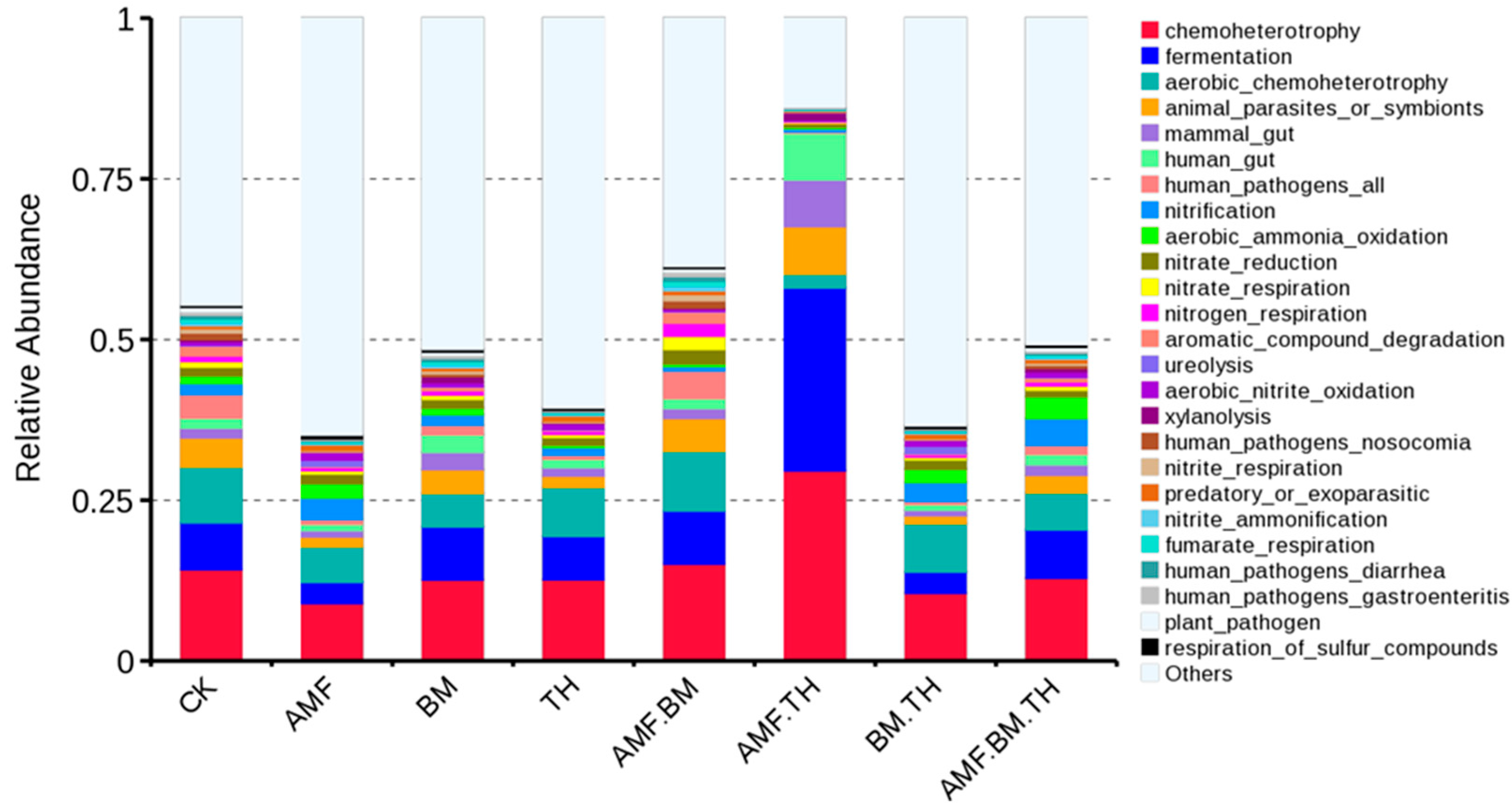
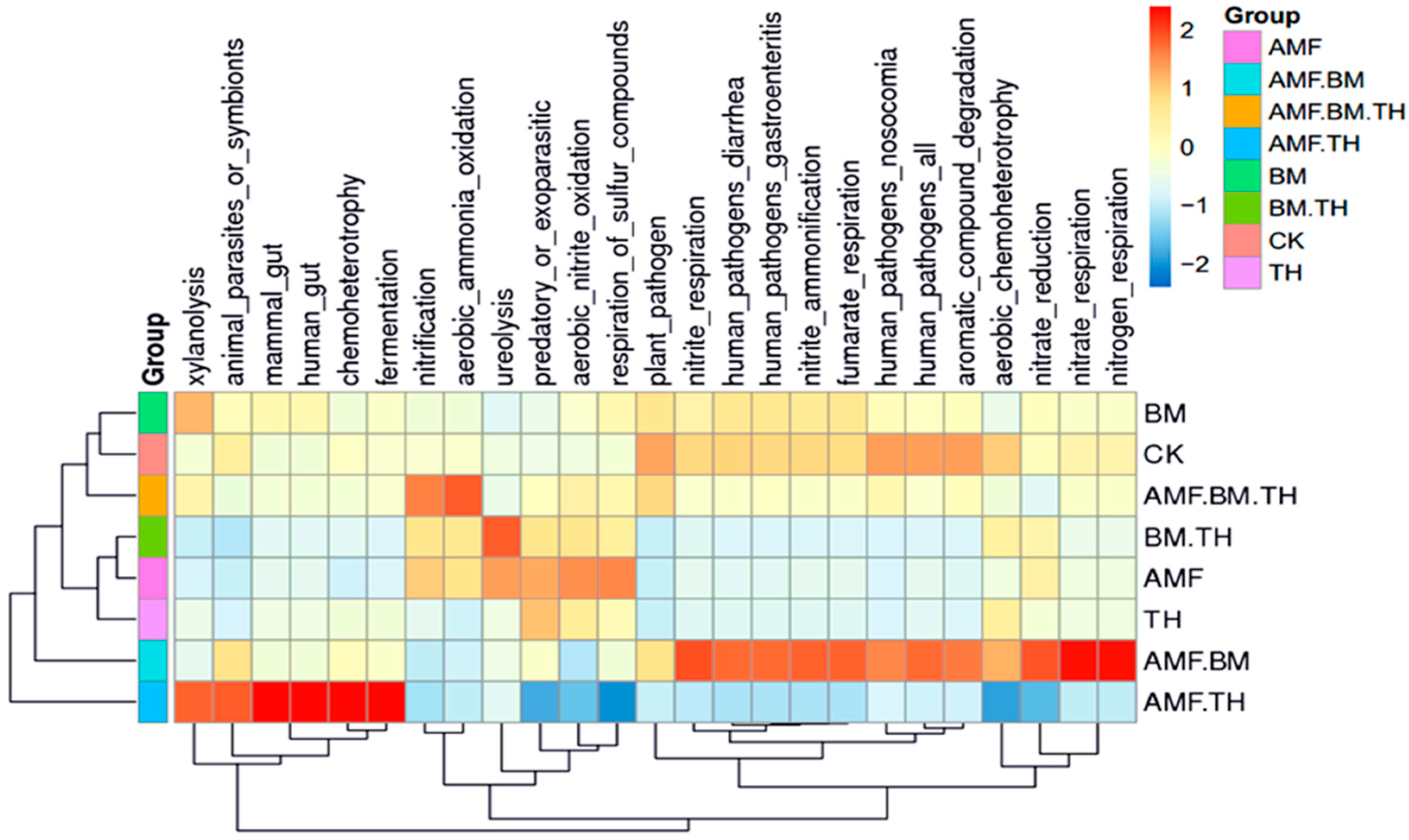
4.3. Effects of Different Bacterial Agents on Ecological Functions of Soil Bacteria
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Lehmann, J.; Bossio, D.A.; Kögel-Knabner, I.; Rillig, M.C. The concept and future prospects of soil health. Nat. Rev. Earth Environ. 2020, 1, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Condron, L.; Stark, C.; O’Callaghan, M.; Clinton, P.; Huang, Z. The Role of Microbial Communities in the Formation and Decomposition of Soil Organic Matter. In Soil Microbiology and Sustainable Crop Production; Dixon, G., Tilston, E., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 81–118. [Google Scholar]
- Rillig, M.C.; Aguilar-Trigueros, C.A.; Camenzind, T.; Cavagnaro, T.R.; Degrune, F.; Hohmann, P.; Lammel, D.R.; Mansour, I.; Roy, J.; van der Heijden, M.G.A.; et al. Why farmers should manage the arbuscular mycorrhizal symbiosis. New Phytol. 2019, 222, 1171–1175. [Google Scholar] [CrossRef]
- Nanjundappa, A.; Bagyaraj, D.J.; Saxena, A.K.; Kumar, M.; Chakdar, H. Interaction between arbuscular mycorrhizal fungi and Bacillus spp. in soil enhancing growth of crop plants. Fungal Biol. Biotechnol. 2019, 6, 23. [Google Scholar] [CrossRef] [PubMed]
- Dacal, M.; García-Palacios, P.; Asensio, S.; Wang, J.; Singh, B.K.; Maestre, F.T. Climate Change Legacies Contrastingly Affect the Resistance and Resilience of Soil Microbial Communities and Multifunctionality to Extreme Drought. Funct. Ecol. 2021, 36, 908–920. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Tian, W.; Xu, X.; He, C. Land use and land cover change in agro-pastoral ecotone in Northern China: A Review. Chin. J. Appl. Ecol. 2018, 29, 3487–3495. [Google Scholar]
- Song, G.; Chen, Y.; Tian, M.; Lv, S.; Zhang, S.; Liu, S. The Ecological Vulnerability Evaluation in Southwestern Mountain Region of China Based on GIS and AHP Method. Procedia Environ. Sci. 2010, 2, 465–475. [Google Scholar] [CrossRef]
- Jia, P.; Liang, J.; Yang, S.; Zhang, S.; Liu, J.; Liang, Z.; Li, F.; Zeng, Q.; Fang, Z.; Liao, B.; et al. Plant diversity enhances the reclamation of degraded lands by stimulating plant–soil feedbacks. J. Appl. Ecol. 2020, 57, 1258–1270. [Google Scholar] [CrossRef]
- Han, Y.; Zhao, W.; Zhou, A.; Pereira, P. Water and Wind Erosion Response to Ecological Restoration Measures in China’s Drylands. Geoderma 2023, 435, 116514. [Google Scholar] [CrossRef]
- Santoyo, G.; Orozco-Mosqueda, M.d.C.; Afridi, M.S.; Mitra, D.; Valencia-Cantero, E.; Macías-Rodríguez, L. Trichoderma and Bacillus: Multifunctional allies for plant growth and health in saline soils: Recent advances and future challenges. Front. Microbiol. 2024, 15, 1423980. [Google Scholar] [CrossRef] [PubMed]
- Philippot, L.; Chenu, C.; Kappler, A.; Rillig, M.C.; Fierer, N. The Interplay between Microbial Communities and Soil Properties. Nat. Rev. Microbiol. 2024, 22, 226–239. [Google Scholar] [CrossRef] [PubMed]
- Coban, O.; De Deyn, G.B.; van der Ploeg, M. Soil microbiota as game-changers in restoration of degraded lands. Science 2022, 375, eabe0725. [Google Scholar] [CrossRef] [PubMed]
- Canfora, L.; Bacci, G.; Pinzari, F.; Papa, G.L.; Dazzi, C.; Benedetti, A. Salinity and Bacterial Diversity: To What Extent Does the Concentration of Salt Affect the Bacterial Community in a Saline Soil? PLoS ONE 2014, 9, e106662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Shi, Z.; Yang, J.; Hao, B.; Hao, L.; Diao, F.; Wang, L.; Bao, Z.; Guo, W. A new strategy for evaluating the improvement effectiveness of degraded soil based on the synergy and diversity of microbial ecological function. Ecol. Indic. 2021, 120, 106917. [Google Scholar] [CrossRef]
- Nicolás, C.; Hermosa, R.; Rubio, B.; Mukherjee, P.K.; Monte, E. Trichoderma genes in plants for stress tolerance-status and prospects. Plant Sci. 2014, 228, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Li, Y.; Fu, C.; Chen, F.; Fu, Q.; Dai, A.; Shinoda, M.; Ma, Z.; Guo, W.; Li, Z.; et al. Dryland climate change: Recent progress and challenges. Rev. Geophys. 2017, 55, 719–778. [Google Scholar] [CrossRef]
- Jiang, S.H.; Zhu, H.Z.; He, P.A.; Chen, C.Z.; Zhou, H.M.; Su, D.C.; Xu, J.M.; Qin, H.Y.; Bao, S.D.; Lu, R.K.; et al. Book review: Analytical methods for soil and agro-chemistry (in Chinese). Eur. J. Soil Sci. 2000, 73, e13280. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis: Part 2 Chemical and Microbiological Properties, 9.2.2, 2nd ed.; ASA-SSSA: Madison, WI, USA, 1982; pp. 539–579. [Google Scholar]
- Ma, Z.; Xu, W.; Chen, Y.; Liu, M.; Wen, J. A Study of the Influence of the Type of Land Use on the Enzymatic Activity of Soils in Southwestern China. Forests 2024, 15, 581. [Google Scholar] [CrossRef]
- Liang, S.; Deng, J.; Jiang, Y.; Wu, S. Functional distribution of bacterial community under different land use patterns based on FaProTax function prediction. Pol. J. Environ. Stud. 2020, 29, 1245–1261. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthi, V.; Meena, S. Quantification of soil organic carbon-Comparison of wet oxidation and dry combustion methods. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 146–154. [Google Scholar] [CrossRef]
- Bremner, J.M. Determination of nitrogen in soil by the Kjeldahl method. J. Agric. Sci. 1960, 55, 11–33. [Google Scholar] [CrossRef]
- Zarcinas, B.A.; Cartwright, B.; Spouncer, L.R. Nitric Acid Digestion and Multi-Element Analysis of Plant Material by Inductively Coupled Plasma Spectrometry. Commun. Soil Sci. Plant Anal. 1987, 18, 131–146. [Google Scholar] [CrossRef]
- Tavares, M.C.; Oliveira, K.A.; de Fátima, Â.; Coltro, W.K.T.; Santos, J.C.C. Paper-based analytical device with colorimetric detection for urease activity determination in soils and evaluation of potential inhibitors. Talanta 2021, 230, 122301. [Google Scholar] [CrossRef] [PubMed]
- Frankeberger, W.T.; Johanson, J.B. Method of measuring invertase activity in soils. Plant Soil 1983, 74, 301–311. [Google Scholar] [CrossRef]
- Tabatabai, M.A. Soil enzymes. In Methods of Soil Analysis: Part 2 Microbiological and Biochemical Properties; SSSA: Madison, WI, USA, 1994; pp. 775–833. [Google Scholar]
- Tabatabai, M.A.; Bremner, J.M. Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem. 1969, 1, 301–307. [Google Scholar] [CrossRef]
- Vance, E.D.; Brookes, P.C.; Jenkinson, D.S. An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 1987, 19, 703–707. [Google Scholar] [CrossRef]
- Zhou, J.; Bruns, M.A.; Tiedje, J.M. DNA recovery from soils of diverse composition. Appl. Environ. Microbiol. 1996, 62, 316–322. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, X.; Liu, M.; Liu, W.; Xu, J.; Li, Y. Comparative evaluation of 16S rRNA primer pairs in identifying nitrifying guilds in soils under long-term organic fertilization and water management. Front. Microbiol. 2024, 15, 1424795. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable, and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Bolat, İ. Microbial Biomass, Basal Respiration, and Microbial Indices of Soil in Diverse Croplands in a Region of Northwestern Turkey (Bartın). Environ. Monit. Assess. 2019, 191, 695. [Google Scholar] [CrossRef] [PubMed]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform Fumigation and the Release of Soil Nitrogen: A Rapid Direct Extraction Method to Measure Microbial Biomass Nitrogen in Soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Geisseler, D.; Horwath, W.R.; Joergensen, R.G.; Ludwig, B. Pathways of Nitrogen Utilization by Soil Microorganisms—A Review. Soil Biol. Biochem. 2010, 42, 2058–2067. [Google Scholar] [CrossRef]
- Chen, L.; Jiang, Y.; Liang, C.; Luo, Y.; Xu, Q.; Han, C.; Zhao, Q.; Sun, B. Competitive Interaction with Keystone Taxa Induced Negative Priming under Biochar Amendments. Microbiome 2019, 7, 35. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Shi, X.; Zheng, C.; Suter, H.; Huang, Z. Different Responses of Soil Bacterial and Fungal Communities to Nitrogen Deposition in a Subtropical Forest. Sci. Total Environ. 2021, 755, 142449. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Maestre, F.T.; Reich, P.B.; Jeffries, T.C.; Gaitan, J.J.; Encinar, D.; Berdugo, M.; Campbell, C.D.; Singh, B.K. Microbial Diversity Drives Multifunctionality in Terrestrial Ecosystems. Nat. Commun. 2016, 7, 10541. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.; Zhang, Y.; Gao, J.; Peng, F.; Gao, P. Long-term combined application of manure and chemical fertilizer sustained higher nutrient status and rhizospheric bacterial diversity in reddish paddy soil of Central South China. Sci. Rep. 2018, 8, 16554. [Google Scholar] [CrossRef] [PubMed]
- Xun, W.; Liu, Y.; Li, W.; Ren, Y.; Xiong, W.; Xu, Z.; Zhang, N.; Miao, Y.; Shen, Q.; Zhang, R. Specialized metabolic functions of keystone taxa sustain soil microbiome stability. Microbiome 2021, 9, 35. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Chen, X.; Jia, Z.; Zhai, L.; Zhang, B.; Grüters, U.; Ma, S.; Qian, J.; Liu, X.; Zhang, J.; et al. Meta-Analysis Reveals the Effects of Microbial Inoculants on the Biomass and Diversity of Soil Microbial Communities. Nat. Ecol. Evol. 2024, 8, 1270–1284. [Google Scholar] [CrossRef] [PubMed]
- Ma, B.; Wang, Y.; Ye, S.; Liu, S.; Stirling, E.; Gilbert, J.A.; Faust, K.; Knight, R.; Jansson, J.K.; Cardona, C.; et al. Earth microbial co-occurrence network reveals interconnection pattern across microbiomes. Microbiome 2020, 8, 82. [Google Scholar] [CrossRef] [PubMed]
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The Current Status of Its Application in Agriculture for the Biocontrol of Fungal Phytopathogens and Stimulation of Plant Growth. Int. J. Mol. Sci. 2022, 23, 2329. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Chen, J.; Wu, H.; Qin, X.; Wang, J.; Wu, Y.; Khan, M.U.; Lin, S.; Xiao, Z.; Luo, X.; et al. Insights into the Regulation of Rhizosphere Bacterial Communities by Application of Bio-organic Fertilizer in Pseudostellaria heterophylla Monoculture Regime. Front. Microbiol. 2016, 7, 1788. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.; Liu, X.; Song, L.; Lin, X.; Zhang, H. Nitrogen fertilization directly affects soil bacterial diversity and indirectly affects bacterial community composition. Soil Biol. Biochem. 2016, 92, 41–49. [Google Scholar] [CrossRef]
- Fierer, N.; Ladau, J.; Clemente, J.C.; Leff, J.W.; Owens, S.M.; Pollard, K.S.; Knight, R.; Gilbert, J.A.; McCulley, R.L. Reconstructing the Microbial Diversity and Function of Pre-Agricultural Tallgrass Prairie Soils in the United States. Science 2013, 342, 621–624. [Google Scholar] [CrossRef] [PubMed]
- Van Berkum, P.; Leibold, J.M.; Eardly, B.D. Proposal for combining Bradyrhizobium spp. (Aeschynomene indica) with Blastobacter denitrificans and to transfer Blastobacter denitrificans (Hirsch and Muller, 1985) to the genus Bradyrhizobium as Bradyrhizobium denitrificans (comb. nov.). Syst. Appl. Microbiol. 2006, 29, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Gadkari, P. Ecology and Physiological Potential of Tundra Soil Bacteria. Ph.D. Thesis, Rutgers University-School of Graduate Studies, New Brunswick, NJ, USA, 2019. [Google Scholar]
- Wang, Z.; Hassan, M.U.; Nadeem, F.; Wu, L.; Zhang, F.; Li, X. Magnesium fertilization improves crop yield in most production systems: A meta-analysis. Front. Plant Sci. 2020, 10, 495191. [Google Scholar] [CrossRef] [PubMed]
- Poomthongdee, N.; Duangmal, K.; Pathom-Aree, W. Acidophilic actinomycetes from rhizosphere soil: Diversity and properties beneficial to plants. J. Antibiot. 2015, 68, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Lv, C.; Fernández-García, V.; Huang, B.; Yao, J.; Ding, W. Biochar and PGPR amendments influence soil enzyme activities and nutrient concentrations in a eucalyptus seedling plantation. BiBiomass Convers. Biorefin. 2021, 11, 1865–1874. [Google Scholar] [CrossRef]
- Liu, Z.; Xie, W.; Yang, Z.; Huang, X.; Zhou, H. Effects of Manure and Chemical Fertilizer on Bacterial Community Structure and Soil Enzyme Activities in North China. Agronomy 2021, 11, 1017. [Google Scholar] [CrossRef]
- Landesman, W.J.; Nelson, D.M.; Fitzpatrick, M.C. Soil properties and tree species drive—Diversity of soil bacterial communities. Soil Biol. Biochem. 2014, 76, 201–209. [Google Scholar] [CrossRef]
- Zhou, H.; Gao, Y.; Jia, X.; Wang, M.; Ding, J.; Cheng, L.; Bao, F.; Wu, B. Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, northwestern China. Soil Biol. Biochem. 2020, 144, 107782. [Google Scholar] [CrossRef]
- Ambrosini, A.; de Souza, R.; Passaglia, L.M.P. Ecological Role of Bacterial Inoculants and Their Potential Impact on Soil Microbial Diversity. Plant Soil 2016, 400, 193–207. [Google Scholar] [CrossRef]
- Ding, J.; Zhang, Y.; Deng, Y.; Cong, J.; Lu, H.; Sun, X.; Yang, C.; Yuan, T.; Van Nostrand, J.D.; Li, D.; et al. Integrated meta-genomics and network analysis of soil microbial community of the forest timberline. Sci. Rep. 2015, 5, 7994. [Google Scholar]


| Soil Microaggregates | 2–0.25 mm | 0.25–0.05 mm | 0.05–0.01 mm | 0.01–0.005 mm | 0.005–0.001 mm | ˂0.001 mm | |
| 2.48% | 51.20% | 39.90% | 3.34% | 0.20% | 2.88% | ||
| Soil mechanical composition | 2.0–1.0 mm | 1.0–0.5 mm | 0.5–0.25 mm | 0.25–0.05 mm | 0.05-0.02 mm | 0.02–0.002 mm | ˂0.002 mm |
| 0.63% | 1.02% | 0.50% | 48.95% | 14.29% | 15.21% | 19.39% | |
| Soil chemical properties | SOM % | TN g·kg−1 | TP g·kg−1 | TK g·kg−1 | Ca g·kg−1 | Mg g·kg−1 | UR mg·g−1 |
| 2.49 | 1.04 | 0.27 | 20.43 | 65.78 | 8.27 | 7.72 | |
| SU mg·g−1 | DH mg·g−1 | CA mg·g−1 | AP mg·g−1 | MBC mg·g−1 | MBN mg·g−1 | ||
| 41.33 | 223.31 | 0.59 | 8.56 | 305.81 | 93.39 | ||
| Soil physical properties | SWC % | pH | EC mS cm−1 | ||||
| 20.51 | 6.83 | 114.63 |
| Sample | CK | AMF | TH | BM | AMF.BM | AMF.TH | BM.TH | AMF.BM.TH |
|---|---|---|---|---|---|---|---|---|
| SOM % | 2.06 abc | 2.91 d | 2.07 abc | 1.83 a | 1.87 ab | 2.18 c | 2.23 c | 2.09 c |
| TN g·kg−1 | 0.85 e | 0.94 de | 0.92 de | 0.97 cde | 1.04 cd | 1.11 bc | 1.35 a | 1.21 ab |
| TP g·kg−1 | 0.22 cd | 0.25 c | 0.25 c | 0.19 d | 0.21 cd | 0.29 b | 0.42 a | 0.20 d |
| TK g·kg−1 | 16.75 b | 17.27 ab | 18.32 ab | 18.86 ab | 17.58 ab | 18.17 ab | 19.65 a | 17.75 ab |
| Ca g·kg−1 | 53.48 d | 51.62 c | 47.31 b | 56.18 e | 57.95 e | 45.97 b | 61.04 f | 41.08 a |
| Mg g·kg−1 | 6.57 cd | 5.68 bc | 5.51 bc | 5.72 bc | 10.09 e | 5.41 b | 4.21 a | 7.29 d |
| UR mg·g−1 | 6.33 a | 6.68 b | 6.99 c | 7.74 d | 8.53 e | 7.04 c | 7.04 c | 7.71 d |
| SU mg·g−1 | 33.07 a | 60.95 d | 56.82 d | 30.68 a | 44.72 c | 39.04 b | 39.76 bc | 28.92 a |
| DH mg·g−1 | 184.55 e | 257.86 g | 188.06 f | 104.92 a | 129.22 b | 138.49 c | 141.63 d | 138.21 c |
| CA mg·g−1 | 0.48 ab | 0.54 b | 0.30 a | 1.01 d | 1.34 e | 1.03 d | 0.72 c | 1.15 d |
| AP mg·g−1 | 6.69 a | 16.83 b | 16.82 b | 19.21 b | 33.71 c | 16.88 b | 9.95 ab | 12.36 ab |
| MBC mg·kg−1 | 250.66 a | 400.13 ab | 690.46 c | 439.17 ab | 907.38 d | 562.94 bc | 339.91 ab | 296.44 a |
| MBN mg·kg−1 | 77.19 a | 183.45 b | 370.00 d | 346.79 cd | 337.49 c | 186.60 b | 92.16 a | 189.01 b |
| SWC % | 16.67 abc | 15.62 a | 16.17 a | 17.94 d | 16.15 a | 17.39 bcd | 16.34 ab | 17.76 cd |
| pH | 6.71 c | 6.93 ab | 6.92 ab | 6.79 bc | 6.67 c | 6.93 ab | 6.82 bc | 7.05 a |
| EC mS cm−1 | 105.28 e | 64.56 a | 66.03 a | 94.71 d | 131.27 g | 74.15 b | 126.36 f | 76.98 c |
| Treatments | Sample Description | Application Rate (g·m−2) |
|---|---|---|
| Blank control | CK | — |
| Mono-microbial | AMF | 50 |
| BM | 10 | |
| TH | 20 | |
| Composite microbial | AMF.BM | 50 + 20 |
| AMF.TH | 50 + 10 | |
| BM.TH | 10 + 20 | |
| AMF.BM.TH | 50 + 10 + 20 |
| Treatments | Shannon | PD_Whole_Tree | Chao1 | ACE | Coverage |
|---|---|---|---|---|---|
| CK | 9.566 cd | 197.551 bc | 3007.171 bc | 3062.13 bc | 0.986 a |
| AMF | 9.533 cd | 232.647 d | 2826.562 b | 2878.51 b | 0.988 a |
| BM | 9.536 cd | 199.09 bc | 2898.360 bc | 2956.51 bc | 0.987 a |
| TH | 9.650 d | 193.08 b | 2952.173 bc | 2996.06 bc | 0.987 a |
| AMF.BM | 9.247 b | 226.75 cd | 3090.625 c | 3075.32 c | 0.982 a |
| AMF.TH | 6.046 a | 131.898 a | 1894.853 a | 2013.79 a | 0.989 a |
| BM.TH | 9.428 bc | 222.012 bcd | 3000.769 bc | 3039.83 bc | 0.983 a |
| AMF.BM.TH | 9.558 cd | 247.878 d | 3006.216 bc | 3063.75 bc | 0.986 a |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, T.; Liu, T.; Gan, Z.; Jin, X.; Xiao, L. Diversity and Functional Differences in Soil Bacterial Communities in Wind–Water Erosion Crisscross Region Driven by Microbial Agents. Agronomy 2025, 15, 1734. https://doi.org/10.3390/agronomy15071734
Kong T, Liu T, Gan Z, Jin X, Xiao L. Diversity and Functional Differences in Soil Bacterial Communities in Wind–Water Erosion Crisscross Region Driven by Microbial Agents. Agronomy. 2025; 15(7):1734. https://doi.org/10.3390/agronomy15071734
Chicago/Turabian StyleKong, Tao, Tong Liu, Zhihui Gan, Xin Jin, and Lin Xiao. 2025. "Diversity and Functional Differences in Soil Bacterial Communities in Wind–Water Erosion Crisscross Region Driven by Microbial Agents" Agronomy 15, no. 7: 1734. https://doi.org/10.3390/agronomy15071734
APA StyleKong, T., Liu, T., Gan, Z., Jin, X., & Xiao, L. (2025). Diversity and Functional Differences in Soil Bacterial Communities in Wind–Water Erosion Crisscross Region Driven by Microbial Agents. Agronomy, 15(7), 1734. https://doi.org/10.3390/agronomy15071734





